Maternal Origins and Haplotype Diversity of Seven Russian Goat Populations Based on the D-loop Sequence Variability
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
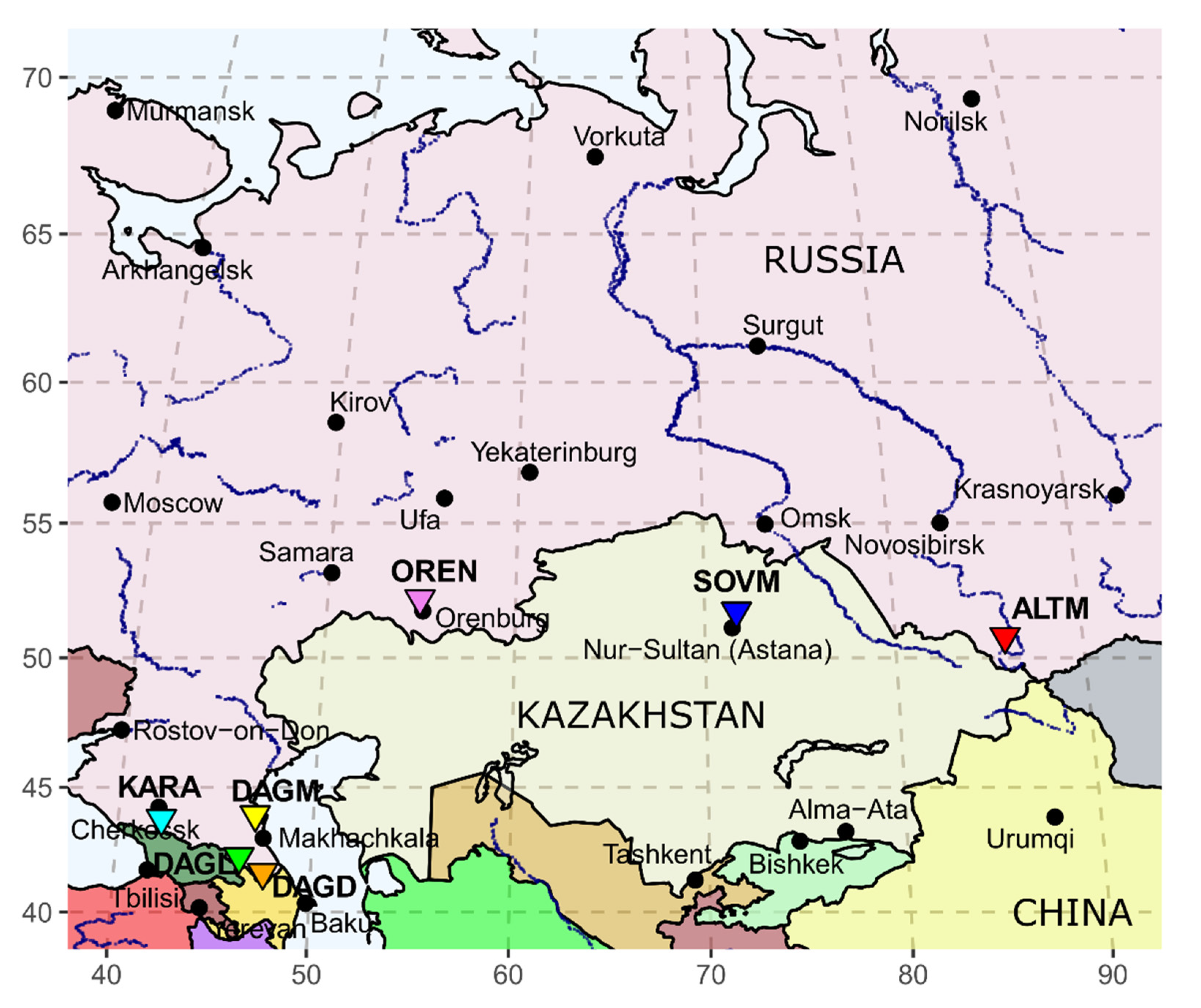
2.1. Sample Collection
2.2. PCR Amplification, Purification, and Sequencing
2.3. Sequence Alignment and Data Analysis
3. Results
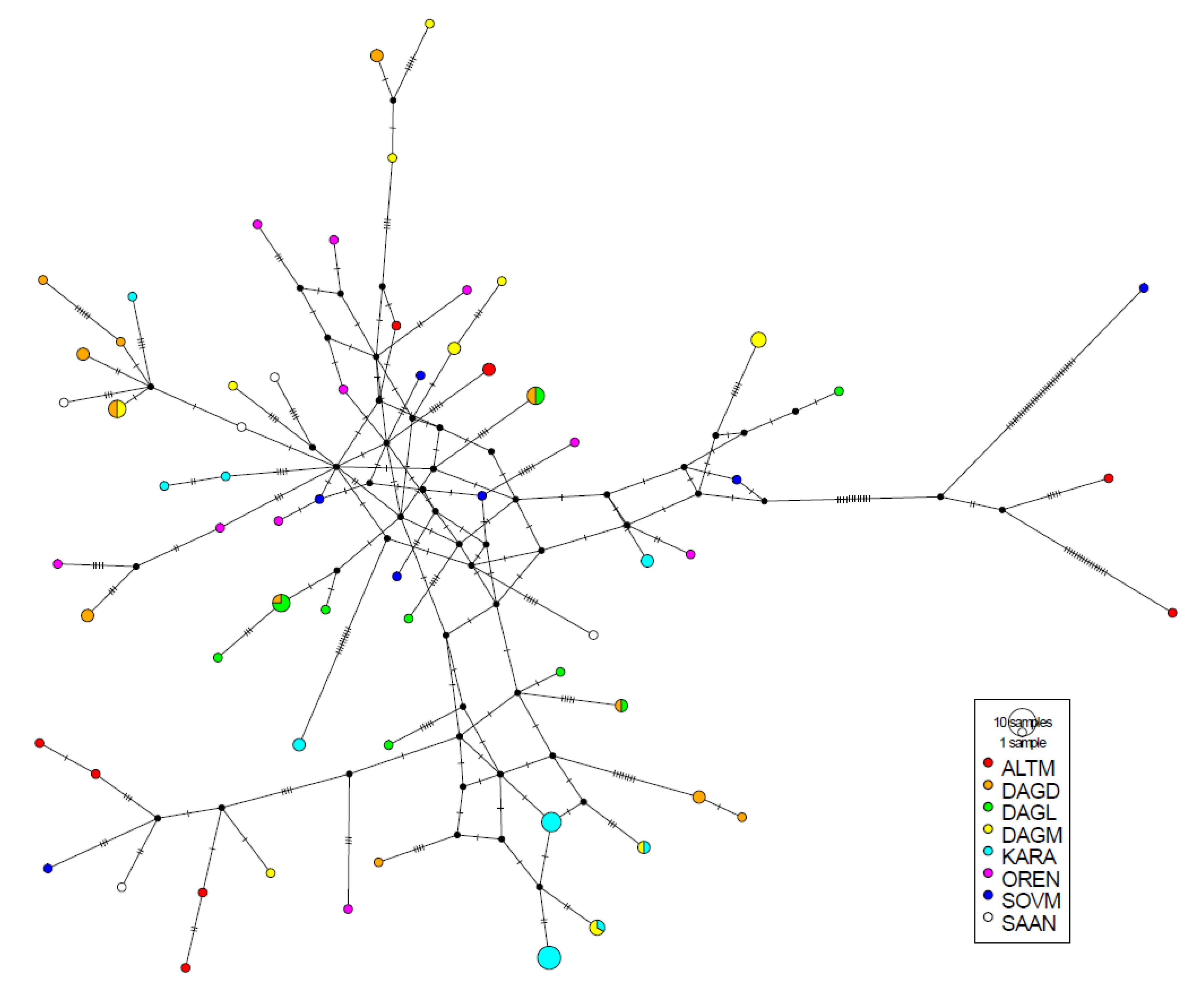
3.1. D-loop MtDNA Sequence Variation and Genetic Diversity
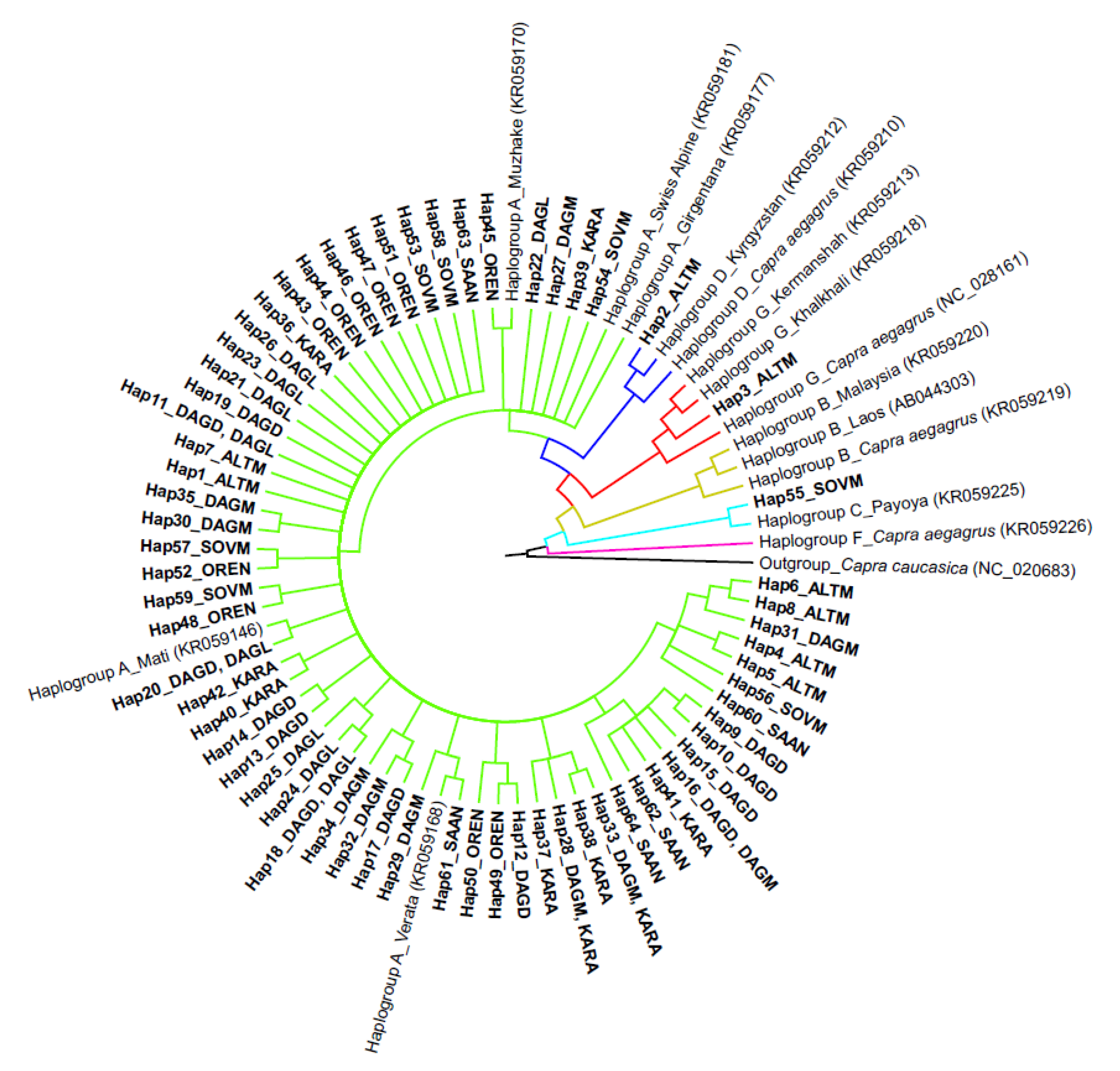
3.2. Analysis of Phylogenetic Links and Haplogroup Assignment of Russian Local Goats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Novopashina, S.I.; Sannikov, M.Y.; Khatataev, S.A.; Kuzmina, T.N.; Khmelevskaya, G.N.; Stepanova, N.G.; Tikhomirov, A.I.; Marinchenko, T.E. Status and Perspective areas for Improving the Genetic Potential of Small Cattle: Scientific and Analytic Overview; Rosinformagrotekh: Moscow, Russia, 2019; pp. 27–39. [Google Scholar]
- Hajitov, A.H.; Stanishevskaya, O.N.; Safarov, T.S. Biologicheskie i hozyajstvennye priznaki mestnyh koz. Izv. Spbgau 2016, 45, 139–145. (In Russian) [Google Scholar]
- Grigoryan, L.N.; Hatataev, S.A.; Sverchkova, S.V. Sostoyanie kozovodstva Rossijskoj Federacii i ego plemennoj bazy. In Ezhegodnik po Plemennoj Rabote v Ovcevodstve i Kozovodstve v Hozyajstvah Rossijskoj Federacii (2005 god); Desyatov, V.G., Ed.; Vserossijskij Nauchno-Issledovatel’skij Institut Plemennogo Dela: Lesnye Polyany, Russia, 2006; pp. 312–313. (In Russian) [Google Scholar]
- Dunin, I.M.; Amerhanov, H.A.; Safina, G.F.; Grigoryan, L.N.; Hatataev, S.A.; Hmelevskaya, G.N.; Pavlov, M.B.; Stepanova, N.G. Kozovodstvo Rossii i ego plemennye resursy. In Ezhegodnik po Plemennoj Rabote v Ovcevodstve i Kozovodstve v Hozyajstvah Rossijskoj Federacii (2019 god); Desyatov, V.G., Ed.; Vserossijskij Nauchno-Issledovatel’skij Institut Plemennogo Dela: Lesnye Polyany, Russia, 2019; pp. 323–325. (In Russian) [Google Scholar]
