Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Exhibits High Predicted Binding Affinity to ACE2 from Lagomorphs (Rabbits and Pikas)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Analysis
2.2. Structure Simulation of the Complex SARS-CoV-2-RBD-ACE2
3. Results
3.1. Sequence Analysis
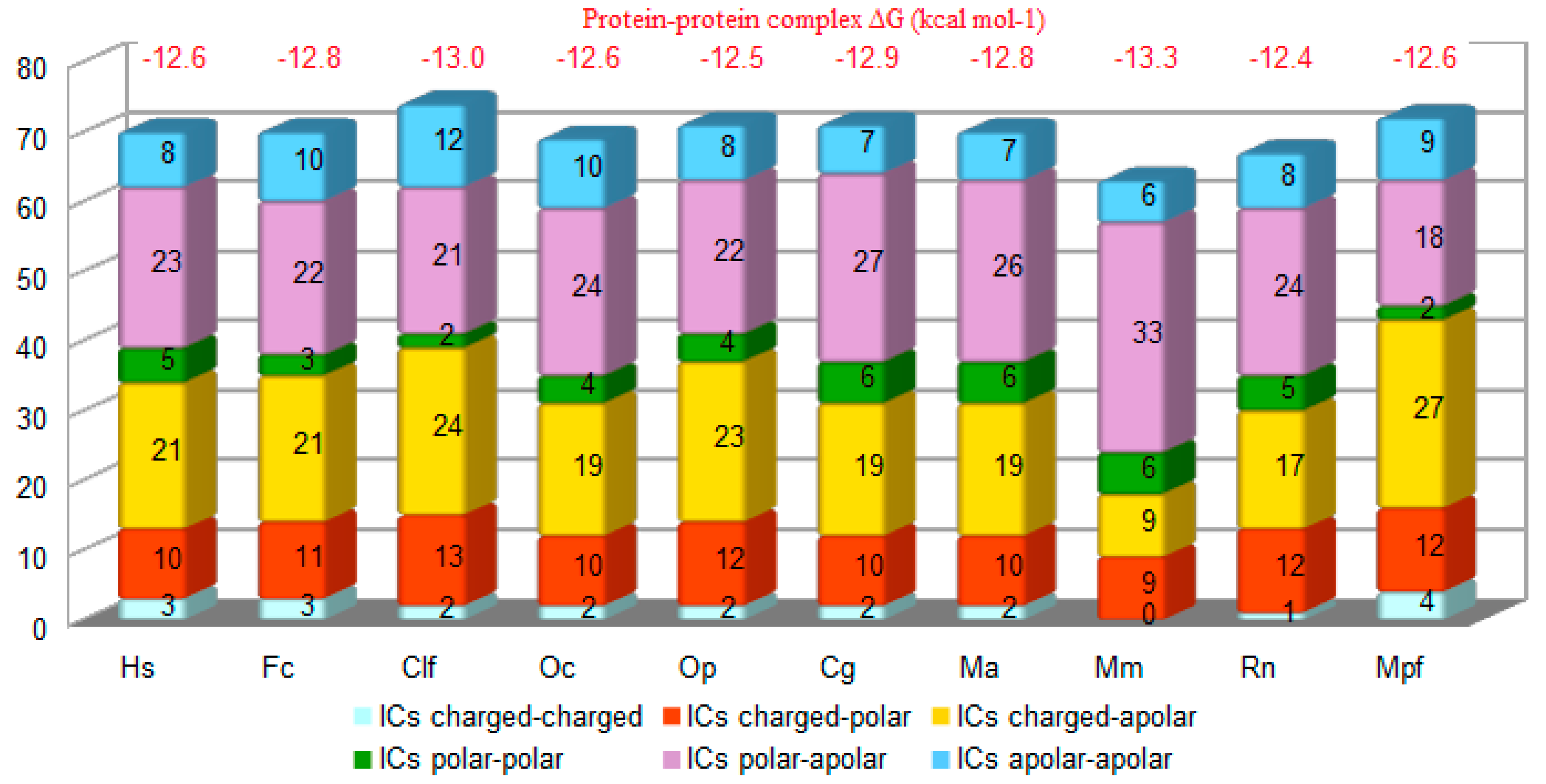
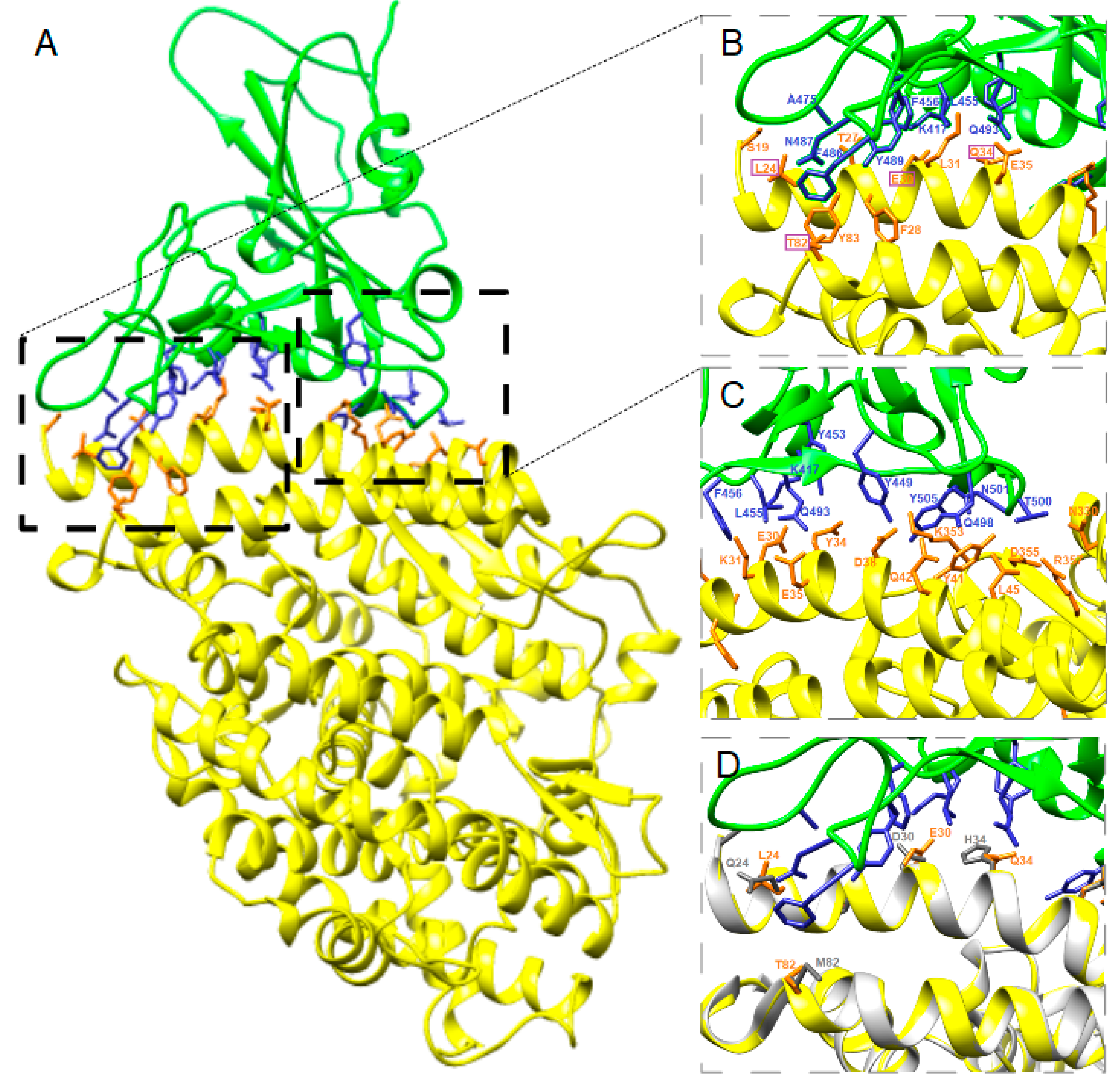
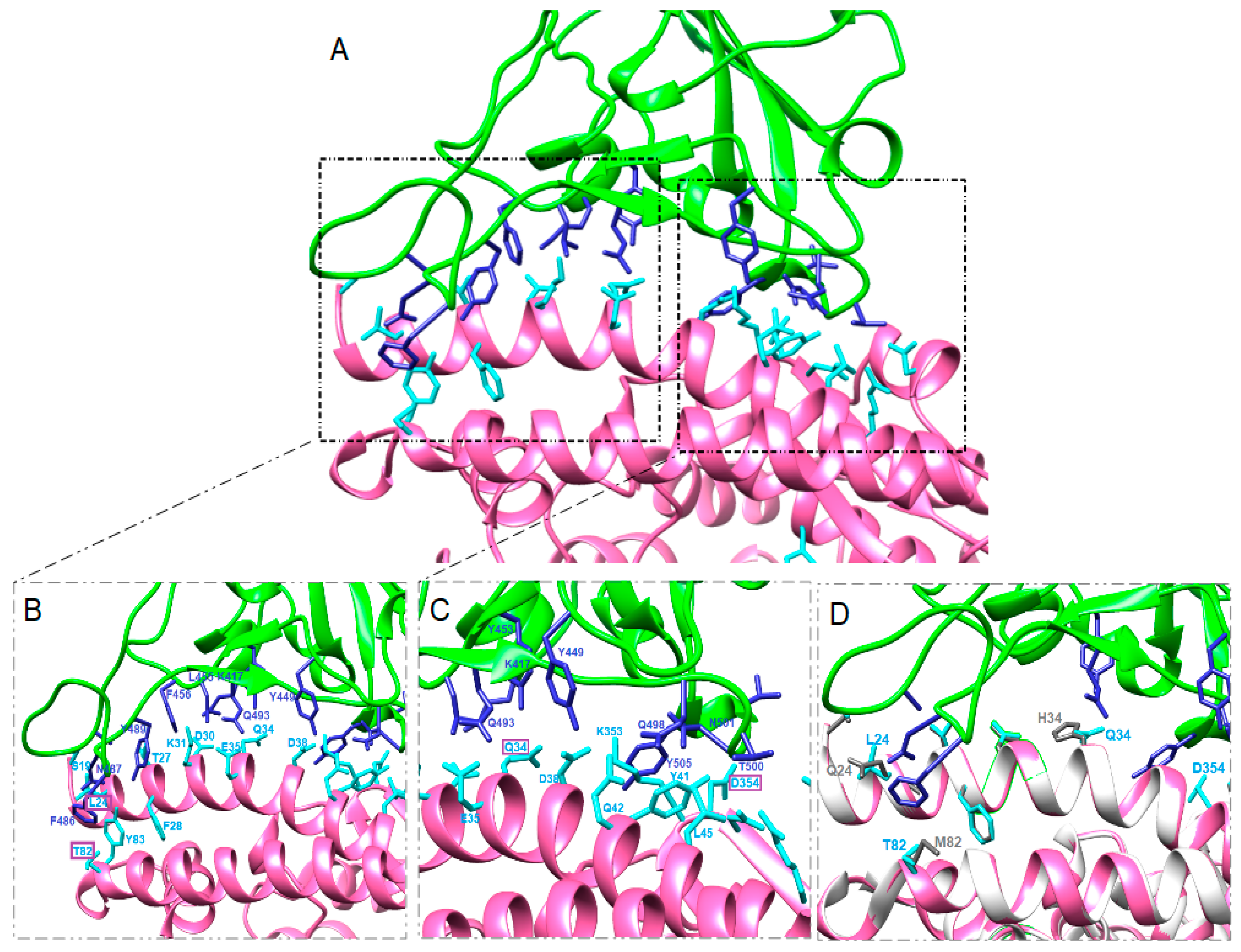
3.2. Structure Simulation of the Complex SARS-CoV-2-RBD-ACE2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yang, Y.; Ren, L. Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect. Genet. Evol. 2020, 82, 104285. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.T.Y.; Shum, M.H.H.; Zhu, H.C.; Tong, Y.G.; Ni, X.B.; Liao, Y.S.; Wei, W.; Cheung, W.Y.M.; Li, W.J.; Li, L.F.; et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Zhai, J.; Feng, Y.; Zhou, N.; Zhang, X.; Zou, J.J.; Li, N.; Guo, Y.; Li, X.; Shen, X.; et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 2020, 583, 286–289. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; Jason, S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Antón-Plágaro, C.; Kavanagh Williamson, M.; Shoemark, D.K.; Simón-Gracia, L.; Klein, K.; Bauer, M.; Hollandi, R.; Greber, U.F.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.D.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; Kallio, K.; Kaya, T.; Anastasina, M.; Smura, T.