E4 Transcription Factor 1 (E4F1) Regulates Sertoli Cell Proliferation and Fertility in Mice
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Fertility Test and Sperm Concentration Analysis
2.3. Histological Analysis
2.4. Meiosis Analysis
2.5. Immunohistochemistry (IHC) and Immunofluorescence (IF)
2.6. TUNEL Labeling
2.7. 5-ethynyl-2′-deoxyuridine (EdU) Assay
2.8. qRT-PCR Analysis
2.9. Statistical Analysis
3. Results
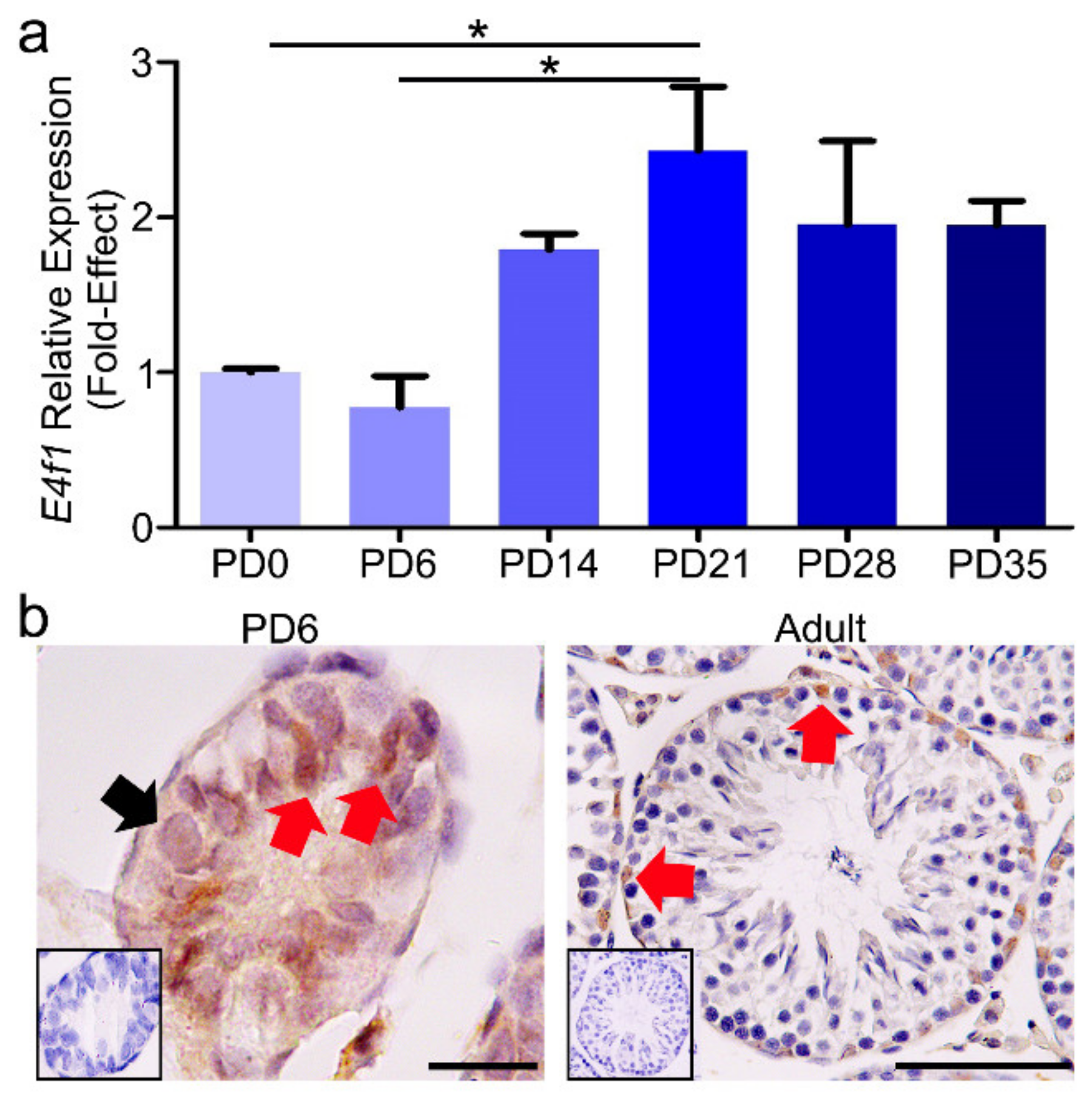
3.1. The Relative mRNA Expression and Protein Localization of E4f1 in Postnatal Mouse Testes
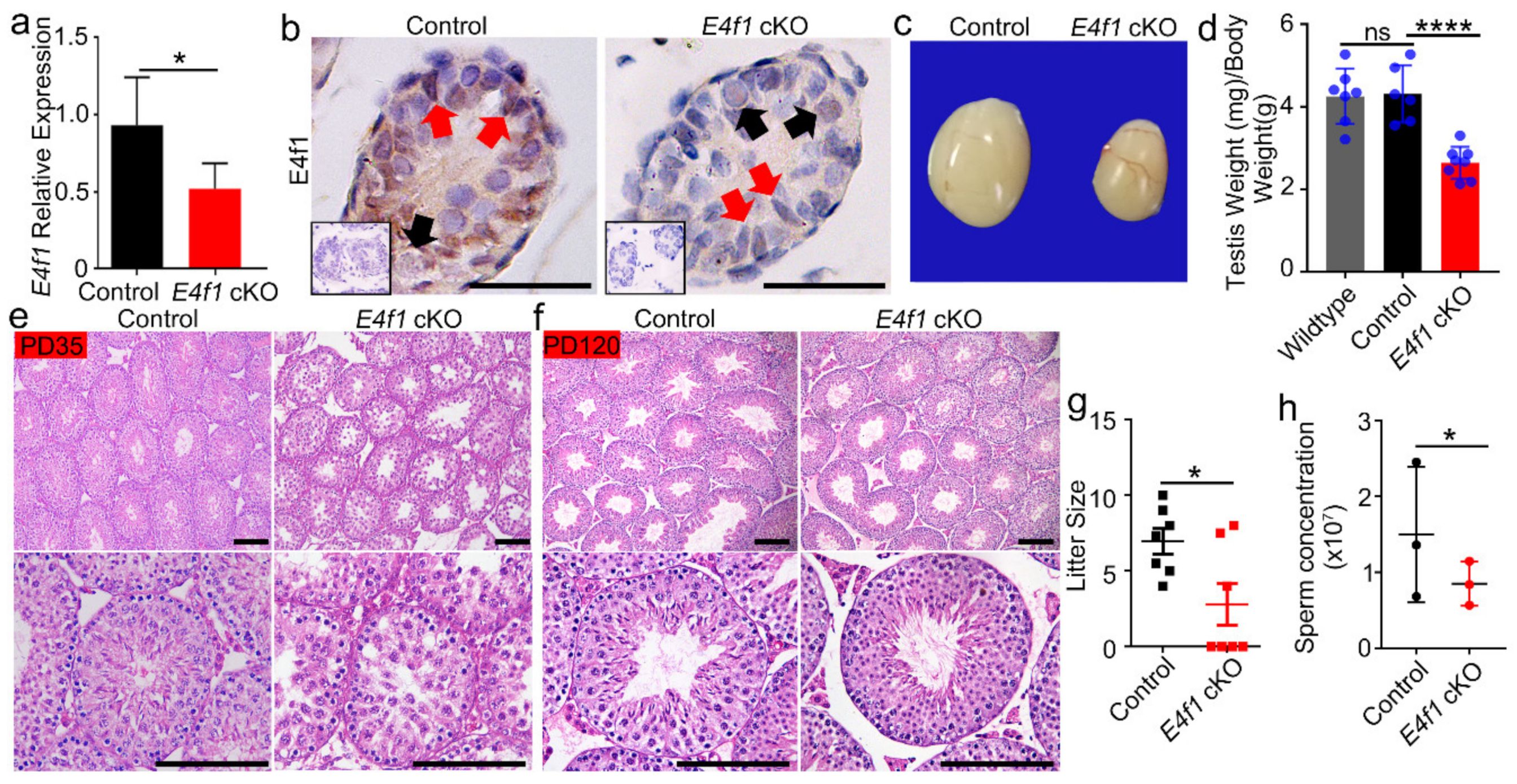
3.2. E4f1 Deletion in Sertoli Cells Leads Reduced Testis Size and Male Subfertility
3.3. E4f1 Deletion in Sertoli Cells Did Not Cause Defects in Meiosis and Spermiogenesis
3.4. E4f1 Deletion Resulted in a Reduction in Sertoli Cell Number
3.5. E4f1 Deletion Affected Sertoli Cell Proliferation and Apoptosis during Neonatal Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Griswold, M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Boil. 1998, 9, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Lindahl, M.; Hyvönen, M.E.; Parvinen, M.; De Rooij, D.G.; Hess, M.W.; Raatikainen-Ahokas, A.; Sainio, K.; Rauvala, H.; Lakso, M.; et al. Regulation of Cell Fate Decision of Undifferentiated Spermatogonia by GDNF. Science 2000, 287, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Eto, K.; Honmyou, A.; Nakao, K.; Kiyonari, H.; Abé, S.-I. Neuregulins are essential for spermatogonial proliferation and meiotic initiation in neonatal mouse testis. Development 2011, 138, 3159–3168. [Google Scholar] [CrossRef] [PubMed]
- Hess, R.A.; Franca, L.R.D. Spermatogenesis and Cycle of the Seminiferous Epithelium. Adv. Exp. Med. Biol. 2008, 636, 1. [Google Scholar]
- Wu, S.; Yan, M.; Ge, R.; Cheng, C.Y. Crosstalk between Sertoli and Germ Cells in Male Fertility. Trends Mol. Med. 2020, 26, 215–231. [Google Scholar] [CrossRef]
- Orth, J.M.; Gunsalus, G.L.; Lamperti, A.A. Evidence from sertoli cell-depleted rats indicates that spermatid number in adults depends on numbers of sertoli cells produced during perinatal development. Endocrinology 1988, 122, 787–794. [Google Scholar] [CrossRef]
