Faecal Microbiota Analysis of Piglets During Lactation
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
- -
- Injection of ceftiofur (3 mg/kg intramuscular injection (IM)) at 7 d and fresh FMT at 13 d (n = 4 litters, n = 6 piglets; cffresh)
- -
- Ceftiofur (3 mg/kg IM) at 7 d and frozen FMT at 13 d (n = 4 litters, n = 7 piglets; cffrozen)
- -
- Ceftiofur (3 mg/kg IM) at 7 d and no FMT (n = 4 litters, n = 8 piglets; CF)
- -
- No ceftiofur and no FMT (n = 3 litters, n = 5 piglets; no CF).
2.2. DNA Extraction and 16S rRNA Amplicon Sequencing
2.3. Statistical Analysis
3. Results
3.1. Body Weight
3.2. Treatment Effects on Diversity Metrics
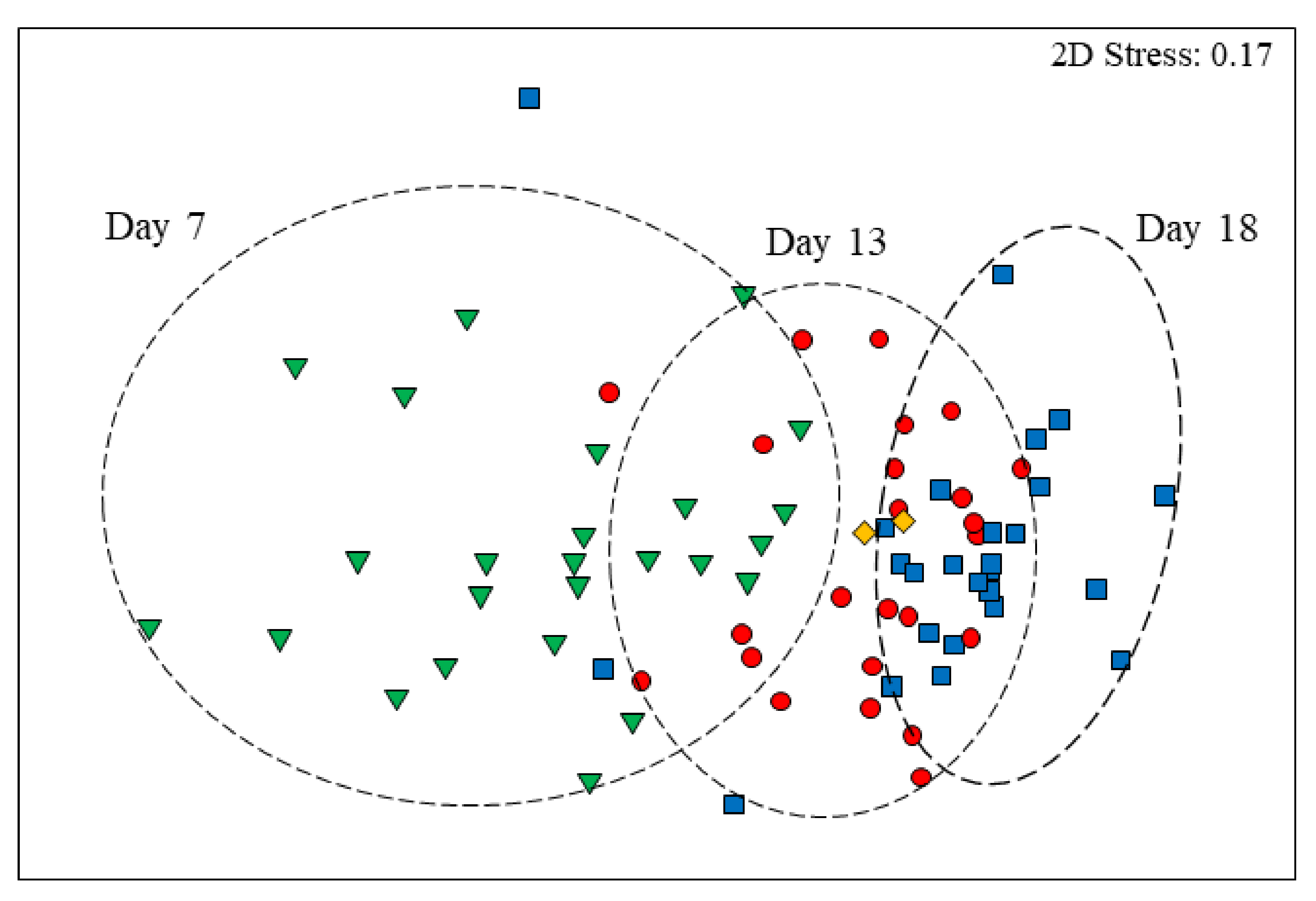
