Cardiac Transcriptomics Reveals That MAPK Pathway Plays an Important Role in Hypoxia Tolerance in Bighead Carp (Hypophthalmichthys nobilis)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Fish and Hypoxia Treatment
2.2. Physiological and Biochemical Responses of Bighead Carp to Hypoxia
2.3. RNA Extraction and Transcriptome Sequencing
2.4. Transcriptome Alignment and Functional Annotation
2.5. Analysis of Differential Gene Expression
2.6. Validation of RNA-Seq Results by qRT-PCR
3. Results
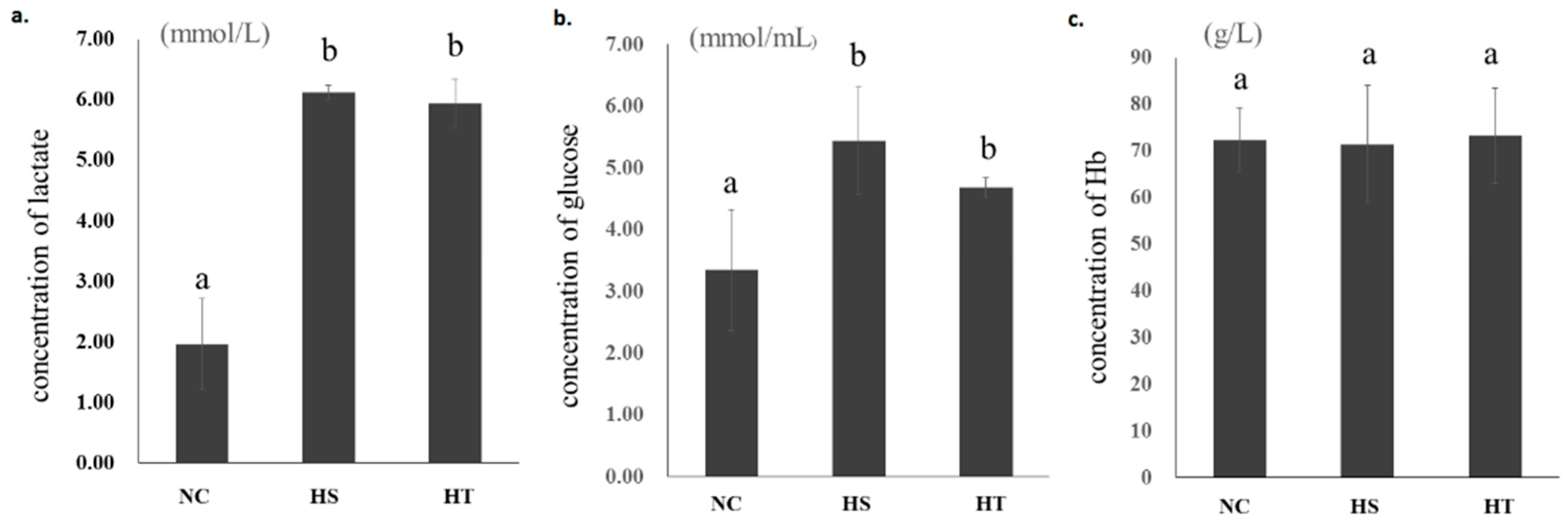
3.1. Changes of Blood Parameters after Hypoxia Stress
3.2. Raw Sequencing Data and De Novo Transcriptome Assembly
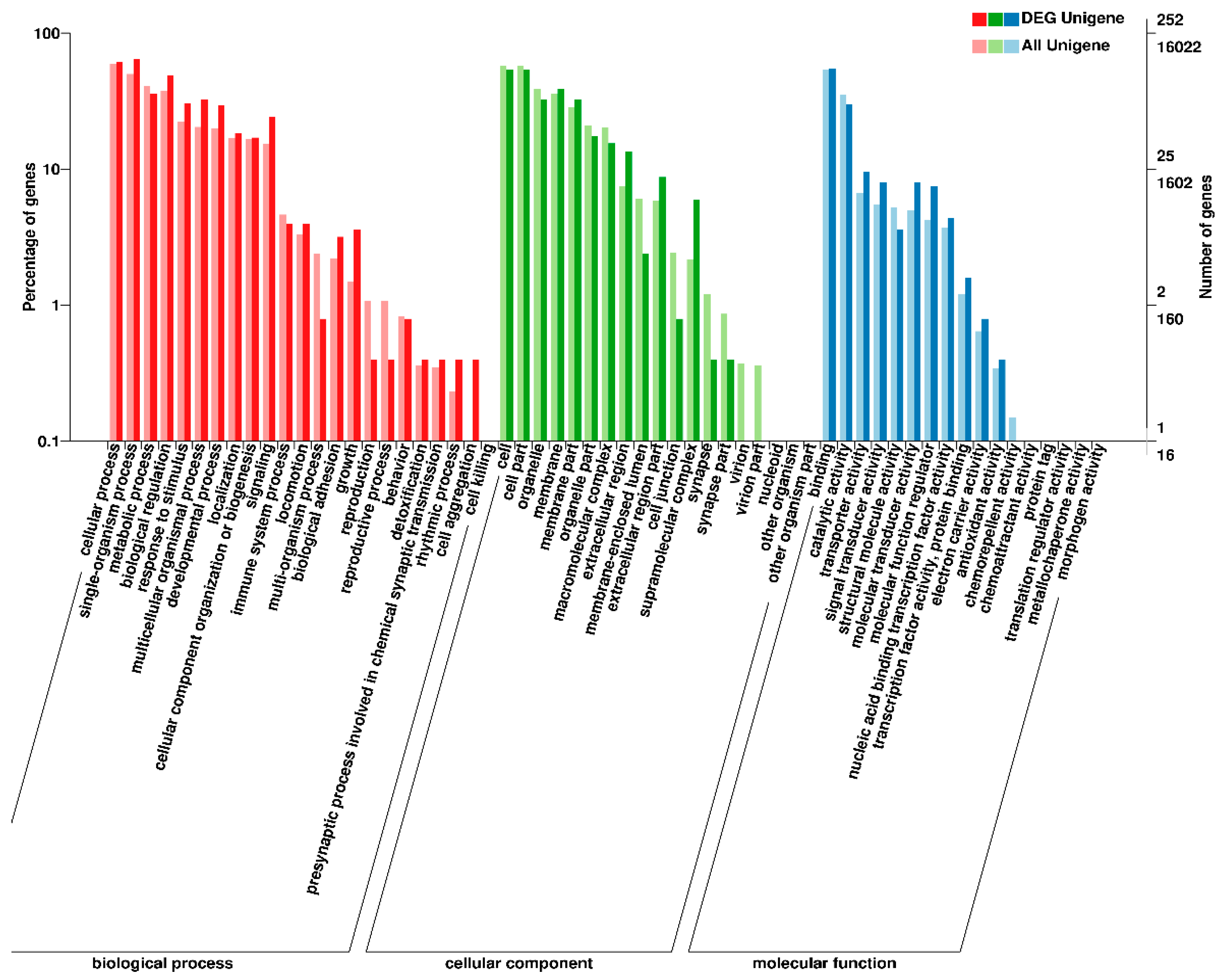
3.3. Functional Annotation and Classification of Unigenes
3.4. Differential Expression Analysis and Functional Annotation
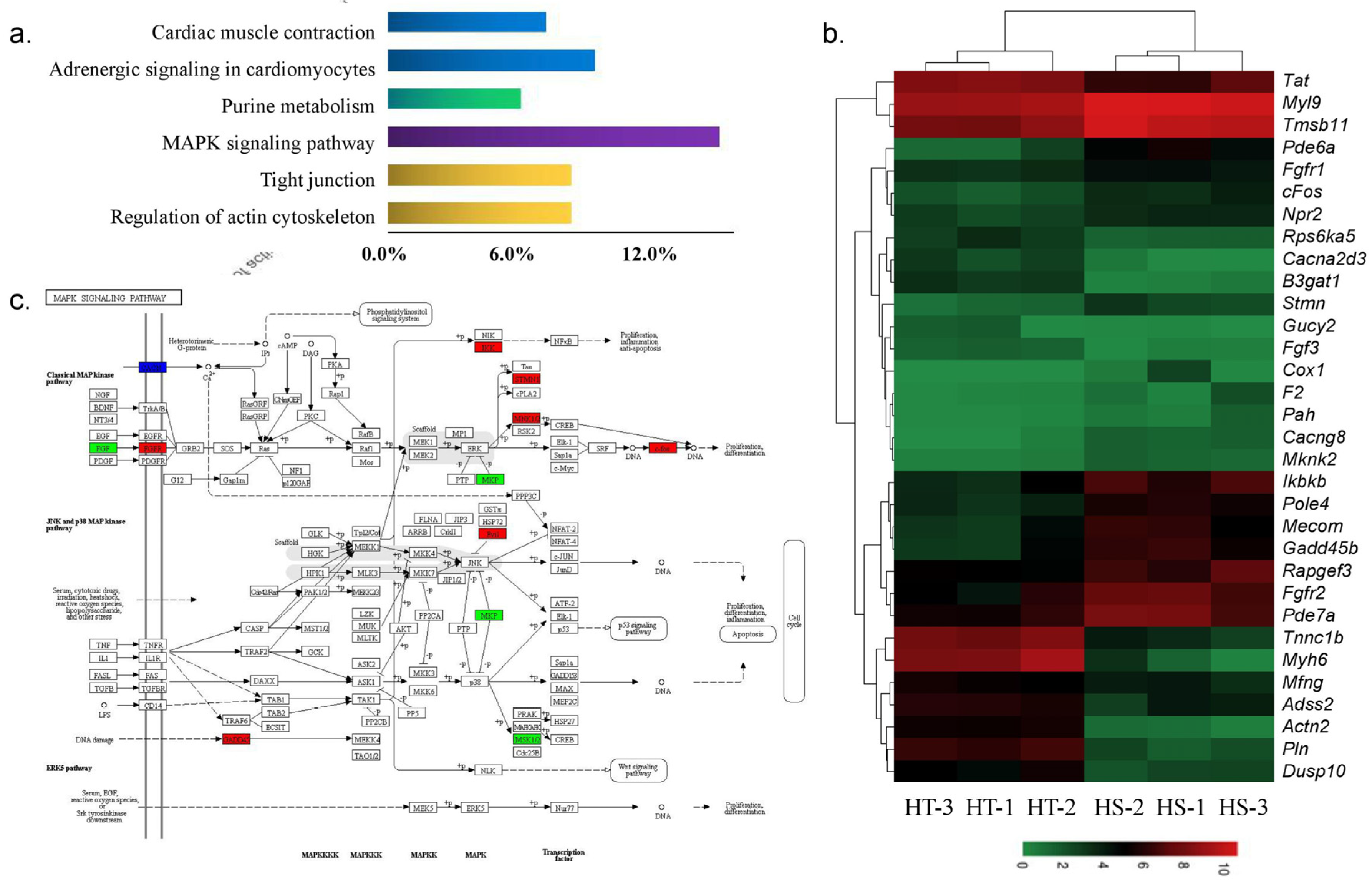
3.5. Critical DEGs Involved in the Response to Hypoxia Stress
3.6. Validation of RNA-Seq Results by qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Val, A.J.; Lessard, J.; Randall, D.J. Effects of hypoxia on rainbow trout (Oncorhynchus mykiss): Intraerythrocytic phosphates. J. Exp. Biol. 1995, 198, 305–310. [Google Scholar] [PubMed]
- Feng, X.; Yu, X.; Pang, M.; Tong, J. Molecular characterization and expression regulation of the factor-inhibiting Hif-1 (Fih-1) gene under hypoxic stress in bighead carp (Aristichthys nobilis). Fish Physiol. Biochem. 2019, 45, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Qin, J.; Zhang, X. Fish adaptation to oxygen variations in aquaculture from hypoxia to hyperoxia. J. Fish Aquat. 2011, 2, 23–28. [Google Scholar]
- Randall, D.; MacKinlay, D. Responses of Fish to Aquatic Hypoxia; University of British Columbia: Vancouver, BC, Canada, 2002. [Google Scholar]
- Richards, J.G. Chapter 10 Metabolic and molecular responses of fish to hypoxia. Fish Physiol. 2009, 27, 443–485. [Google Scholar]
- Timmerman, C.M.; Chapman, L.J. Behavioral and physiological compensation for chronic hypoxia in the sailfin molly (Poecilia latipinna). Physiol. Biochem. Zool. 2004, 77, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Gracey, A.Y.; Troll, J.V.; Somero, G.N. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc. Natl. Acad. Sci. USA 2001, 98, 1993–1998. [Google Scholar] [CrossRef]