- Rebourcet, D.; Wu, J.; Cruickshanks, L.; Smith, S.E.; Milne, L.; Fernando, A.; Wallace, R.J.; Gray, C.D.; Hadoke, P.W.F.; Mitchell, R.T.; et al. Sertoli Cells Modulate Testicular Vascular Network Development, Structure, and Function to Influence Circulating Testosterone Concentrations in Adult Male Mice. Endocrinology 2016, 157, 2479–2488. [Google Scholar] [CrossRef]
- Rosati, L.; Di Fiore, M.M.; Prisco, M.; Russo, F.D.G.; Venditti, M.; Andreuccetti, P.; Baccari, G.C.; Santillo, A. Seasonal expression and cellular distribution of star and steroidogenic enzymes in quail testis. J. Exp. Zool. Part B Mol. Dev. Evol. 2019, 332, 198–209. [Google Scholar] [CrossRef]
- Prisco, M.; Agnese, M.; De Marino, A.; Andreuccetti, P.; Rosati, L. Spermatogenic Cycle and Steroidogenic Control of Spermatogenesis in Mytilus galloprovincialis Collected in the Bay of Naples. Anat. Rec. 2017. [Google Scholar] [CrossRef]
- Sekido, R.; Lovell-Badge, R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008, 453, 930–934. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, M.; Lau, Y.-F.C. The Sex-Determining Factors SRY and SOX9 Regulate Similar Target Genes and Promote Testis Cord Formation during Testicular Differentiation. Cell Rep. 2014, 8, 723–733. [Google Scholar] [CrossRef] [PubMed]
- França, L.R.; Hess, R.A.; Dufour, J.M.; Hofmann, M.-C.; Griswold, M.D. The Sertoli cell: One hundred fifty years of beauty and plasticity. Andrology 2016, 4, 189–212. [Google Scholar] [CrossRef] [PubMed]
- Vergouwen, R.P.F.A.; Jacobs, S.G.P.M.; Huiskamp, R.; Davids, J.A.G.; De Rooij, D.G. Proliferative activity of gonocytes, Sertoli cells and interstitial cells during testicular development in mice. J. Reprod. Fertil. 1991, 93, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Holsberger, D.R.; Kiesewetter, S.E.; Cooke, P.S. Regulation of neonatal Sertoli cell development by thyroid hormone receptor alpha1. Biol. Reprod. 2005, 73, 396–403. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, P.; Adeniran, S.O.; Ma, M.; Huang, F.; Adegoke, E.O.; Zhang, G. Thyroid hormone (T3) is involved in inhibiting the proliferation of newborn calf Sertoli cells via the PI3K/Akt signaling pathway in vitro. Theriogenology 2019, 133, 1–9. [Google Scholar] [CrossRef]