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and provides a possible pathway into the central nervous system. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A fifirst step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xiao, X.; Wei, X.; Li, J.; Yang, J.; Tan, H.; Zhu, J.; Zhang, Q.; Wu, J.; Liu, L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020, 92, 595–601. [Google Scholar] [CrossRef]
- Luan, J.; Lu, Y.; Jin, X.; Zhang, L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020, 526, 165–169. [Google Scholar] [CrossRef]
- Abdel-Moneim, A.S.; Abdelwhab, E.M. Evidence for SARS-CoV-2 Infection of Animal Hosts. Pathogens 2020, 9, 529. [Google Scholar] [CrossRef]
- Hossain, M.G.; Javed, A.; Akter, S.; Saha, S. SARS-CoV-2 host diversity: An update of natural infections and experimental evidence. Microbiol. Immunol. Infect. 2020. [Google Scholar] [CrossRef]
- Chan, J.F.; Zhang, A.J.; Yuan, S.; Poon, V.K.; Chan, C.C.; Lee, A.C.; Chan, W.M.; Fan, Z.; Tsoi, H.W.; Wen, L.; et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020, ciaa325. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of dogs with SARS-CoV-2. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Sailleau, C.; Dumarest, M.; Vanhomwegen, J.; Delaplace, M.; Caro, V.; Kwasiborski, A.; Hourdel, V.; Chevaillier, P.; Barbarino, A.; Comtet, L.; et al. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.A.P.M.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Vangone, A.; Bonvin, A.M. Contacts-based prediction of binding affinity in protein–protein complexes. eLife 2015, 4, e07454. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]