- Ton, C.; Stamatiou, D.; Liew, C.C. Gene expression profile of zebrafish exposed to hypoxia during development. Physiol. Genom. 2003, 13, 97–106. [Google Scholar] [CrossRef]
- Bickler, P.E.; Buck, L.T. Hypoxia tolerance in reptiles, amphibians, and fishes: Life with variable oxygen availability. Annu. Rev. Physiol. 2007, 69, 145–170. [Google Scholar] [CrossRef]
- Zhang, G.; Yin, S.; Mao, J.; Liang, F.; Tang, Z. Integrated analysis of mRNA-seq and miRNA-seq in the liver of Pelteobagrus vachelli in response to hypoxia. Sci. Rep. 2016, 6, 22907. [Google Scholar] [CrossRef]
- Wang, Q.F.; Shen, W.L.; Hou, C.C.; Liu, C.; Wu, X.F.; Zhu, J.Q. Physiological responses and changes in gene expression in the large yellow croaker Larimichthys crocea following exposure to hypoxia. Chemosphere 2017, 169, 418–427. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factor 1: Oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 2001, 7, 345–350. [Google Scholar] [CrossRef]
- Merante, F.; Mickle, D.A.G.; Weisel, R.D.; Li, R.K.; Tumiati, L.C.; Rao, V.; Williams, W.G.; Robonson, B.H. Myocardial aerobic metabolism is impaired in a cell culture model of cyanotic heart disease. Am. J. Physiol. 1998, 275, H1673–H1681. [Google Scholar] [CrossRef] [PubMed]
- Marques, I.J.; Leito, J.T.D.; Spaink, H.P.; Testerink, J.; Jaspers, R.T.; Witte, F.; van Den Berg, S.; Bagowski, C.P. Transcriptome analysis of the response to chronic constant hypoxia in zebrafish hearts. J. Comp. Physiol. B 2008, 178, 77–92. [Google Scholar] [CrossRef]
- Yan, T.T.; Fan, W.W.; He, J.P. The effect of hypoxia tolerance on cardiac muscle structure of gansu zokor (Myospalax cansus). J. Shaanxi Norm. Univ. 2012, 40, 62–66. [Google Scholar]
- Szarszoi, O.; Asemu, G.; Ostadal, B.; Kolar, F. The role of reactive oxygen species and nitric oxide in ischemia/reperfusion injury of chronically hypoxic rat heart. Eur. Heart Fail. 2003, 2, 53. [Google Scholar] [CrossRef]
- Xia, H.; Li, H.L.; Li, B.J.; Gu, X.H.; Lin, H.R. Acute hypoxia stress induced abundant differential expression genes and alternative splicing events in heart of tilapia. Gene 2018, 639, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Liu, J. Practical success of biomanipulation using filter-feeding fish to control cyanobacteria blooms: A synthesis of decades of research and application in a subtropical hypereutrophic lake. Sci. World J. 2014, 1, 337–356. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.B.; Chen, W.X.; Fan, Z.T. Advancement of the study on respiratory metabolism of fishes. Chin. J. Fish. 2004, 17, 82–89. [Google Scholar]