- Bardi, G.; Bottazzi, C.; Demori, I.; Palmero, S. Thyroid hormone and retinoic acid induce the synthesis of insulin-like growth factor-binding protein-4 in prepubertal pig sertoli cells. Eur. J. Endocrinol. 1999, 141, 637–643. [Google Scholar] [CrossRef][Green Version]
- Buzzard, J.J.; Wreford, N.G.; Morrison, J.R. Thyroid Hormone, Retinoic Acid, and Testosterone Suppress Proliferation and Induce Markers of Differentiation in Cultured Rat Sertoli Cells. Endocrinology 2003, 144, 3722–3731. [Google Scholar] [CrossRef]
- Boitani, C.; Stefanini, M.; Fragale, A.; Morena, A.R. Activin stimulates Sertoli cell proliferation in a defined period of rat testis development. Endocrinology 1995, 136, 5438–5444. [Google Scholar] [CrossRef]
- Marchlewska, K.; Kula, K.; Walczak-Jędrzejowska, R.; Oszukowska, E.; Filipiak, E.; Slowikowska-Hilczer, J. Role of FSH and triiodothyronine in Sertoli cell development expressed by formation of connexin 43-based gap junctions. J. Exp. Zool. Part A Ecol. Genet. Physiol. 2011, 315, 329–336. [Google Scholar] [CrossRef]
- Puglisi, R.; Montanari, M.; Chiarella, P.; Stefanini, M.; Boitani, C. Regulatory role of BMP2 and BMP7 in spermatogonia and Sertoli cell proliferation in the immature mouse. Eur. J. Endocrinol. 2004, 151, 511–520. [Google Scholar] [CrossRef]
- Petersen, C.; Boitani, C.; Fröysa, B.; Söder, O. Interleukin-1 is a potent growth factor for immature rat Sertoli cells. Mol. Cell. Endocrinol. 2002, 186, 37–47. [Google Scholar] [CrossRef]
- Froment, P.; Vigier, M.; Nègre, D.; Fontaine, I.; Beghelli, J.; Cosset, F.-L.; Holzenberger, M.; Durand, P. Inactivation of the IGF-I receptor gene in primary Sertoli cells highlights the autocrine effects of IGF-I. J. Endocrinol. 2007, 194, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Holsberger, D.R.; Buchold, G.M.; Leal, M.C.; Kiesewetter, S.E.; O’Brien, D.A.; Hess, R.A.; França, L.R.; Kiyokawa, H.; Cooke, P.S. Cell-Cycle Inhibitors p27Kip1 and p21Cip1 Regulate Murine Sertoli Cell Proliferation1. Boil. Reprod. 2005, 72, 1429–1436. [Google Scholar] [CrossRef] [PubMed]
- Nalam, R.L.; Andreu-Vieyra, C.; Braun, R.E.; Akiyama, H.; Matzuk, M.M. Retinoblastoma Protein Plays Multiple Essential Roles in the Terminal Differentiation of Sertoli Cells. Mol. Endocrinol. 2009, 23, 1900–1913. [Google Scholar] [CrossRef] [PubMed]
