Genome-Wide SNP Analysis Reveals the Population Structure and the Conservation Status of 23 Italian Chicken Breeds
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Genotyping
2.2. Admixture and Genetic Relationship
2.3. Runs of Homozygosity
3. Results
3.1. Analysis of Whole-Genome Diversity
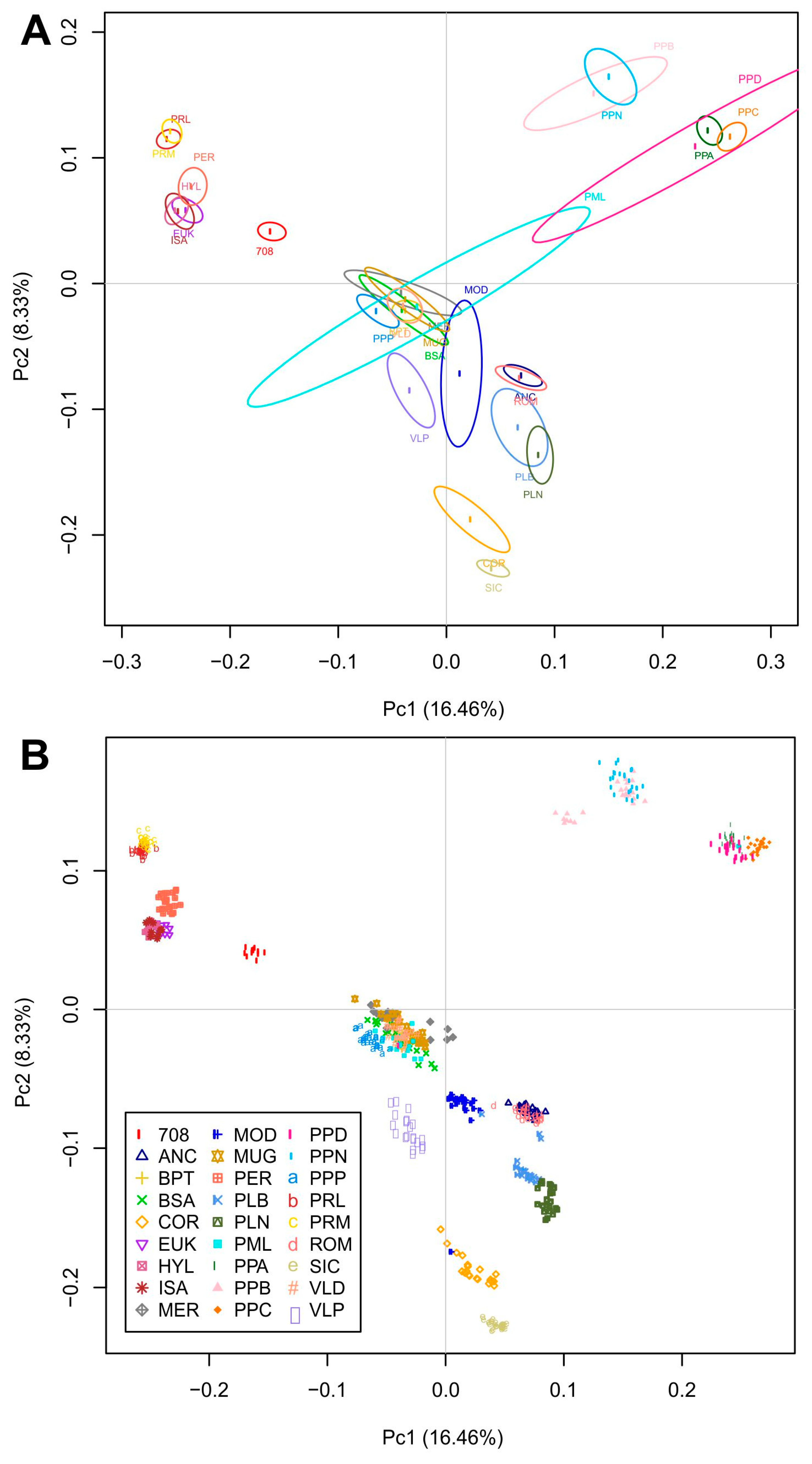
3.2. Analysis of Genetic Distance and Population Structure
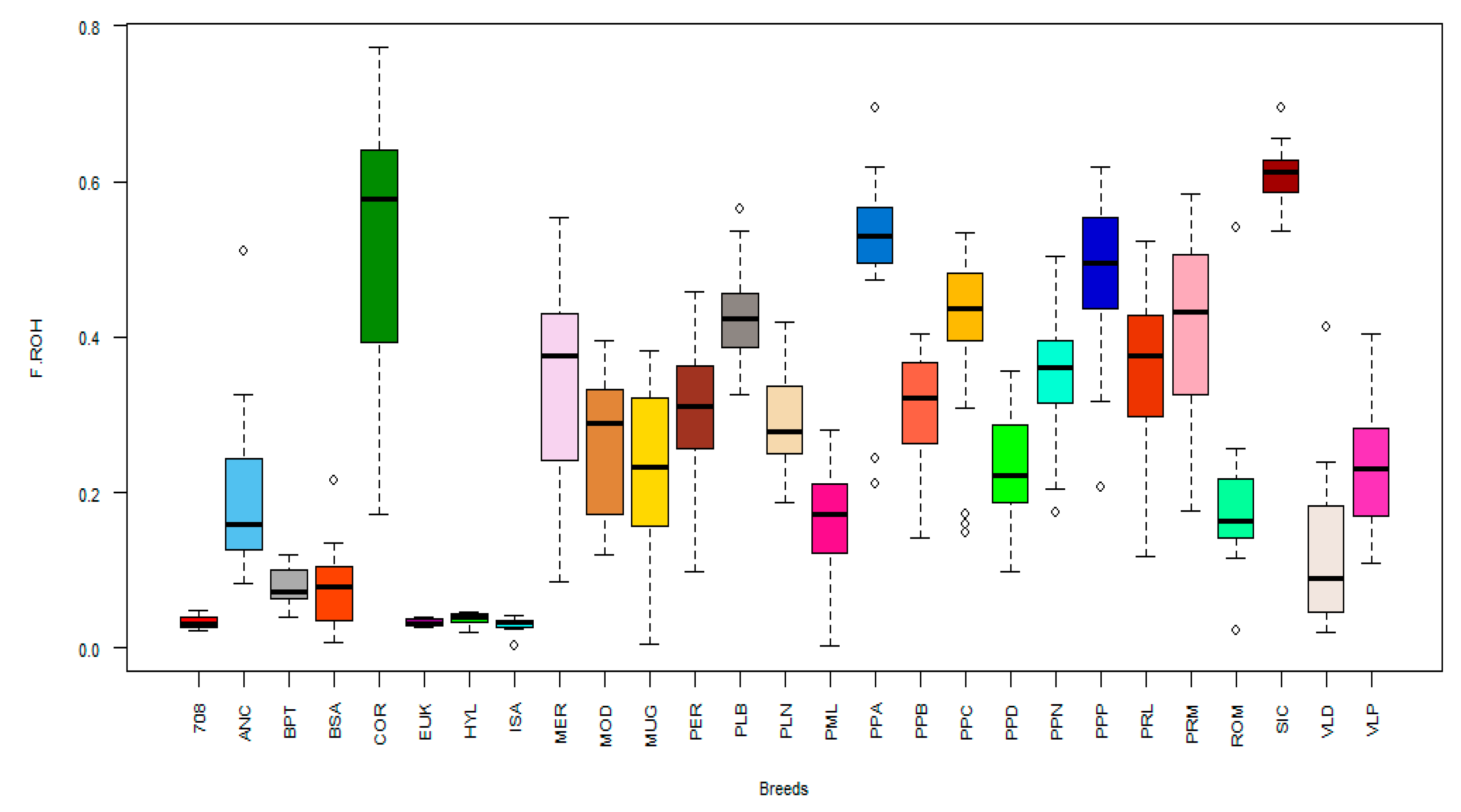
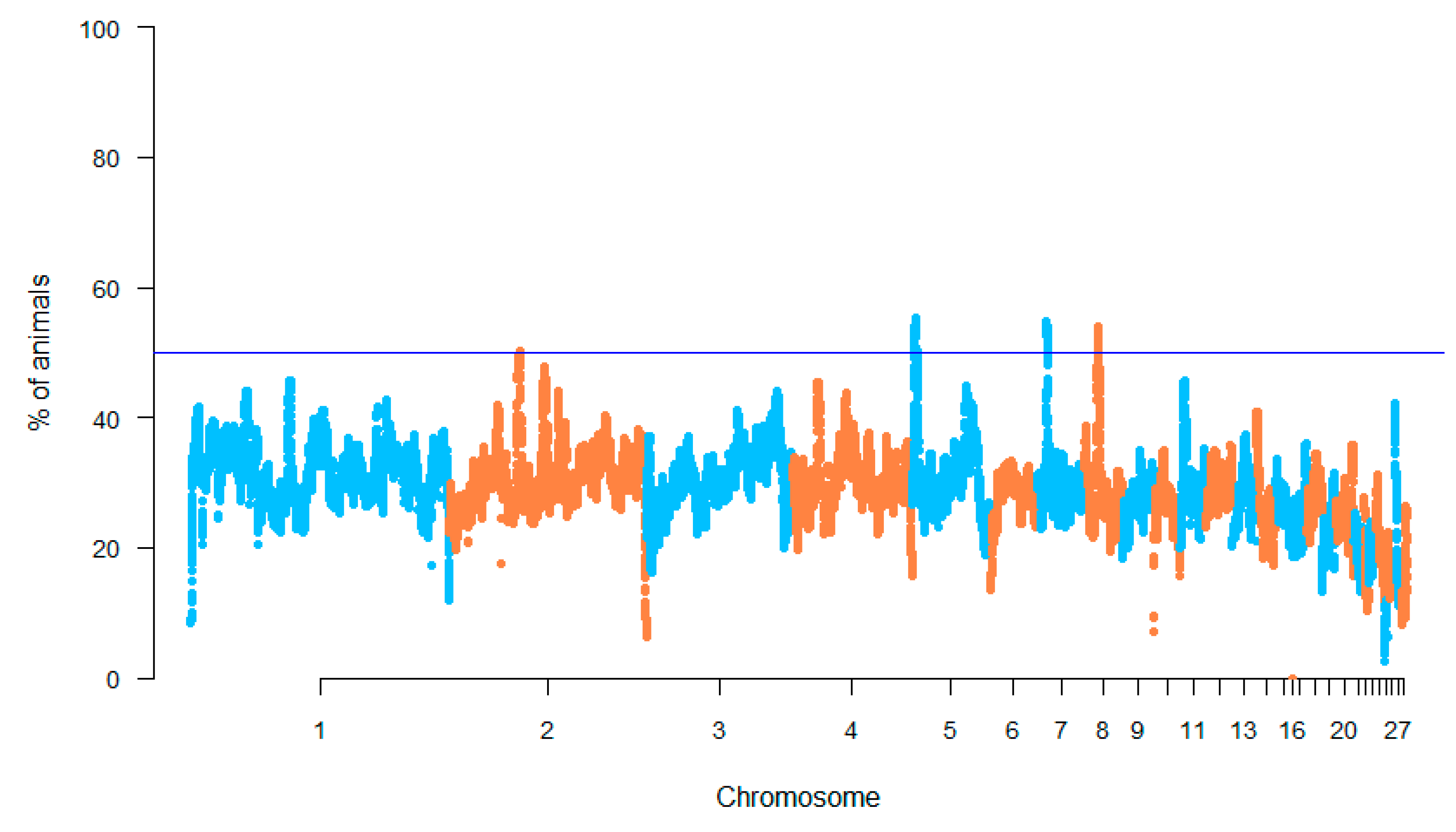
3.3. Run of Homozygosity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Delany, M.E. Genetic variants for chick biology research: From breeds to mutants. Mech. Dev. 2004, 121, 1169–1177. [Google Scholar] [CrossRef]
- Rischkowsky, B.; Pilling, D. The State of the World’s Animal Genetic Resources for Food and Agriculture. In Food & Agriculture Org.; FAO Document: Rome, Italy, 2007. [Google Scholar]
- Zanetti, E.; de Marchi, M.; Dalvit, C.; Cassandro, M. Genetic characterization of local Italian breeds of chickens undergoing in situ conservation. Poult. Sci. 2010, 89, 420–427. [Google Scholar] [CrossRef]
- Hoffmann, I. The global plan of action for animal genetic resources and the conservation of poultry genetic resources. Worlds. Poult. Sci. J. 2009, 65, 286–297. [Google Scholar] [CrossRef]