- Kastritis, P.L.; Moal, I.H.; Hwang, H.; Weng, Z.; Bates, P.A.; Bonvin, A.M.; Janin, J. A structure-based benchmark for protein-protein binding affinity. Protein. Sci. 2011, 20, 482–491. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020. [Google Scholar] [CrossRef]
- Luan, J.; Jin, X.; Lu, Y.; Zhang, L. SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J. Med. Virol. 2020, 92, 1649–1656. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Wood, J.; Subbarao, K.; Ferguson, M.; Wood, D.; Cherian, T. Animal models and antibody assays for evaluating candidate SARS vaccines: Summary of a technical meeting 25–26 August 2005, London, UK. Vaccine 2006, 24, 7056–7065. [Google Scholar] [CrossRef] [PubMed]
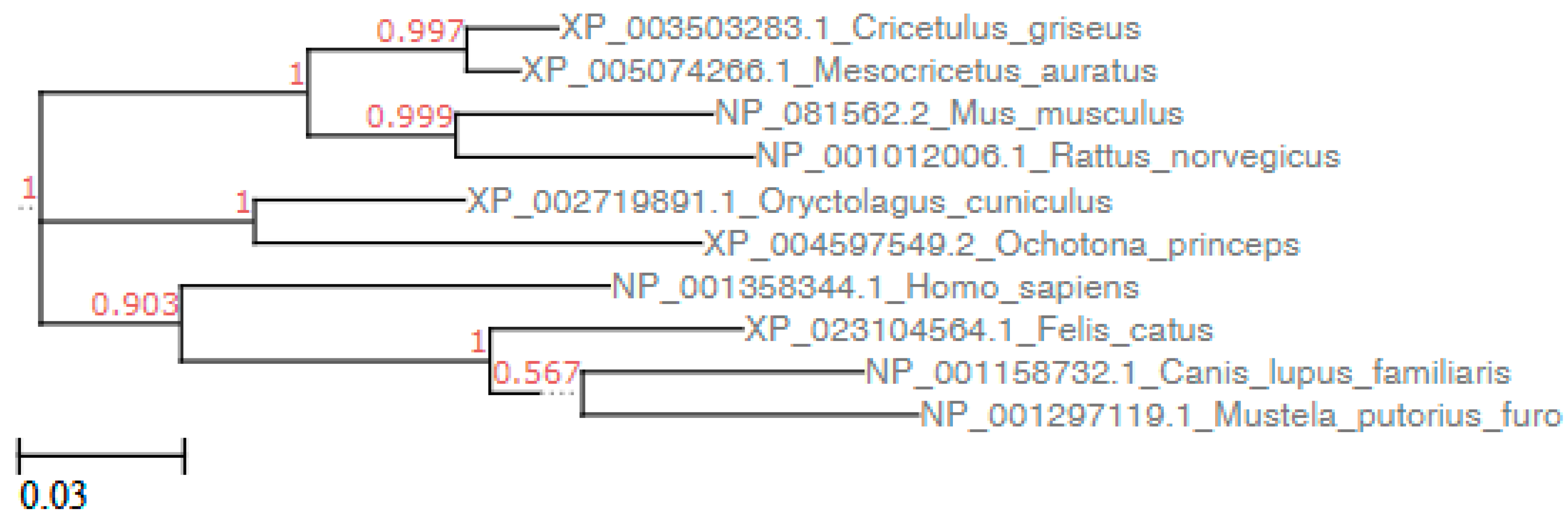
| ACE2 Sequence | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NP_001358344.1_Homo_sapiens | - | ||||||||
| 2 | XP_023104564.1_Felis_catus | 0.170 | - | |||||||
| 3 | NP_001158732.1_Canis_lupus_familiaris | 0.188 | 0.105 | - | ||||||
| 4 | XP_002719891.1_Oryctolagus_cuniculus | 0.164 | 0.179 | 0.187 | - | |||||
| 5 | XP_004597549.2_Ochotona_princeps | 0.219 | 0.224 | 0.220 | 0.126 | - | ||||
| 6 | XP_003503283.1_Cricetulus_griseus | 0.174 | 0.195 | 0.198 | 0.149 | 0.178 | - | |||