- Boswell, M.G.; Wells, M.C.; Kirk, L.M.; Ju, Z.; Zhang, Z.; Booth, R.E.; Walter, R.B. Comparison of gene expression responses to hypoxia in viviparous (Xiphophorus) and oviparous (Oryzias) fishes using a medaka microarray. Comp. Biochem. Phys. C 2009, 149, 258–265. [Google Scholar] [CrossRef]
- Leveelahti, L.; Leskinen, P.; Leder, E.H.; Waser, W.; Nikinmaa, M. Responses of threespine stickleback (Gasterosteus aculeatus, L) transcriptome to hypoxia. Comp. Biochem. Physiol. D 2011, 6, 370–381. [Google Scholar] [CrossRef]
- Olsvik, P.A.; Kristensen, T.; Waagb, R.; Rosseland, B.O.; Tollefsen, K.E.; Baeverfjord, G.; Berntssen, M.H.G. mRNA expression of antioxidant enzymes (SOD, CAT and GSH-Px) and lipid peroxidative stress in liver of Atlantic salmon (Salmo salar) exposed to hyperoxic water during smoltification. Comp. Biochem. Physiol. C 2005, 141, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.B.; Liu, Z.Y.; Li, F.G.; Chen, J.; Jiang, X.Y.; Zou, S.M. Gill remodeling in response to hypoxia and temperature occurs in the hypoxia sensitive blunt snout bream (Megalobrama amblycephala). Aquaculture 2017, 479, 479–486. [Google Scholar] [CrossRef]
- Qi, D.L.; Chao, Y.; Wu, R.R.; Xia, M.Z.; Chen, Q.C.; Zheng, Z.Q. Transcriptome analysis provides insights into the adaptive responses to hypoxia of a schizothoracine Fish (Gymnocypris eckloni). Front. Physiol. 2018, 9, 1326. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Wang, X.H.; Yu, X.M.; Pang, M.X.; Tong, J.G. cDNA-AFLP analysis of differentially expressed genes related to hypoxia stress in bighead carp (Hypophthalmichthys nobilis). J. Agric. Biotechnol. 2017, 25, 1326–1335. [Google Scholar]
- Zhu, C.D.; Wang, Z.H.; Yan, B. Strategies for hypoxia adaptation in fish species: A review. J. Comp. Physiol. B 2013, 183, 1005–1013. [Google Scholar] [CrossRef]
- Storz, J.F.; Scott, G.R.; Cheviron, Z.A. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J. Exp. Biol. 2010, 213, 4125–4136. [Google Scholar] [CrossRef]