- Strillacci, M.G.; Marelli, S.P.; Cozzi, M.C.; Colombo, E.; Polli, M.; Gualtieri, M.; Cristalli, A.; Pignattelli, P.; Longeri, M.; Cavalchini, L.G. Italian autochthonous chicken breeds conservation: Evaluation of biodiversity in Valdarnese Bianca breed (Gallus gallus domesticus). Avian Biol. Res. 2009, 2, 229–233. [Google Scholar] [CrossRef]
- De Marchi, M.; Dalvit, C.; Targhetta, C.; Cassandro, M. Assessing genetic diversity in indigenous Veneto chicken breeds using AFLP markers. Anim. Genet. 2006, 37, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Malomane, D.K.; Simianer, H.; Weigend, A.; Reimer, C.; Schmitt, A.O.; Weigend, S. The SYNBREED chicken diversity panel: A global resource to assess chicken diversity at high genomic resolution. BMC Genom. 2019, 20, 345. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, S.; Cendron, F.; Sottile, G.; Niero, G.; Portolano, B.; Biscarini, F.; Cassandro, M. Genome-Wide Analyses Identifies Known and New Markers Responsible of Chicken Plumage Color. Animals 2020, 10, 493. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; De Marchi, M.; Barcaccia, G.; Zanon, A.; Sabbioni, A. Genetic similarities among Modenese, Romagnola and three Veneto chicken breeds. Ital. J. Anim. Sci. 2005, 4 (Suppl. S2), 106–108. [Google Scholar] [CrossRef]
- Warren, W.C.; Hillier, L.D.W.; Tomlinson, C.; Minx, P.; Kremitzki, M.; Graves, T.; Markovic, C.; Bouk, N.; Pruitt, K.D.; Thibaud-Nissen, F.; et al. A new chicken genome assembly provides insight into avian genome structure. G3 Genes Genomes Genet. 2017, 7, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, s13742-015-0047-8. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinforma. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Milanesi, M.; Capomaccio, S.; Vajana, E.; Bomba, L.; Garcia, J.F.; Ajmone-Marsan, P.; Colli, L. BITE: An R package for biodiversity analyses. BioRxiv 2017, 181610. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 1 July 2020).
- Mastrangelo, S.; Ciani, E.; Sardina, M.T.; Sottile, G.; Pilla, F.; Portolano, B. Runs of homozygosity reveal genome-wide autozygosity in Italian sheep breeds. Anim. Genet. 2018, 49, 71–81. [Google Scholar] [CrossRef]
- Sartore, S.; Sacchi, P.; Soglia, D.; Maione, S.; Schiavone, A.; De Marco, M.; Ceccobelli, S.; Lasagna, E.; Rasero, R. Genetic variability of two Italian indigenous chicken breeds inferred from microsatellite marker analysis. Br. Poult. Sci. 2016, 57, 435–443. [Google Scholar] [CrossRef]
- Manunza, A.; Noce, A.; Serradilla, J.M.; Goyache, F.; Martínez, A.; Capote, J.; Delgado, J.V.; Jordana, J.; Muñoz, E.; Molina, A.; et al. A genome-wide perspective about the diversity and demographic history of seven Spanish goat breeds. Genet. Sel. Evol. 2016, 48, 1–9. [Google Scholar] [CrossRef]
- Chagunda, M.G.; Mujibi, F.D.; Dusingizimana, T.; Kamana, O.; Cheruiyot, E.; Mwai, O.A. Use of high-density single nucleotide polymorphism (SNP) arrays to assess genetic diversity and population structure of dairy cattle in smallholder dairy systems: The case of Girinka Programme in Rwanda. Frontiers 2018, 9, 438. [Google Scholar] [CrossRef]