| 7 | XP_005074266.1_Mesocricetus_auratus | 0.172 | 0.200 | 0.204 | 0.147 | 0.175 | 0.029 | - | ||
| 8 | NP_081562.2_Mus_musculus | 0.204 | 0.213 | 0.219 | 0.183 | 0.205 | 0.114 | 0.109 | - | |
| 9 | NP_001012006.1_Rattus_norvegicus | 0.201 | 0.219 | 0.229 | 0.183 | 0.210 | 0.118 | 0.112 | 0.103 | - |
| 10 | NP_001297119.1_Mustela_putorius_furo | 0.199 | 0.115 | 0.112 | 0.195 | 0.232 | 0.201 | 0.206 | 0.216 | 0.235 |
| ACE2 | Amino Acid Position | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 19 | 24 | 27 | 28 | 30 | 31 | 34 | 35 | 37 | 38 | 41 | |
| Homo sapiens | S | Q | T | F | D | K | H | E | E | D | Y |
| Felis catus | . | L | . | . | E | . | . | . | . | E | . |
| Canis lupus familiaris | . | L | . | . | E | . | Y | . | . | E | . |
| Oryctolagus cuniculus | . | L | . | . | E | . | Q | . | . | . | . |
| Ochotona princeps | . | L | . | . | . | . | Q | . | . | . | . |
| Cricetulus griseus | . | . | . | . | . | . | Q | . | . | . | . |
| Mesocricetus auratus | . | . | . | . | . | . | Q | . | . | . | . |
| Mus musculus | . | N | . | . | N | N | Q | . | . | . | . |
| Rattus norvegicus | . | K | S | . | N | . | Q | . | . | . | . |
| Mustela putorius furio | . | L | . | E | . | Y | . | . | E | . | |
| ACE2 | Amino Acid Position | ||||||||||
| 42 | 45 | 79 | 82 | 83 | 330 | 353 | 354 | 355 | 357 | 393 | |
| Homo sapiens | Q | L | L | M | Y | N | K | G | D | R | R |
| Felis catus | . | . | . | T | . | . | . | . | . | . | . |
| Canis lupus familiaris | . | . | . | T | . | . | . | . | . | . | . |