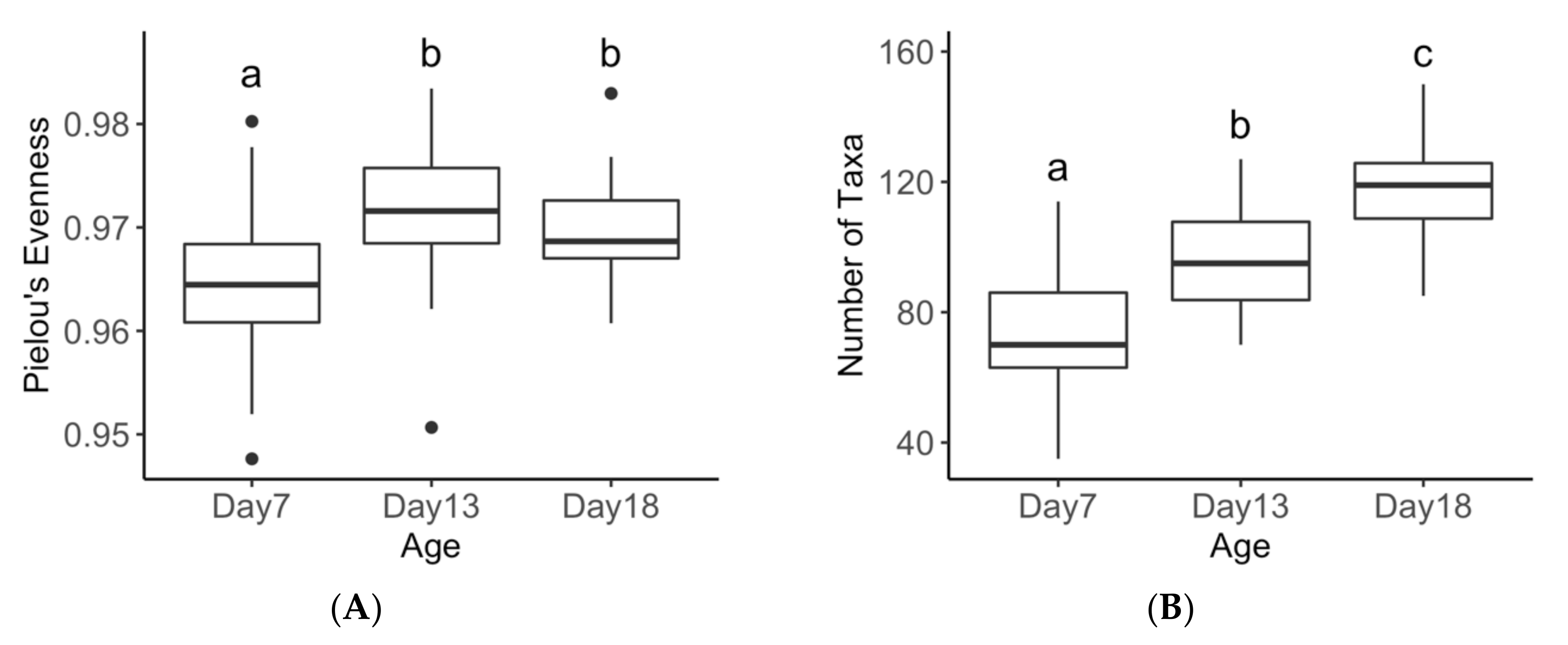
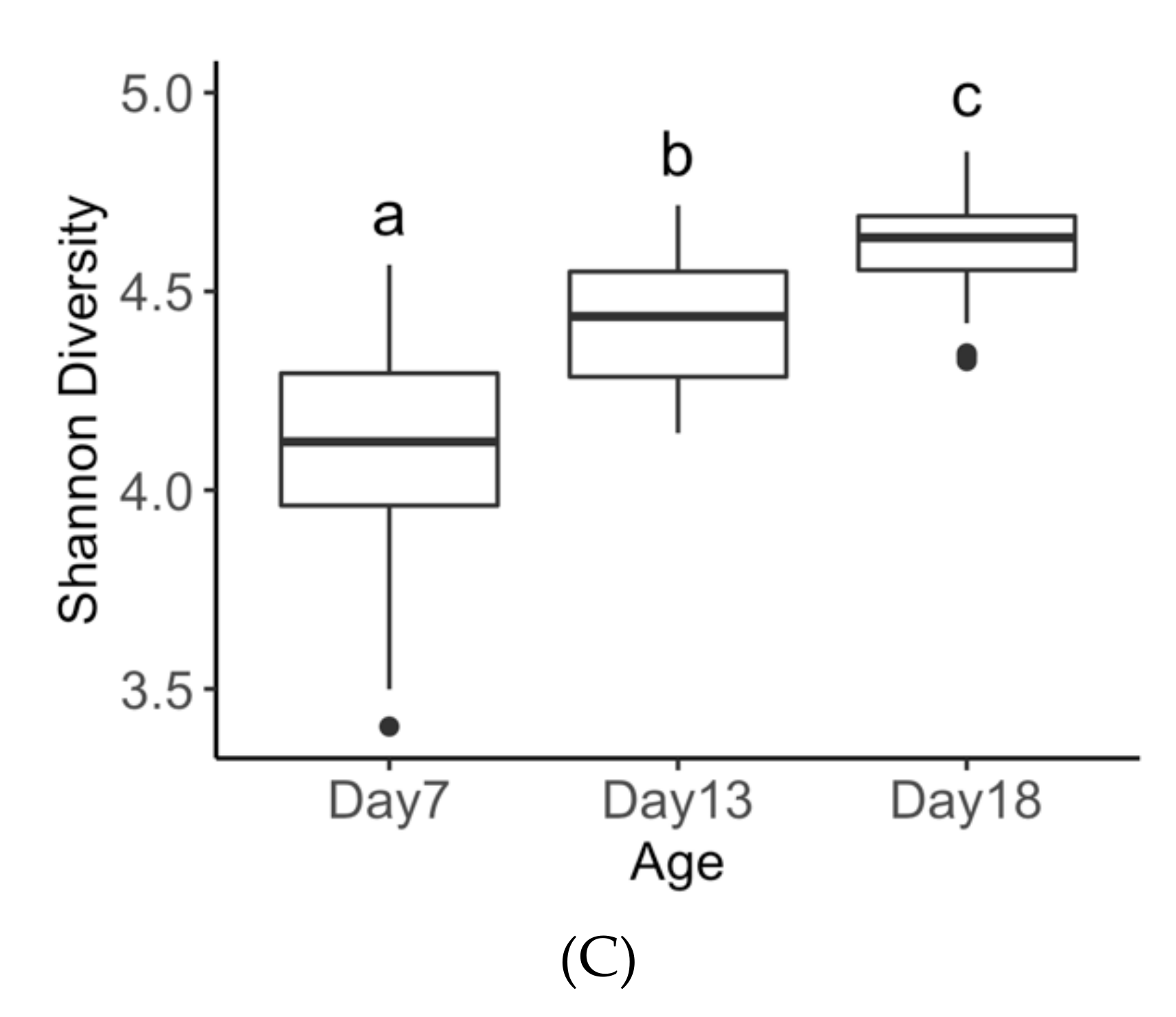
3.3. Age Effects on Diversity Metrics
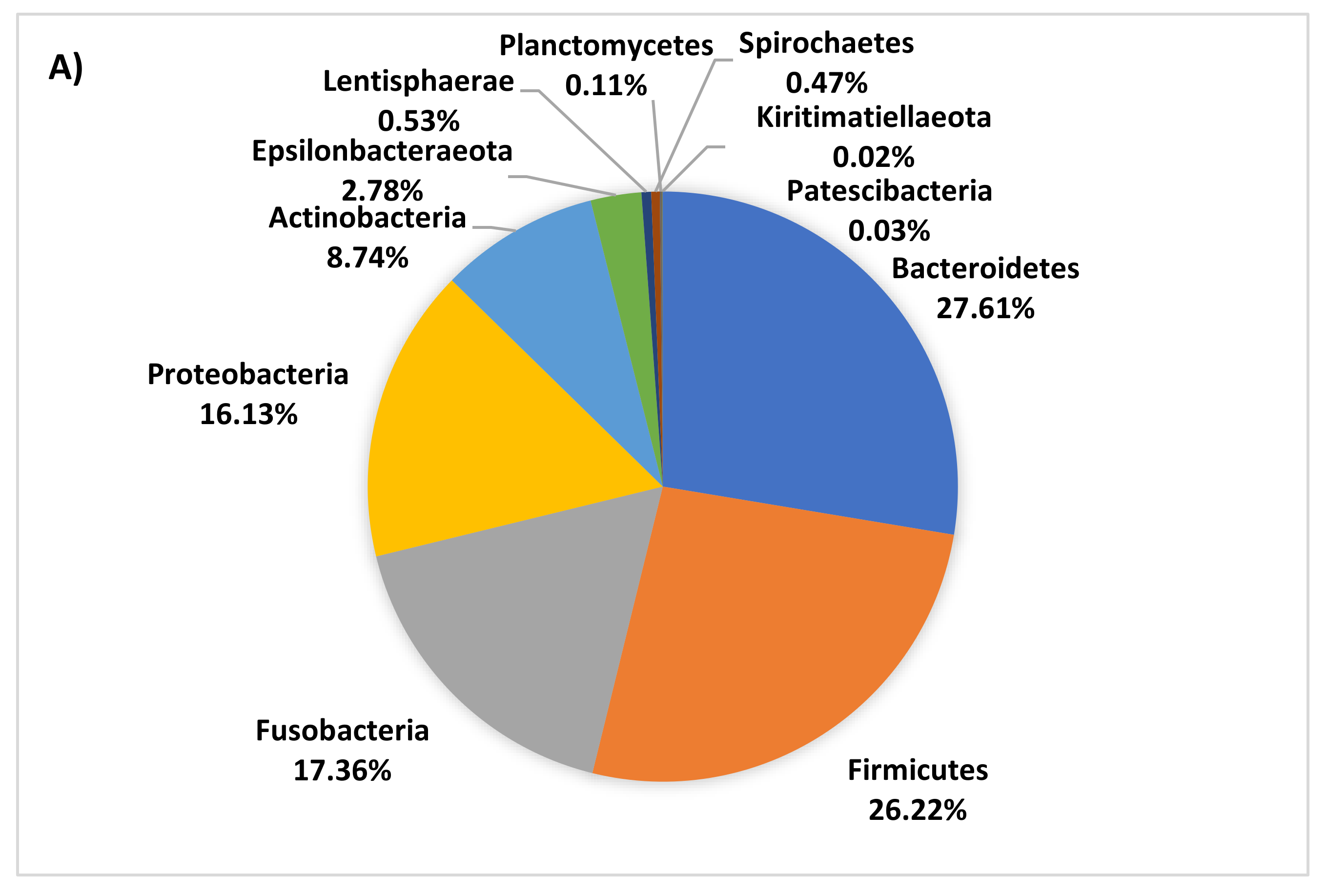
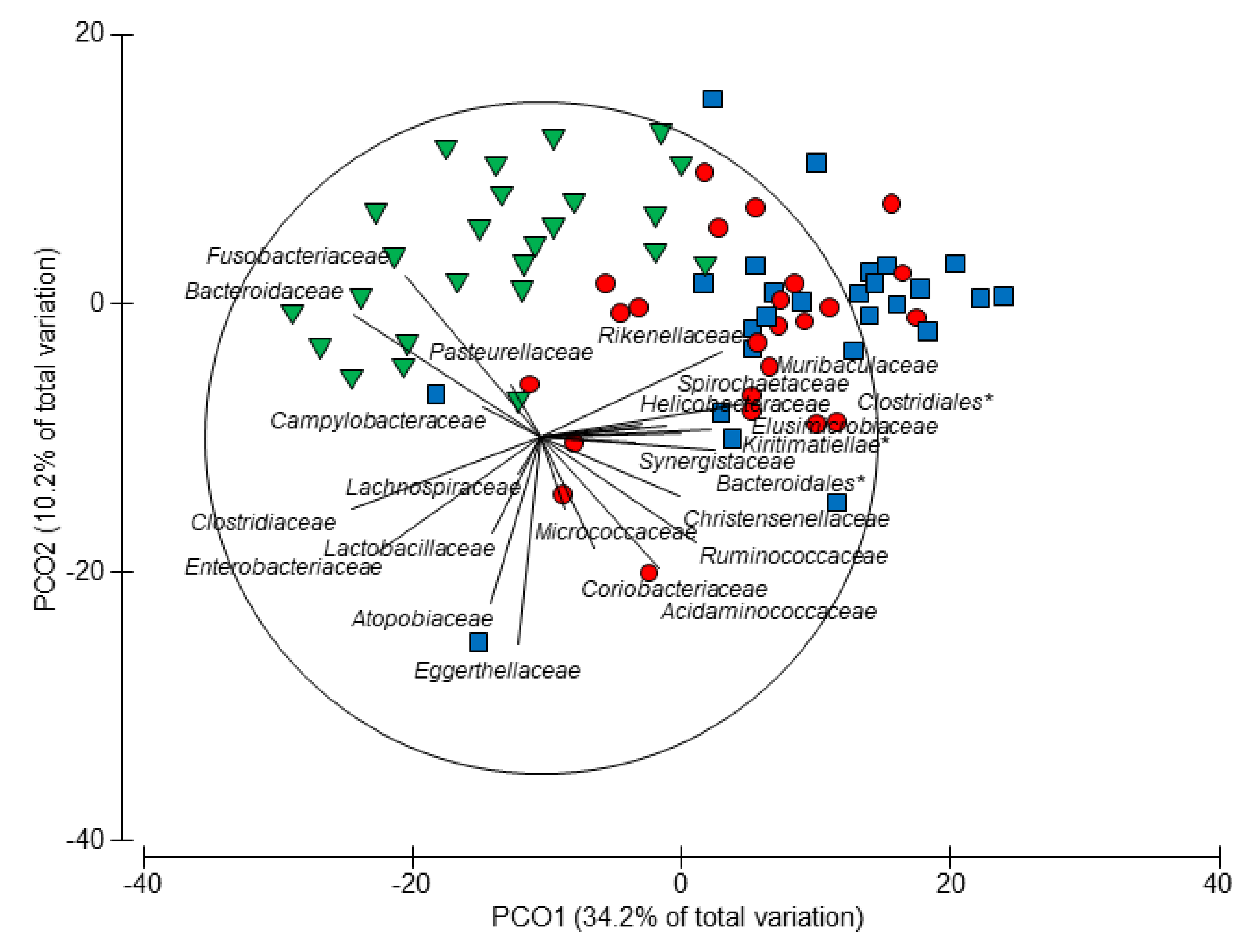
3.4. Age-Related Taxonomic Composition of Bacterial Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Borody, T.J.; Khoruts, A. Fecal microbiota transplantation and emerging applications. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 88–96. [Google Scholar]
- Rea, M.C.; Sit, C.S.; Clayton, E.; O’Connor, P.M.; Whittal, R.M.; Zheng, J.; Vederas, J.; Ross, R.P.; Hill, C. Thuricin cd, a posttranslationally modified bacteriocin with a narrow spectrum