- Strillacci, M.G.; Cozzi, M.C.; Gorla, E.; Mosca, F.; Schiavini, F.; Román-Ponce, S.I.; Ruiz López, F.J.; Schiavone, A.; Marzoni, M.; Cerolini, S.; et al. Genomic and genetic variability of six chicken populations using single nucleotide polymorphism and copy number variants as markers. Animal 2017, 11, 737–745. [Google Scholar] [CrossRef]
- Bortoluzzi, C.; Crooijmans, R.P.M.A.; Bosse, M.; Hiemstra, S.J.; Groenen, M.A.M.; Megens, H.J. The effects of recent changes in breeding preferences on maintaining traditional Dutch chicken genomic diversity. Heredity 2018, 121, 564–578. [Google Scholar] [CrossRef]
- Ceccobelli, S.; Di Lorenzo, P.; Lancioni, H.; Castellini, C.; Ibáñez, L.V.M.; Sabbioni, A.; Sarti, F.M.; Weigend, S.; Lasagna, E. Phylogeny, genetic relationships and population structure of five Italian local chicken breeds. Ital. J. Anim. Sci. 2013, 12, e66. [Google Scholar] [CrossRef]
- Muir, W.M.; Wong, G.K.S.; Zhang, Y.; Wang, J.; Groenen, M.A.M.; Crooijmans, R.P.M.A.; Megens, H.J.; Zhang, H.; Okimoto, R.; Vereijken, A. Genome-wide assessment of worldwide chicken SNP genetic diversity indicates significant absence of rare alleles in commercial breeds. Proc. Natl. Acad. Sci. USA 2008, 105, 17312–17317. [Google Scholar] [CrossRef] [PubMed]
- FAO DAD-IS. Available online: http://www.fao.org/dad-is/browse-by-country-and-species/en/ (accessed on 1 July 2020).