| Oryctolagus cuniculus | . | . | . | T | . | . | . | . | . | . | . |
| Ochotona princeps | . | . | . | T | . | . | . | D | . | . | . |
| Cricetulus griseus | . | . | . | N | . | . | . | . | . | . | . |
| Mesocricetus auratus | . | . | . | N | . | . | . | . | . | . | . |
| Mus musculus | . | . | T | S | F | . | H | . | . | . | . |
| Rattus norvegicus | . | . | I | N | F | . | H | . | . | . | . |
| Mustela putorius furio | . | . | H | T | . | . | . | R | . | . | . |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Preziuso, S. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Exhibits High Predicted Binding Affinity to ACE2 from Lagomorphs (Rabbits and Pikas). Animals 2020, 10, 1460. https://doi.org/10.3390/ani10091460
Preziuso S. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Exhibits High Predicted Binding Affinity to ACE2 from Lagomorphs (Rabbits and Pikas). Animals. 2020; 10(9):1460. https://doi.org/10.3390/ani10091460
Chicago/Turabian StylePreziuso, Silvia. 2020. "Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Exhibits High Predicted Binding Affinity to ACE2 from Lagomorphs (Rabbits and Pikas)" Animals 10, no. 9: 1460. https://doi.org/10.3390/ani10091460
APA StylePreziuso, S. (2020). Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Exhibits High Predicted Binding Affinity to ACE2 from Lagomorphs (Rabbits and Pikas). Animals, 10(9), 1460. https://doi.org/10.3390/ani10091460