- Musalaev, K.K.; Palaganova, G.A.; Abdullabekov, R.A. Rezultaty nauchnykh issledovanii po ovtsevodstvu i kozovodstvu Dagestana. Gorn. Sel’skoe Hozyajstvo 2015, 2, 121–124. (In Russian) [Google Scholar]
- Mamontova, T.V.; Gadzhiev, Z.K.; Aybazov, A.-M.M. Produktivnye i vosproizvoditelnye osobennosti mestnykh karachaevskikh koz. Sb. Nauchnyh Tr. Stavropol’skogo Nauchno-Issledovatel’skogo Inst. Zhivotnovodstva I Kormoproizvod. 2011, 1, 15–17. (In Russian) [Google Scholar]
- Colli, L.; Lancioni, H.; Cardinali, I.; Olivieri, A.; Capodiferro, M.R.; Pellecchia, M.; Rzepus, M.; Zamani, W.; Naderi, S.; Gandini, F.; et al. Whole mitochondrial genomes unveil the impact of domestication on goat matrilineal variability. BMC Genom. 2015, 16, 1115. [Google Scholar] [CrossRef]
- Luikart, G.; Gielly, L.; Excoffier, L.; Vigne, J.D.; Bouvet, J.; Taberlet, P. Multiple maternal origins and weak phylogeographic structure in domestic goat. Proc. Natl. Acad. Sci. USA 2001, 98, 5927–5932. [Google Scholar] [CrossRef]
- Naderi, S.; Rezaei, H.-R.; Taberlet, P.; Zundel, S.; Rafat, S.-A.; Naghash, H.R.; El-Barody, M.A.; Ertugrul, O.; Pompanon, F.; Econogene Consortium. Large-Scale Mitochondrial DNA Analysis of the Domestic Goat Reveals Six Haplogroups with High Diversity. PLoS ONE 2007, 2, e1012. [Google Scholar] [CrossRef]
- Naderi, S.; Rezaei, H.R.; Pompanon, F.; Blum, M.G.B.; Negrini, R.; Naghash, H.R.; Balkiz, O.; Mashkour, M.; Gaggiotti, O.E.; Ajmone-Marsan, P.; et al. The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals. Proc. Natl. Acad. Sci. USA 2008, 105, 17659–17664. [Google Scholar] [CrossRef]
- Piras, D.; Doro, M.G.; Casu, G.; Melis, P.M.; Vaccargiu, S.; Piras, I.; Parracciani, D.; Stradoni, R.; Frongia, B.; Lai, G.; et al. Haplotype affinities resolve a major component of goat (Capra hircus) MtDNA D-loop diversity and reveal specific features of the Sardinian stock. PLoS ONE 2012, 7, e30785. [Google Scholar] [CrossRef]