- Mastrangelo, S.; Sardina, M.T.; Tolone, M.; Di Gerlando, R.; Sutera, A.M.; Fontanesi, L.; Portolano, B. Genome-wide identification of runs of homozygosity islands and associated genes in local dairy cattle breeds. Animal 2018, 12, 2480–2488. [Google Scholar] [CrossRef]
- Nicoloso, L.; Bomba, L.; Colli, L.; Negrini, R.; Milanesi, M.; Mazza, R.; Chessa, S. Genetic diversity of Italian goat breeds assessed with a medium-density SNP chip. Genet. Sel. Evol. 2015, 47, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ciani, E.; Crepaldi, P.; Nicoloso, L.; Lasagna, E.; Sarti, F.M.; Moioli, B.; Marletta, D. Genome-wide analysis of Italian sheep diversity reveals a strong geographic pattern and cryptic relationships between breeds. Anim. Genet. 2014, 45, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Cepollina, S. Standard Italiano Delle Razze Avicole; FIAV Publisher: Sacile (PN), Italy, 2015. [Google Scholar]
- Cerolini, S.; Madeddu, M.; Zaniboni, L.; Cassinelli, C.; Mangiagalli, M.G.; Marelli, S.P. Breeding performance in the Italian chicken breed Mericanel della Brianza. Ital. J. Anim. Sci. 2009, 9, e72. [Google Scholar] [CrossRef]
- Baruchello, M.; Cassandro, M. Avicoli Veneti CO.VA Project. Available online: https://www.venetoagricoltura.org/2007/01/editoria/avicoli-veneti-progetto-co-va-2012-ristampa-aggiornataschedacod-e476/ (accessed on 1 July 2020).
- Bosse, M.; Megens, H.J.; Madsen, O.; Paudel, Y.; Frantz, L.A.F.; Schook, L.B.; Crooijmans, R.P.M.A.; Groenen, M.A.M. Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape. PLoS Genet. 2012, 8, e1003100. [Google Scholar] [CrossRef] [PubMed]
- Howrigan, D.P.; Simonson, M.A.; Keller, M.C. Detecting autozygosity through runs of homozygosity: A comparison of three autozygosity detection algorithms. BMC Genom. 2011, 12, 460. [Google Scholar] [CrossRef]
- Qanbari, S.; Gianola, D.; Hayes, B.; Schenkel, F.; Miller, S.; Moore, S.; Thaller, G.; Simianer, H. Application of site and haplotype-frequency based approaches for detecting selection signatures in cattle. BMC Genom. 2011, 12, 318. [Google Scholar] [CrossRef]
- Kehua, C.G.L.F.W.; Kuanwei, W.X.C. Comparison on contents of muscle thiamine in chinese native chickens. J. Jiangsu Agric. Coll. 1998, 2. [Google Scholar]
- Sun, M.; Sui, Y.; Li, L.; Su, W.; Hao, F.; Zhu, Q.; Di, W.; Gao, H.; Ma, T. Anoctamin 1 calcium-activated chloride channel downregulates estrogen production in mouse ovarian granulosa cells. Endocrinology 2014, 155, 2787–2796. [Google Scholar] [CrossRef] [PubMed]