- Urbina, M.A.; Glover, C.N. Should I stay or should I go? Physiological, metabolic and biochemical consequences of voluntary emersion upon aquatic hypoxia in the scaleless fish Galaxias maculatus. J. Comp. Physiol. B 2012, 182, 1057–1067. [Google Scholar] [CrossRef]
- McKenzie, D.J.; Steffensen, J.F.; Korsmeyer, K.; Whiteley, N.M.; Bronzi, P.; Taylor, E.W. Swimming alters responses to hypoxia in the Adriatic sturgeon Acipenser naccarii. J. Fish Biol. 2007, 70, 651–658. [Google Scholar] [CrossRef]
- Portner, H.O.; Heisler, N.; Grieshaber, M.K. Oxygen consumption and mode of energy production in the intertidal worm Sipunculus nudus L: Definition and characterization of the critical PO2 for an oxyconformer. Res. Physiol. 1985, 59, 361–377. [Google Scholar] [CrossRef]
- Cooper, R.U.; Clough, L.M.; Farwell, M.A.; West, T.L. Hypoxia-induced metabolic and antioxidant enzymatic activities in the estuarine fish Leiostomus xanthurus. J. Exp. Mar. Biol. Ecol. 2002, 279, 1–20. [Google Scholar] [CrossRef]
- Rimoldi, S.; Terova, G.; Ceccuzzi, P.; Marelli, S.; Antonini, M.; Saroglia, M. HIF-1a mRNA levels in Eurasian perch (Perca fluviatilis) exposed to acute and chronic hypoxia. Mol. Biol. Rep. 2012, 39, 4009–4015. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.J.; Fu, C.; Yan, G.J.; Cao, Z.D.; Zhang, A.J.; Pang, X. Interspecific variation in hypoxia tolerance, swimming performance and plasticity in cyprinids that prefer different habitats. J. Exp. Biol. 2014, 217, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.X.; Yi, S.K.; Wang, W.F.; He, Y.; Huang, Y.; Gao, Z.X.; Liu, H.; Wang, W.M.; Wang, H.L. Transcriptome comparison reveals insights into muscle response to hypoxia in blunt snout bream (Megalobrama amblycephala). Gene 2017, 624, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Nikinmaa, V. Oxygen-dependent cellular functions–why fishes and their aquatic environment are a prime choice of study. Comp. Biochem. Physiol. A 2002, 133, 1–16. [Google Scholar] [CrossRef]
- Preiser, M.J.C.; Ichai, C.; Orban, J.C.; Groeneveld, A.B. Metabolic response to the stress of critical illness. Br. J. Anaesth. 2014, 113, 945–954. [Google Scholar] [CrossRef]