- Rotgers, E.; Cisneros-Montalvo, S.; Nurmio, M.; Toppari, J. Retinoblastoma protein represses E2F3 to maintain Sertoli cell quiescence in mouse testis. J. Cell Sci. 2019, 132, jcs229849. [Google Scholar] [CrossRef]
- Faisal, I.; Cisneros-Montalvo, S.; Hamer, G.; Tuominen, M.M.; Laurila, P.-P.; Tumiati, M.; Jauhiainen, M.; Kotaja, N.; Toppari, J.; Mäkelä, J.-A.; et al. Transcription Factor USF1 Is Required for Maintenance of Germline Stem Cells in Male Mice. Endocrinology 2019, 160, 1119–1136. [Google Scholar] [CrossRef]
- Godmann, M.; Kosan, C.; Behr, R. Krüppel-like factor 4 is widely expressed in the mouse male and female reproductive tract and responds as an immediate early gene to activation of the protein kinase A in TM4 Sertoli cells. Reproduction 2010, 139, 771–782. [Google Scholar] [CrossRef]
- Lucas, T.F.G.; Lazari, M.F.M.; Porto, C.S. Differential role of the estrogen receptors ESR1 and ESR2 on the regulation of proteins involved with proliferation and differentiation of Sertoli cells from 15-day-old rats. Mol. Cell. Endocrinol. 2014, 382, 84–96. [Google Scholar] [CrossRef]
- Gorga, A.; Rindone, G.; Regueira, M.; Riera, M.F.; Pellizzari, E.H.; Cigorraga, S.B.; Meroni, S.B.; Galardo, M.N. HIF involvement in the regulation of rat Sertoli cell proliferation by FSH. Biochem. Biophys. Res. Commun. 2018, 502, 508–514. [Google Scholar] [CrossRef]
- Fernandes, E.R.; Rooney, R.J. The adenovirus E1A-regulated transcription factor E4F is generated from the human homolog of nuclear factor phiAP3. Mol. Cell. Boil. 1997, 17, 1890–1903. [Google Scholar] [CrossRef]
- Lee, K.A.; Green, M.R. A cellular transcription factor E4F1 interacts with an E1a-inducible enhancer and mediates constitutive enhancer function in vitro. EMBO J. 1987, 6, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Matthieu, L.; Julie, C.; Perrine, G.R.; Linares, L.K.; Soline, E.; Elodie, H.; Geneviève, R.; Gwendaline, L.; Carine, D.B.; Amélie, T. Transcription factor E4F1 is essential for epidermal stem cell maintenance and skin homeostasis. Proc. Nat. Acad. Sci. USA 2010, 107, 21076–21081. [Google Scholar]
- Le Cam, L.; Lacroix, M.; Ciemerych, M.A.; Sardet, C.; Sicinski, P. The E4F protein is required for mitotic progression during embryonic cell cycles. Mol. Cell. Biol. 2004, 24, 6467–6475. [Google Scholar] [CrossRef] [PubMed]