- Elferink, M.G.; Megens, H.J.; Vereijken, A.; Hu, X.; Crooijmans, R.P.M.A.; Groenen, M.A.M. Signatures of selection in the genomes of commercial and non-commercial chicken breeds. PLoS ONE 2012, 7, e32720. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gilbert, E.R.; Zhang, Y.; Crasta, O.; Emmerson, D.; Webb, K.E.; Wong, E.A. Expression profiling of the solute carrier gene family in chicken intestine from the late embryonic to early post-hatch stages. Anim. Genet. 2008, 39, 407–424. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Karnuah, A.B.; Rekaya, R.; Anthony, N.B.; Aggrey, S.E. Transcriptomic analysis to elucidate the molecular mechanisms that underlie feed efficiency in meat-type chickens. Mol. Genet. Genom. 2015, 290, 1673–1682. [Google Scholar] [CrossRef]
- Izadnia, H.R.; Tahmoorespur, M.; Bakhtiarizadeh, M.R.; Nassiri, M.; Esmaeilkhanien, S. Gene expression profile analysis of residual feed intake for Isfahan native chickens using RNA-SEQ data. Ital. J. Anim. Sci. 2019, 18, 246–260. [Google Scholar] [CrossRef]
- Walugembe, M.; Bertolini, F.; Dematawewa, C.M.B.; Reis, M.P.; Elbeltagy, A.R.; Schmidt, C.J.; Lamont, S.J.; Rothschild, M.F. Detection of selection signatures among Brazilian, Sri Lankan, and Egyptian chicken populations under different environmental conditions. Front. Genet. 2019, 9, 737. [Google Scholar] [CrossRef]


| Breed | Acronym | N | MAF | Ho | He | FHOM | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |||
| Ancona | ANC | 24 | 0.267 | 0.242 | 0.263 | 0.181 | 0.274 | 0.187 | 0.284 | 0.100 |
| Bianca di Saluzzo | BSA | 24 | 0.286 | 0.190 | 0.339 | 0.172 | 0.336 | 0.151 | 0.076 | 0.059 |
| Bionda Piemontese | BPT | 22 | 0.283 | 0.210 | 0.325 | 0.186 | 0.317 | 0.164 | 0.116 | 0.025 |
| Cornuta Caltanissetta | COR | 22 | 0.267 | 0.301 | 0.167 | 0.162 | 0.210 | 0.178 | 0.545 | 0.180 |
| Ermellinata di Rovigo | PER | 23 | 0.309 | 0.321 | 0.199 | 0.192 | 0.220 | 0.198 | 0.459 | 0.044 |
| Livorno Bianca | PLB | 24 | 0.269 | 0.295 | 0.205 | 0.196 | 0.218 | 0.186 | 0.465 | 0.061 |
| Livorno Nera | PLN | 24 | 0.263 | 0.279 | 0.233 | 0.211 | 0.231 | 0.195 | 0.365 | 0.062 |
| Mericanel della Brianza | MER | 24 | 0.282 | 0.268 | 0.232 | 0.180 | 0.261 | 0.186 | 0.368 | 0.127 |
| Millefiori di Lonigo | PML | 23 | 0.281 | 0.238 | 0.293 | 0.199 | 0.291 | 0.178 | 0.202 | 0.080 |
| Modenese | MOD | 24 | 0.273 | 0.252 | 0.260 | 0.197 | 0.27 | 0.181 | 0.296 | 0.083 |
| Mugellese | MUG | 24 | 0.284 | 0.231 | 0.281 | 0.182 | 0.300 | 0.175 | 0.236 | 0.115 |
| Padovana Argenta | PPA | 24 | 0.241 | 0.331 | 0.151 | 0.198 | 0.146 | 0.185 | 0.588 | 0.098 |
| Padovana Camosciata | PPC | 24 | 0.238 | 0.303 | 0.169 | 0.191 | 0.179 | 0.193 | 0.538 | 0.095 |
| Padovana Dorata | PPD | 24 | 0.247 | 0.264 | 0.219 | 0.194 | 0.232 | 0.187 | 0.404 | 0.081 |
| Pepoi | PPP | 24 | 0.277 | 0.341 | 0.154 | 0.191 | 0.168 | 0.196 | 0.579 | 0.039 |