of activity against clostridium difficile. Proc. Natl. Acad. Sci. USA 2010, 107, 9352–9357. [Google Scholar] [PubMed]
- Bauer, E.; Williams, B.A.; Smidt, H.; Verstengen, M.W.; Mosenthin, R. Infuence of the gastrointestinal microbiota on development of the immune system in young animals. Curr. Issues Intest. Microbiol. 2006, 7, 35–52. [Google Scholar]
- Schachtschneider, K.M.; Yeoman, C.J.; Isaacson, R.E.; White, B.A.; Schook, L.B.; Pieters, M. Modulation of systemic immune responses through commensal gastrointestinal microbiota. PLoS ONE 2013, 8, 16. [Google Scholar]
- La Fata, G.; Weber, P.; Mohajeri, M.H. Probiotics and the gut immune system: Indirect regulation. Probiotics Antimicrob. Prot. 2018, 10, 11–21. [Google Scholar]
- Willing, B.P.; Gill, N.; Finlay, B.B. The role of the immune system in regulating the microbiota. Gut Microbes 2010, 1, 213–223. [Google Scholar]
- Wlodarska, M.; Willing, B.; Keeney, K.M.; Menendez, A.; Bergstrom, K.S.; Gill, N.; Russell, S.L.; Vallance, B.A.; Finlay, B.B. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated citrobacter rodentium-induced colitis. Infect. Immun. 2011, 79, 1536–1545. [Google Scholar]
- Janczyk, P.; Pieper, R.; Souffrant, W.B.; Bimczok, D.; Rothkotter, H.J.; Smidt, H. Parenteral long-acting amoxicillin reduces intestinal bacterial community diversity in piglets even 5 weeks after the administration. ISME J. 2007, 1, 180–183. [Google Scholar]
- Bakken, J.S.; Borody, T.; Brandt, L.J.; Brill, J.V.; Demarco, D.C.; Franzos, M.A.; Kelly, C.; Khoruts, A.; Louie, T.; Martinelli, L.P.; et al. Treating clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011, 9, 1044–1049. [Google Scholar]
- Brandt, L.J.; Aroniadis, O.C. An overview of fecal microbiota transplantation: Techniques, indications, and outcomes. Gastrointest. Endosc. 2013, 78, 240–249. [Google Scholar]
- Illumina. 16s metagenomic sequencing library preparation (15044223 b). Available online: https://www.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 1 September 2019).