- Chagraoui, J.; Niessen, S.L.; Lessard, J.; Girard, S.; Coulombe, P.; Sauvageau, M.; Meloche, S.; Sauvageau, G. E4F1: A novel candidate factor for mediating BMI1 function in primitive hematopoietic cells. Genes Dev. 2006, 20, 2110–2120. [Google Scholar] [CrossRef][Green Version]
- Fajas, L.; Paul, C.; Vié, A.; Estrach, S.; Medema, R.; Blanchard, J.M.; Sardet, C.; Vignais, M.-L. Cyclin A Is a Mediator of p120E4F-Dependent Cell Cycle Arrest in G1. Mol. Cell. Boil. 2001, 21, 2956–2966. [Google Scholar] [CrossRef]
- Fajas, L.; Paul, C.; Zugasti, O.; Le Cam, L.; Polanowska, J.; Fabbrizio, E.; Medema, R.; Vignais, M.L.; Sardet, C. pRB binds to and modulates the transrepressing activity of the E1A-regulated transcription factor p120E4F. Proc. Nat. Acad. Sci. USA 2000, 97, 7738–7743. [Google Scholar] [CrossRef]
- Fernandes, E.R.; Zhang, J.Y.; Rooney, R.J. Adenovirus E1A-Regulated Transcription Factor p120E4F Inhibits Cell Growth and Induces the Stabilization of the cdk Inhibitor p21WAF1. Mol. Cell. Boil. 1998, 18, 459–467. [Google Scholar] [CrossRef]
- Grote, D.; Moison, C.; Duhamel, S.; Chagraoui, J.; Girard, S.; Yang, J.; Mayotte, N.; Coulombe, Y.; Masson, J.Y.; Brown, G.W.; et al. E4F1 is a master regulator of CHK1-mediated functions. Cell Rep. 2015, 11, 210–219. [Google Scholar] [CrossRef]
- Goguet-Rubio, P.; Seyran, B.; Gayte, L.; Bernex, F.; Sutter, A.; Delpech, H.; Linares, L.K.; Riscal, R.; Repond, C.; Rodier, G.; et al. E4F1-mediated control of pyruvate dehydrogenase activity is essential for skin homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 11004–11009. [Google Scholar] [CrossRef]
- He, Z.; Yan, R.G.; Zhang, X.N.; Yang, Q.E. Ring 1 and YY1 Binding Protein Expressed in Murine Spermatocytes but Dispensable for Spermatogenesis. Genes 2020, 11, 84. [Google Scholar] [CrossRef]
- Joyce, K.L.; Porcelli, J.; Cooke, P.S. Neonatal goitrogen treatment increases adult testis size and sperm production in the mouse. J. Androl. 1993, 14. [Google Scholar] [CrossRef]
- Cai, K.; Hua, G.; Ahmad, S.; Liang, A.; Han, L.; Wu, C.; Yang, F.; Yang, L. Action Mechanism of Inhibin α-Subunit on the Development of Sertoli Cells and First Wave of Spermatogenesis in Mice. PLoS ONE 2011, 6, e25585. [Google Scholar] [CrossRef] [PubMed]