- Lebelo, S.L.; Saunders, D.K.; Crawford, T.G. Observations on blood viscosity in striped bass, Morone saxatilis (Walbaum) associated with fish hatchery conditions. Trans. Kans. Acad. Sci. 2001, 104, 183–194. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Mohindra, V.; Singh, A. Physiological responses to acute experimental hypoxia in the air-breathing Indian catfish, Clarias batrachus (Linnaeus, 1758). J. Biosci. 2013, 38, 373–383. [Google Scholar] [CrossRef]
- Brahimi-Horn, M.C.; Pouysségur, J. Hif at a glance. J. Cell Sci. 2009, 122, 1055–1057. [Google Scholar] [CrossRef]
- Pelster, B.; Egg, M. Hypoxia-inducible transcription factors in fish: Expression, function and interconnection with the circadian clock. J. Exp. Biol. 2018, 211. [Google Scholar] [CrossRef]
- Brusselmans, K.; Bono, F.; Maxwell, P.; Dor, Y.; Dewerchin, M.; Collen, D.; Herbert, J.M.; Carmeliet, P. Hypoxia-inducible factor-2alpha (HIF-2alpha) is involved in the apoptotic response to hypoglycemia but not to hypoxia. J. Biol. Chem. 2001, 276, 39192–39196. [Google Scholar] [CrossRef]
- Rahman, M.S.; Thomas, P. Molecular cloning, characterization and expression of two hypoxia-inducible factor alpha subunits, hif-1α and hif-2α, in a hypoxia-tolerant marine teleost, atlantic croaker (Micropogonias undulatus). Gene 2007, 396, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Ohneda, K.; Nagano, M.; Miyoshi, C.; Kaneko, N.; Miwa, Y. Hypoxia-inducible transcription factor-2α in endothelial cells regulates tumor neovascularization through activation of ephrin a1. J. Biol. Chemis. 2008, 283, 18926. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Y.; Munday, J.S.; Perrott, M.; Wang, G.; Liu, X. Association of Age with the Expression of Hypoxia-Inducible Factors HIF-1α, HIF-2α, HIF-3α and VEGF in Lung and Heart of Tibetan Sheep. Animals 2019, 9, 673. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Peng, M.; Luo, Q.; Jiang, M.; Zhang, X.L.; Zong, X.C.; Meng, F.J.; Li, Y. Isolation and detection of transcript-derived fragments (tdfs) in nacl-stressed black locust (Robinia pseudoacacia L.) using cDNA-AFLP analysis. Acta Physiol. Plant. 2015, 37, 168. [Google Scholar] [CrossRef]
- Krishnan, J.; Ahuja, P.; Bodenmann, S.; Knapik, D.; Perriard, E.; Krek, W.; Perriard, J.C. Essential Role of Developmentally Activated Hypoxia-Inducible Factor 1? For Cardiac Morphogenesis and Function. Circ. Res. 2008, 103, 1139–1146. [Google Scholar] [CrossRef]