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. Pear: A fast and accurate illumina paired-end read merger. Bioinformatics 2014, 30, 614–620. [Google Scholar] [PubMed]
- Caporaso, J.G.; Kucztnski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. Uchime improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar]
- Edgar, R.C. Uparse: Highly accurate otu sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16s rrna gene database and workbench compatible with arb. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [PubMed]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of southern wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar]
- Kruskal, J.B. Multidimensional scaling by optimizing a goodness of fit to a nonmetric hypothesis. Phsychometrics 1964, 29, 1–28. [Google Scholar]
- Shepard, R.N. The analysis of proximities: Multidimensional scaling with an unknown distance function. I. Psychometrika 1962, 27, 125–140. [Google Scholar]
- Shepard, R.N. The analysis of proximities: Multidimensional scaling with an unknown distance function. II. Psychometrika 1962, 27, 219–246. [Google Scholar]
- Gower, J.C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrics 1966, 53, 325–338. [Google Scholar]
- Clarke, K.R.; Warwick, R.M. Quantifying structural redundancy in ecological communities. Oecology 1998, 113, 278–289. [Google Scholar]
- Hu, J.; Nie, Y.F.; Chen, J.W.; Zhang, Y.; Wang, Z.C.; Fan, Q.W.; Yan, X.H. Gradual changes of gut microbiota in weaned miniature piglets. Front. Microbiol. 2016, 7, 15. [Google Scholar]
- Guevarra, R.B.; Hong, S.H.; Cho, J.H.; Kim, B.R.; Shin, J.; Lee, J.H.; Kang, B.N.; Kim, Y.H.; Wattanaphansak, S.; Isaacson, R.E.; et al. The dynamics of the piglet gut microbiome during the weaning transition in association with health and nutrition. J. Anim. Sci. Biotechnol. 2018, 9, 54. [Google Scholar] [PubMed]
- Arrieta, M.C.; Stiemsma, L.T.; Amenyogbe, N.; Brown, E.M.; Finlay, B. The intestinal microbiome in early life: Health and disease. Front. Immunol. 2014, 5, 18. [Google Scholar]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar]
- Murphy, K.; O’Shea, C.A.; Ryan, C.A.; Dempsey, E.M.; O’Toole, P.W.; Stanton, C.; Ross, R.P. The gut microbiota composition in dichorionic triplet sets suggests a role for host genetic factors. PLoS ONE 2015, 10, e0122561. [Google Scholar]
- Nowland, T.L.; Plush, K.J.; Barton, M.; Kirkwood, R.N. Development and function of the intestinal microbiome and potential implications for pig production. Animals Basel 2019, 9, 76. [Google Scholar]
- Frese, S.A.; Parker, K.; Calvert, C.C.; Mills, D.A. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 2015, 3, 28. [Google Scholar]
- Wattanakul, W.; Bulman, C.A.; Edge, H.L.; Edwards, S.A. The effect of creep feed presentation method on feeding behaviour, intake and performance of suckling piglets. Appl. Anim. Behav. Sci. 2005, 92, 27–36. [Google Scholar]
- Guevarra, R.B.; Lee, J.H.; Lee, S.H.; Seok, M.J.; Kim, D.W.; Kang, B.N.; Johnson, T.J.; Isaacson, R.E.; Kim, H.B. Piglet gut microbial shifts early in life: Causes and effects. J. Anim. Sci. Biotechnol. 2019, 10, 1. [Google Scholar] [PubMed]