| Polverara Bianca | PPB | 24 | 0.260 | 0.261 | 0.216 | 0.179 | 0.248 | 0.187 | 0.411 | 0.052 |
| Polverara Nera | PPN | 24 | 0.257 | 0.290 | 0.201 | 0.193 | 0.213 | 0.194 | 0.454 | 0.062 |
| Robusta Lionata | PRL | 23 | 0.305 | 0.345 | 0.181 | 0.199 | 0.185 | 0.195 | 0.508 | 0.039 |
| Robusta Maculata | PRM | 24 | 0.304 | 0.358 | 0.157 | 0.190 | 0.166 | 0.193 | 0.572 | 0.032 |
| Romagnola | ROM | 24 | 0.271 | 0.241 | 0.281 | 0.197 | 0.278 | 0.182 | 0.235 | 0.091 |
| Siciliana | SIC | 24 | 0.259 | 0.361 | 0.129 | 0.205 | 0.123 | 0.189 | 0.648 | 0.034 |
| Valdarnese | VLD | 24 | 0.283 | 0.204 | 0.321 | 0.181 | 0.322 | 0.160 | 0.127 | 0.098 |
| Valplatani | VLP | 20 | 0.281 | 0.268 | 0.280 | 0.224 | 0.261 | 0.184 | 0.239 | 0.086 |
| 708 Broiler Ross | 708 | 13 | 0.317 | 0.234 | 0.369 | 0.219 | 0.324 | 0.162 | −0.005 | 0.009 |
| Eureka | EUK | 9 | 0.329 | 0.261 | 0.374 | 0.260 | 0.305 | 0.177 | −0.018 | 0.013 |
| Hy-lyne white eggs | HYL | 10 | 0.333 | 0.278 | 0.375 | 0.286 | 0.289 | 0.285 | −0.020 | 0.008 |
| Isa Brown | ISA | 9 | 0.332 | 0.261 | 0.378 | 0.276 | 0.298 | 0.182 | −0.028 | 0.017 |
| Breed | FROH | SD | Mean ROH | SD | Total Number ROH |
|---|---|---|---|---|---|
| Ancona (ANC) | 0.201 | 0.099 | 56.21 | 14.01 | 1351 |
| Bianca di Saluzzo (BSA) | 0.081 | 0.057 | 20.53 | 9.22 | 492 |
| Bionda Piemontese (BPT) | 0.081 | 0.024 | 31.52 | 6.82 | 694 |
| Cornuta di Caltanissetta (COR) | 0.507 | 0.184 | 80.01 | 29.72 | 1761 |
| Ermellinata di Rovigo (PER) | 0.305 | 0.082 | 133.71 | 24.83 | 3077 |
| Livorno Bianca (PLB) | 0.427 | 0.059 | 77.72 | 6.15 | 1865 |
| Livorno Nera (PLN) | 0.296 | 0.063 | 68.18 | 7.94 | 1636 |
| Mericanel della Brianza (MER) | 0.326 | 0.135 | 65.15 | 15.97 | 1563 |
| Millefiori di Lonigo (PML) | 0.166 | 0.073 | 56.01 | 20.79 | 1289 |
| Modenese (MOD) | 0.264 | 0.086 | 54.14 | 9.42 | 1299 |
| Mugellese (MUG) | 0.225 | 0.112 | 39.64 | 16.29 | 951 |
| Padovana Argentata (PPA) | 0.509 | 0.118 | 96.76 | 12.71 | 2323 |
| Padovana Camosciata (PPC) | 0.410 | 0.109 | 103.52 | 17.74 | 2485 |
| Padovana Dorata (PPD) | 0.230 | 0.070 | 100.42 | 20.66 | 2410 |
| Pepoi (PPP) | 0.482 | 0.096 | 151.81 | 30.76 | 3645 |
| Polverara Bianca (PPB) | 0.310 | 0.068 | 113.85 | 22.07 | 2732 |
| Polverara Nera (PPN) | 0.353 | 0.087 | 127.80 | 21.91 | 3069 |
| Robusta Lionata (PRL) | 0.353 | 0.109 | 135.11 | 26.44 | 3109 |
| Robusta Maculata (PRM) | 0.410 | 0.113 | 157.58 | 22.06 | 3782 |
| Romagnola (ROM) | 0.187 | 0.091 | 43.17 | 10.04 | 1054 |
| Siciliana (SIC) | 0.607 | 0.037 | 96.09 | 6.90 | 2305 |
| Valdarnese (VLD) | 0.121 | 0.095 | 30.76 | 15.60 | 737 |
| Valplatani (VLP) | 0.236 | 0.087 | 41.55 | 5.91 | 830 |
| 708 Broiler ROSS (708) | 0.034 | 0.009 | 17.24 | 4.31 | 224 |