- Holdcraft, R.W.; Braun, R.E. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development 2003, 131, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, R.M.; McKinnell, C.; Kivlin, C.; Fisher, J.S. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 2003, 125, 769–784. [Google Scholar] [CrossRef]
- Manku, G.; Culty, M. Mammalian gonocyte and spermatogonia differentiation: Recent advances and remaining challenges. Reproduction 2015, 149, R139–R157. [Google Scholar] [CrossRef]
- De Rooij, D.G. The spermatogonial stem cell niche. Microsc. Res. Tech. 2009, 72, 580–585. [Google Scholar] [CrossRef]
- Rebourcet, D.; O’Shaughnessy, P.J.; Pitetti, J.-L.; Monteiro, A.; O’Hara, L.; Milne, L.; Tsai, Y.T.; Cruickshanks, L.; Riethmacher, D.; Guillou, F.; et al. Sertoli cells control peritubular myoid cell fate and support adult Leydig cell development in the prepubertal testis. Development 2014, 141, 2139–2149. [Google Scholar] [CrossRef]
- Kumar, T.R.; Wang, Y.; Lu, N.; Matzuk, M.M. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat. Genet. 1997, 15, 201–204. [Google Scholar] [CrossRef]
- Nascimento, A.R.; Macheroni, C.; Lucas, T.F.G.; Porto, C.S.; Lazari, M.F.M. Crosstalk between FSH and relaxin at the end of the proliferative stage of rat Sertoli cells. Reproduction 2016, 152, 613–628. [Google Scholar] [CrossRef]
- Pimenta, M.T.; Porto, C.S.; Lazari, M.F. Effects of relaxin in a co-culture of Sertoli and germ cells. Arch. Ital. Anat. Embriol. 2013, 118, 29. [Google Scholar]
- Lim, K.; Hwang, B.-D. Follicle-stimulating hormone transiently induces expression of protooncogene c-myc in primary Sertoli cell cultures of early pubertal and prepubertal rat. Mol. Cell. Endocrinol. 1995, 111, 51–56. [Google Scholar] [CrossRef]
- Godmann, M.; Katz, J.P.; Guillou, F.; Simoni, M.; Kaestner, K.H.; Behr, R. Krüppel-like factor 4 is involved in functional differentiation of testicular Sertoli cells. Dev. Boil. 2008, 315, 552–566. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Igarashi, K.; Suzuki, T.; Kanno, J.; Inoue, T.; Tazawa, C.; Saruta, M.; Ando, T.; Moriyama, N.; Furukawa, T.; et al. E4F1, a Novel Estrogen-Responsive Gene in Possible Atheroprotection, Revealed by Microarray Analysis. Am. J. Pathol. 2004, 165, 2019–2031. [Google Scholar] [CrossRef][Green Version]