- Piltti, J.; Bygdell, J.; Qu, C.J.; Lammi, J.M. Effects of long-term hypoxia in human chondrosarcoma cells. J. Cell. Biochem. 2017, 119, 2320–2332. [Google Scholar] [CrossRef]
- Zhang, Q.L.; Cui, B.R.; Li, H.Y.; Li, P.; Hong, L.; Liu, L.P.; Ding, D.Z.; Xun, C. Mapk and pi3k pathways regulate hypoxia-induced atrial natriuretic peptide secretion by controlling hif-1 alpha expression in beating rabbit atria. Biochem. Biophys. Res. Commun. 2013, 438, 507–512. [Google Scholar] [CrossRef]
- Ostadal, B.; Kolar, F. Cardiac adaptation to chronic high-altitude hypoxia: Beneficial and adverse effects. Respir. Physiol. Neurobi. 2007, 158, 224–236. [Google Scholar] [CrossRef]
- Risbud, M.V.; Guttapalli, A.; Albert, T.J.; Shapiro, I.M. Hypoxia activates MAPK activity in rat nucleus pulposus cells: Regulation of integrin expression and cell survival. Spine 2005, 30, 2503–2509. [Google Scholar] [CrossRef]
- Nam, K.; Lee, K.W.; Chung, O.; Yim, H.S.; Cha, S.S.; Lee, S.W.; Jun, J.; Cho, Y.S.; Bhak, J.; Magalhaes, J.P.; et al. Analysis of the FGF gene family provides insights into aquatic adaptation in cetaceans. Sci. Rep. 2017, 7, 40233. [Google Scholar] [CrossRef]
- Deindl, E.; Kolar, F.; Neubauer, E.; Vogel, S.; Schaper, W.; Ostadal, B. Effect of intermittent high altitude hypoxia on gene expression in rat heart and lung. Physiol. Res. 2003, 52, 47–57. [Google Scholar]
- Webster, K.A.; Discher, D.J.; Bishopric, N.H. Induction and nuclear accumulation of fos and jun proto-oncogenes in hypoxic cardiac myocytes. J. Biol. Chem. 1993, 268, 16852–16858. [Google Scholar] [PubMed]
- Kim, M.Y.; Seo, E.J.; Lee, D.H.; Kim, E.J.; Kim, H.S.; Cho, H.Y.; Chung, E.Y.; Lee, S.H.; Baik, E.J.; Moon, C.H.; et al. Gadd45beta is a novel mediator of cardiomyocyte apoptosis induced by ischaemia/hypoxia. Cardiovasc. Res. 2010, 87, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Takekawa, M.; Saito, H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell 1998, 95, 521–530. [Google Scholar] [CrossRef]


| Category | Count |
|---|---|
| KOG classified sequences | 14,613 |
| Swiss-Prot | 13,208 |
| GO classified sequences | 13,996 |
| KEGG classified sequences | 12,071 |
| Nr annotations | 25,295 |
| All annotations | 25,892 |