- Fernández, H.; Hughes, S.; Vigne, J.-D.; Helmer, D.; Hodgins, G.; Miquel, C.; Miquel, C.; Hänni, C.; Luikart, G.; Taberlet, P. Divergent mtDNA lineages of goats in an Early Neolithic site, far from the initial domestication areas. Proc. Natl. Acad. Sci. USA 2006, 103, 15375–15379. [Google Scholar] [CrossRef]
- Nomura, K.; Yonezawa, T.; Mano, S.; Kawakami, S.; Shedlock, A.M.; Hasegawa, M.; Amano, T. Domestication Process of the Goat Revealed by an Analysis of the Nearly Complete Mitochondrial Protein-Encoding Genes. PLoS ONE 2013, 8, e67775. [Google Scholar] [CrossRef]
- Doro, M.G.; Piras, D.; Leoni, G.G.; Casu, G.; Vaccargiu, S.; Parracciani, D.; Naitana, S.; Pirastu, M.; Novelletto, A. Phylogeny and Patterns of Diversity of Goat mtDNA Haplogroup A Revealed by Resequencing Complete Mitogenomes. PLoS ONE 2014, 9, e95969. [Google Scholar] [CrossRef] [PubMed]
- Orekhov, A.A. Goats. Animal Genetic Resources of the USSR; Dmitriev, N.G., Ernst, L.K., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 1989; pp. 344–365. [Google Scholar]
- Dunin, I.M.; Dankvert, A.G. Breeds and Types of Farm Animals in the Russian Federation; Fgbnu Vniiplem: Moscow, Russia, 2013; pp. 234–242. [Google Scholar]
- Tabata, R.; Kawaguchi, F.; Sasazaki, S.; Yamamoto, Y.; Bakhtin, M.; Kazymbet, P.; Meldevekob, A.; Suleimenov, M.Z.; Nishibori, M.; Mannen, H. The Eurasian Steppe is an important goat propagation route: A phylogeographic analysis using mitochondrial DNA and Y-chromosome sequences of Kazakhstani goats. Anim. Sci. J. 2018, 90, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Wagner, A.P.; Taper, M.L. ML-Relate: A computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 2006, 6, 576–579. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Bodenhofer, U.; Bonatesta, E.; Horejs-Kainrath, C.; Hochreiter, S. msa: An R package for multiple sequence alignment. Bioinformatics 2015, 31, 3997–3999. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Akaike, H. A new look at statistical model identification. IEEE Trans Auto Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Molecular Evolution, Phylogenetics and Epidemiology. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 30 August 2020).
- Becker, R.A.; Wilks, A.R.; Brownrigg, R.; Minka, T.P.; Deckmyn, A. Maps: Draw Geographical Maps. R Package Version 3.2.0; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Kharzinova, V.R.; Petrov, S.N.; Dotsev, A.V.; Bezborodova, N.A.; Zinovieva, N.A. Populiatsionno-geneticheskaia kharakteristika nekotorykh porod koz na osnove analiza mikrosatellitov. Ovcy Kozy Sherstyanoe Delo 2019, 3, 7–11. (In Russian) [Google Scholar] [CrossRef]
- Deniskova, T.; Dotsev, A.; Fornara, M.; Selionova, M.; Reyer, H.; Wimmers, K.; Brem, G.; Zinovieva, N. Whole-genome assessment of goat breeds in Russia. In Proceedings of the 37th International Conference on Animal Genetics, Lleida, Spain, 7–12 July 2019; p. 132. [Google Scholar]
- Petrov, N.I. Sokhranenie genofonda koz orenburgskoi porody. Izv. Orenb. Gos. Agrar. Univ. 2016, 4, 157–159. (In Russian) [Google Scholar]
- Zagirov, N.G. Nauchnoe obespechenie—Zalog effektivnogo i konkurentosposobnogo zhivotnovodstva Dagestana. Sel’skohozyajstvennyj Zhurnal 2014, 3, 3–11. (In Russian) [Google Scholar]
- Gadzhiev, Z.K.; Bobryshova, G.T. Grubosherstnye ovtsy Dagestana. Sel’skohozyajstvennyj Zhurnal 2017, 2, 3–10. (In Russian) [Google Scholar]
- Wang, G.Z.; Pi, X.S.; Ji, Z.B.; Qin, Z.J.; Hou, L.; Chao, T.L.; Wang, J.M. Investigation of the diversity and origins of Chinese dairy goats via the mitochondrial DNA D-loop. J. Anim. Sci. 2015, 93, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Kamalakkannan, R.; Jose, J.; Thomas, S.; Prabhu, V.R.; Nagarajan, M. Genetic diversity and maternal lineages of south Indian goats. Mol. Biol. Rep. 2018, 45, 2741–2748. [Google Scholar] [CrossRef]