- Rotgers, E.; Rivero-Müller, A.; Nurmio, M.; Parvinen, M.; Guillou, F.; Huhtaniemi, I.; Kotaja, N.; Bourguiba-Hachemi, S.; Toppari, J. Retinoblastoma protein (RB) interacts with E2F3 to control terminal differentiation of Sertoli cells. Cell Death Dis. 2014, 5, e1274. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.-C.; Jiang, M.; Beaudet, A.L.; Wu, M.-Y. ARID4A and ARID4B regulate male fertility, a functional link to the AR and RB pathways. Proc. Natl. Acad. Sci. USA 2013, 110, 4616–4621. [Google Scholar] [CrossRef]
- Lacroix, M.; Rodier, G.; Kirsh, O.; Houles, T.; Delpech, H.; Seyran, B.; Gayte, L.; Casas, F.; Pessemesse, L.; Heuillet, M.; et al. E4F1 controls a transcriptional program essential for pyruvate dehydrogenase activity. Proc. Natl. Acad. Sci. USA 2016, 113, 10998–11003. [Google Scholar] [CrossRef]
- Thomas, K.; Del Mazo, J.; Eversole, P.; Bellvé, A.; Hiraoka, Y.; Li, S.S.; Simon, M. Developmental regulation of expression of the lactate dehydrogenase (LDH) multigene family during mouse spermatogenesis. Development 1990, 109, 483–493. [Google Scholar]
- Tan, Q.; Li, J.; Peng, J.; Liu, Z.; Liu, J.; Zhang, H.; Yuan, Q.; Pan, Z.; Liu, L. E4F1 silencing inhibits the cell growth through cell-cycle arrest in malignant transformed cells induced by hydroquinone. J. Biochem. Mol. Toxicol. 2018, 33, e22269. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, R.-G.; Yang, Q.-L.; Yang, Q.-E. E4 Transcription Factor 1 (E4F1) Regulates Sertoli Cell Proliferation and Fertility in Mice. Animals 2020, 10, 1691. https://doi.org/10.3390/ani10091691
Yan R-G, Yang Q-L, Yang Q-E. E4 Transcription Factor 1 (E4F1) Regulates Sertoli Cell Proliferation and Fertility in Mice. Animals. 2020; 10(9):1691. https://doi.org/10.3390/ani10091691
Chicago/Turabian StyleYan, Rong-Ge, Qi-Lin Yang, and Qi-En Yang. 2020. "E4 Transcription Factor 1 (E4F1) Regulates Sertoli Cell Proliferation and Fertility in Mice" Animals 10, no. 9: 1691. https://doi.org/10.3390/ani10091691
APA StyleYan, R.-G., Yang, Q.-L., & Yang, Q.-E. (2020). E4 Transcription Factor 1 (E4F1) Regulates Sertoli Cell Proliferation and Fertility in Mice. Animals, 10(9), 1691. https://doi.org/10.3390/ani10091691