| Regulation | Name of Pathway | Pathway ID | Genes |
|---|---|---|---|
| up | MAPK signaling pathway | ko04010 | Cacng8, Gadd45, cFos, Mknk2, Stmn, Ikbkb, Fgfr2, Mecom, Fgfr1a |
| Insulin signaling pathway | ko04910 | Cbl, Mknk2, Flot1, Ikbkb, G6pc | |
| Regulation of actin cytoskeleton | ko04810 | Tmsb11, F2, Myl9, Fgfr2, Fgfr1a | |
| Neuroactive ligand-receptor interaction | ko01230 | Adra2a, Htr1a, F2, Plg | |
| Purine metabolism | ko01200 | Pde6a, Pole4, Npr2, Pde7a | |
| down | Tight junction | ko04530 | Actn2, Myh7, Myh6 |
| Adrenergic signaling in cardiomyocytes | ko04261 | Tnnc1b, Pln, Rps6ka5, Myh7, Cacna2d3 | |
| Cardiac muscle contraction | ko04260 | Tnnc1b, Myh7, Cacna2d3 | |
| Other types of O-glycan biosynthesis | ko00514 | B3gat1, Mfng | |
| MAPK signaling pathway | ko04010 | Fgf3, Dusp10, Rps6ka5, Cacna2d3 |
| RNA-Seq | cDNA-AFLP | Regulation | Groups |
|---|---|---|---|
| Stbp2 | Stbp2 | up | HS vs. HT |
| Titin | Titin | down | HS vs. HT |
| Map3k15 | - | up | NC vs. HS |
| - | Mapk6 | up | HS vs. HT |
| Kcnh2 | - | down | HS vs. HT |
| - | Kcnh1 | down | HS vs. HT |
| Tnfrsf12 | - | up | HS vs. HT |
| - | Tnfrsf14 | up | HS vs. HT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Luo, W.; Yu, X.; Wang, J.; Feng, Y.; Tong, J. Cardiac Transcriptomics Reveals That MAPK Pathway Plays an Important Role in Hypoxia Tolerance in Bighead Carp (Hypophthalmichthys nobilis). Animals 2020, 10, 1483. https://doi.org/10.3390/ani10091483
Zhou Y, Luo W, Yu X, Wang J, Feng Y, Tong J. Cardiac Transcriptomics Reveals That MAPK Pathway Plays an Important Role in Hypoxia Tolerance in Bighead Carp (Hypophthalmichthys nobilis). Animals. 2020; 10(9):1483. https://doi.org/10.3390/ani10091483
Chicago/Turabian StyleZhou, Ying, Weiwei Luo, Xiaomu Yu, Junru Wang, Yizhao Feng, and Jingou Tong. 2020. "Cardiac Transcriptomics Reveals That MAPK Pathway Plays an Important Role in Hypoxia Tolerance in Bighead Carp (Hypophthalmichthys nobilis)" Animals 10, no. 9: 1483. https://doi.org/10.3390/ani10091483
APA StyleZhou, Y., Luo, W., Yu, X., Wang, J., Feng, Y., & Tong, J. (2020). Cardiac Transcriptomics Reveals That MAPK Pathway Plays an Important Role in Hypoxia Tolerance in Bighead Carp (Hypophthalmichthys nobilis). Animals, 10(9), 1483. https://doi.org/10.3390/ani10091483