- Ganbold, O.; Lee, S.H.; Paek, W.K.; Munkhbayar, M.; Seo, D.; Manjula, P.; Khujuu, T.; Purevee, E.; Lee, J.H. Mitochondrial DNA variation and phylogeography of native Mongolian goats. Asian Australas. J. Anim. Sci. 2020, 33, 902–912. [Google Scholar] [CrossRef]
- Tarekegn, G.M.; Tesfaye, K.; Mwai, O.A.; Djikeng, A.; Dessie, T.; Birungi, J.; Osama, S.; Zergaw, N.; Alemu, A.; Achieng, G.; et al. Mitochondrial DNA variation reveals maternal origins and demographic dynamics of Ethiopian indigenous goats. Ecol. Evol. 2018, 8, 1543–1553. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, R.; Zhao, Z.; Xu, H.; Zhao, E.; Zhang, J. Genetic diversity and molecular phylogeography of Chinese domestic goats by large-scale mitochondrial DNA analysis. Mol. Biol. Rep. 2014, 41, 3695–3704. [Google Scholar] [CrossRef] [PubMed]
- Sultana, S.; Mannen, H.; Tsuji, S. Mitochondrial DNA diversity of Pakistani goats. Anim. Genet. 2003, 34, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.B.; Rout, P.K.; Mandal, A.K.; Tyler-Smith, C.; Singh, L.; Thangaraj, K. Phylogeography and origin of Indian domestic goats. Mol. Biol. Evol. 2004, 21, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Akis, I.; Oztabak, K.; Mengi, A.; Un, C. Mitochondrial DNA diversity of Anatolian indigenous domestic goats. J. Anim. Breed. Genet. 2014, 131, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Barbato, M.; Traspov, A.A.; Brem, G.; et al. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. GSE 2018, 50, 29. [Google Scholar] [CrossRef]
| Breed/Group | Code | N 1 | S 2 | H 3 | HD 4 | K 5 | π 6 |
|---|---|---|---|---|---|---|---|
| Altai Mountain | ALTM | 9 | 45 | 8 | 0.972 ± 0.064 | 16.222 | 0.02266 ± 0.00466 |
| Dagestan Downy | DAGD | 18 | 45 | 12 | 0.961 ± 0.026 | 11.608 | 0.01619 ± 0.00137 |
| Dagestan Local | DAGL | 12 | 28 | 9 | 0.939 ± 0.058 | 7.364 | 0.01027 ± 0.00138 |
| Dagestan Milk | DAGM | 15 | 38 | 10 | 0.943 ± 0.040 | 10.781 | 0.01542 ± 0.00114 |