| Eureka (EUK) | 0.033 | 0.005 | 17.74 | 2.74 | 160 |
| Hy-lyne white eggs (HYL) | 0.038 | 0.008 | 19.23 | 3.59 | 192 |
| IsaBrown (ISA) | 0.030 | 0.011 | 16.22 | 5.95 | 146 |
| GGA | No. of SNPs | Start | End | Length (bp) | Genes | QTL |
|---|---|---|---|---|---|---|
| 2 | 18 | 53,138,767 | 53,202,574 | 63,807 | TPK1, LOC107051643 | - |
| 5 | 315 | 2,124,338 | 3,730,724 | 1,606,386 | NELL1, SLC6A5, LOC107053351, LOC107053349, LOC107053350, LOC107053348, ANO5, SLC17A6, FANCF, GAS2, SVIP, ANO3, SLC5A12, BBOX1, SLC5A12, FIBIN, CCDC34, LGR4, LIN7B | Body weight (28 days) QTL (95,416) Body weight (28 days) QTL (95,415) |
| 7 | 273 | 6,771,434 | 7,892,629 | 1,121,195 | COL6A2, LOC107053768, LOC107053769, LOC107053763, FTCD, MCM3AP, YBEY, LOC107053762, MCM3AP, YBEY, POFUT2, LOC107053766, CD163L1, LSS, S100B, DIP2A, PCNT, KMO, FAM207a, ITGB3, ADARB1 | Feed conversion ratio QTL (139,597) Feed conversion ratio QTL (139,472) Feed conversion ratio QTL (139,435) Feed conversion ratio QTL (139,598) |
| 8 | 371 | 9,506,680 | 10,604,288 | 1,097,608 | LOC101751732, PLA2G4A, PTGS2, PDC, C8H10RF27, TPR, LOC100859371, HMCN1, LOC107053953, LOC101750397, LOC107053952, INVS1ABP, SWT1, TRMT1L, LOC107053951 | Feed conversion ratio QTL (139,596) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cendron, F.; Perini, F.; Mastrangelo, S.; Tolone, M.; Criscione, A.; Bordonaro, S.; Iaffaldano, N.; Castellini, C.; Marzoni, M.; Buccioni, A.; et al. Genome-Wide SNP Analysis Reveals the Population Structure and the Conservation Status of 23 Italian Chicken Breeds. Animals 2020, 10, 1441. https://doi.org/10.3390/ani10081441
Cendron F, Perini F, Mastrangelo S, Tolone M, Criscione A, Bordonaro S, Iaffaldano N, Castellini C, Marzoni M, Buccioni A, et al. Genome-Wide SNP Analysis Reveals the Population Structure and the Conservation Status of 23 Italian Chicken Breeds. Animals. 2020; 10(8):1441. https://doi.org/10.3390/ani10081441
Chicago/Turabian StyleCendron, Filippo, Francesco Perini, Salvatore Mastrangelo, Marco Tolone, Andrea Criscione, Salvatore Bordonaro, Nicolaia Iaffaldano, Cesare Castellini, Margherita Marzoni, Arianna Buccioni, and et al. 2020. "Genome-Wide SNP Analysis Reveals the Population Structure and the Conservation Status of 23 Italian Chicken Breeds" Animals 10, no. 8: 1441. https://doi.org/10.3390/ani10081441
APA StyleCendron, F., Perini, F., Mastrangelo, S., Tolone, M., Criscione, A., Bordonaro, S., Iaffaldano, N., Castellini, C., Marzoni, M., Buccioni, A., Soglia, D., Schiavone, A., Cerolini, S., Lasagna, E., & Cassandro, M. (2020). Genome-Wide SNP Analysis Reveals the Population Structure and the Conservation Status of 23 Italian Chicken Breeds. Animals, 10(8), 1441. https://doi.org/10.3390/ani10081441