| Karachaev | KARA | 21 | 36 | 9 | 0.843 ± 0.057 | 7.848 | 0.01121 ± 0.00197 |
| Orenburg | OREN | 10 | 30 | 10 | 1.000 ± 0.045 | 8.844 | 0.01234 ± 0.00147 |
| Soviet Mohair | SOVM | 7 | 57 | 7 | 1.000 ± 0.076 | 18.667 | 0.02607 ± 0.01078 |
| Saanen | SAAN | 5 | 25 | 5 | 1.000 ± 0.126 | 10.600 | 0.01478 ± 0.00297 |
| Source of Variation | Russian Breeds | Russian Breeds Clustered with Samples Carrying Haplogroup A | ||||||
|---|---|---|---|---|---|---|---|---|
| d.f. | SS | VC | V% | d.f. | SS | VC | V% | |
| Among breeds | 6 | 194.954 | 1.99876 | 22.76 | 6 | 187.719 | 2.03601 | 24.98 |
| Within breeds | 85 | 576.611 | 6.78366 | 77.24 | 82 | 501.405 | 6.11469 | 75.02 |
| Total | 91 | 771.565 | 8.78242 | 88 | 689.124 | 8.15070 | ||
| Haplotype | Number of Variable Sites 1 | Distribution of Haplotypes 2 among Studied Breeds | Haplotype | Number of Variable Sites 1 | Distribution of Haplotypes 3 among Studied Breeds | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Altai Mountain | Dagestan Downy | Dagestan Local | Dagestan Milk | Karachaev | Orenburg | Soviet Mohair | Saanen | Altai Mountain | Dagestan Downy | Dagestan Local | Dagestan Milk | Karachaev | Orenburg | Soviet Mohair | Saanen | ||||
| H-1 | 5 | 2 | H-33 | 8 4 | 2 | 1 | |||||||||||||
| H-2 | 22 | 1 | H-34 | 8 | 1 | ||||||||||||||
| H-3 | 24 3 | 1 | H-35 | 6 | 1 | ||||||||||||||
| H-4 | 8 | 1 | H-36 | 12 | 2 | ||||||||||||||
| H-5 | 9 | 1 | H-37 | 5 4 | 5 | ||||||||||||||
| H-6 | 9 | 1 | H-38 | 8 4 | 7 | ||||||||||||||
| H-7 | 3 | 1 | H-39 | 7 | 2 | ||||||||||||||
| H-8 | 7 | 1 | H-40 | 5 | 1 | ||||||||||||||
| H-9 | 10 | 1 | H-41 | 8 | 1 | ||||||||||||||
| H-10 | 4 | 1 | H-42 | 7 | 1 | ||||||||||||||
| H-11 | 6 | 2 | 2 | H-43 | 7 | 1 | |||||||||||||
| H-12 | 7 | 2 | H-44 | 3 | 1 | ||||||||||||||
| H-13 | 14 | 1 | H-45 | 8 | 1 | ||||||||||||||
| H-14 | 13 | 2 | H-46 | 1 | 1 | ||||||||||||||
| H-15 | 5 | 2 | H-47 | 6 | 1 | ||||||||||||||
| H-16 | 4 | 2 | 2 | H-48 | 3 | 1 | |||||||||||||
| H-17 | 8 | 2 | H-49 | 8 | 1 | ||||||||||||||
| H-18 | 3 | 1 | 3 | H-50 | 4 | 1 | |||||||||||||
| H-19 | 7 | 1 | H-51 | 3 | 1 | ||||||||||||||
| H-20 | 9 | 1 | 1 | H-52 | 9 | 1 | |||||||||||||
| H-21 | 5 | 1 | H-53 | 3 | 1 | ||||||||||||||
| H-22 | 6 | 1 | H-54 | 6 | 1 | ||||||||||||||
| H-23 | 6 | 1 | H-55 | 46 | 1 | ||||||||||||||
| H-24 | 6 | 1 | H-56 | 8 5 | 1 | ||||||||||||||
| H-25 | 2 | 1 | H-57 | 3 | 1 | ||||||||||||||
| H-26 | 8 | 1 | H-58 | 3 | 1 | ||||||||||||||
| H-27 | 12 | 3 | H-59 | 2 | 1 | ||||||||||||||
| H-28 | 9 4 | 1 | 1 | H-60 | 7 | 1 | |||||||||||||
| H-29 | 7 | 1 | H-61 | 7 | 1 | ||||||||||||||
| H-30 | 3 | 2 | H-62 | 6 | 1 | ||||||||||||||
| H-31 | 7 5 | 1 | H-63 | 8 | 1 | ||||||||||||||
| H-32 | 6 | 1 | H-64 | 2 | 1 | ||||||||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deniskova, T.; Bakoev, N.; Dotsev, A.; Selionova, M.; Zinovieva, N. Maternal Origins and Haplotype Diversity of Seven Russian Goat Populations Based on the D-loop Sequence Variability. Animals 2020, 10, 1603. https://doi.org/10.3390/ani10091603
Deniskova T, Bakoev N, Dotsev A, Selionova M, Zinovieva N. Maternal Origins and Haplotype Diversity of Seven Russian Goat Populations Based on the D-loop Sequence Variability. Animals. 2020; 10(9):1603. https://doi.org/10.3390/ani10091603
Chicago/Turabian StyleDeniskova, Tatiana, Nekruz Bakoev, Arsen Dotsev, Marina Selionova, and Natalia Zinovieva. 2020. "Maternal Origins and Haplotype Diversity of Seven Russian Goat Populations Based on the D-loop Sequence Variability" Animals 10, no. 9: 1603. https://doi.org/10.3390/ani10091603
APA StyleDeniskova, T., Bakoev, N., Dotsev, A., Selionova, M., & Zinovieva, N. (2020). Maternal Origins and Haplotype Diversity of Seven Russian Goat Populations Based on the D-loop Sequence Variability. Animals, 10(9), 1603. https://doi.org/10.3390/ani10091603