- Aviles-Rosa, E.O.; Rakhshandeh, A.; McGlone, J.J. Preliminary study: Depriving piglets of maternal feces for the first seven days post-partum changes piglet physiology and performance before and after weaning. Animals Basel 2019, 9. [Google Scholar]
- McCormack, U.M.; Curiao, T.; Wilkinson, T.; Metzler-Zebeli, B.U.; Reyer, H.; Ryan, T.; Calderon-Diaz, J.A.; Crispie, F.; Cotter, P.D.; Creevey, C.J.; et al. Fecal microbiota transplantation in gestating sows and neonatal offspring alters lifetime intestinal microbiota and growth in offspring. Am. Soc. Microbiol. 2018, 3, 268. [Google Scholar]
- Dou, S.; Gadonna-Widehem, P.; Rome, V.; Hamoudi, D.; Rhazi, L.; Lakhal, L.; Larcher, T.; Bahi-Jaber, N.; Pinon-Quintana, A.; Guyonvarch, A.; et al. Characterisation of early-life fecal microbiota in susceptible and healthy pigs to post-weaning diarrhoea. PLoS ONE 2017, 12, e0169851. [Google Scholar]
- Gao, K.; Pi, Y.; Peng, Y.; Mu, C.L.; Zhu, W.Y. Time-course responses of ileal and fecal microbiota and metabolite profiles to antibiotics in cannulated pigs. Appl. Microbiol. Biotechnol. 2018, 102, 2289–2299. [Google Scholar]
- Ruczizka, U.; Metzler-Zebeli, B.; Unterweger, C.; Mann, E.; Schwarz, L.; Knecht, C.; Hennig-Pauka, I. Early parenteral administration of ceftiofur has gender-specific short- and long-term effects on the fecal microbiota and growth in pigs from the suckling to growing phase. Animals Basel 2019, 10, 17. [Google Scholar]
- Khoruts, A.; Dicksved, J.; Jansson, J.K.; Sadowsky, M.J. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent clostridium difficile-associated diarrhea. J. Clin. Gastroenterol. 2010, 44, 354–360. [Google Scholar] [PubMed]
- Hu, L.S.; Geng, S.J.; Li, Y.; Cheng, S.S.; Fu, X.F.; Yue, X.J.; Han, X.Y. Exogenous fecal microbiota transplantation from local adult pigs to crossbred newborn piglets. Front. Microbiol. 2018, 8, 2663. [Google Scholar]

| Day 7 | Day 18 | ||
|---|---|---|---|
| Family | Average Abundance | Average Abundance | % |
| Uncharacterised Clostridiales | 0.22 | 1.21 | 4.27 |
| Christensenellaceae | 0.38 | 1.37 | 8.47 |
| Fusobacteriaceae | 1.71 | 0.88 | 12.59 |
| Muribaculaceae | 0.86 | 1.52 | 16.25 |
| Clostridiaceae | 1.35 | 0.61 | 19.56 |
| Prevotellaceae | 1.32 | 1.45 | 22.5 |
| Uncharacterised Bacteroidales | 0.06 | 0.7 | 25.32 |
| Synergistaceae | 0.02 | 0.69 | 28.11 |
| Bacteroidaceae | 2.1 | 1.63 | 30.86 |
| Lactobacillaceae | 1.4 | 1.24 | 33.52 |
| Campylobacteraceae | 0.58 | 0.83 | 36.13 |
| Enterobacteriaceae | 1.37 | 1.09 | 38.62 |
| Rikenellaceae | 0.88 | 1.24 | 41.03 |
| Marinifilaceae | 0.83 | 0.9 | 43.36 |
| Ruminococcaceae | 1.61 | 1.94 | 45.44 |
| Actinomycetaceae | 0.58 | 0.28 | 47.37 |
| Spirochaetaceae | 0.17 | 0.53 | 49.29 |
| Victivallaceae | 0.16 | 0.5 | 51 |
| Enterococcaceae | 0.36 | 0.31 | 52.7 |
| Helicobacteraceae | 0.12 | 0.41 | 54.31 |
| Veillonellaceae | 0.88 | 0.68 | 55.87 |
| Lachnospiraceae | 1.7 | 1.75 | 58.98 |
| Oligosphaeraceae | 0.04 | 0.38 | 60.52 |
| Coriobacteriaceae | 0.51 | 0.61 | 62.05 |
| Acidaminococcaceae | 0.86 | 1.13 | 63.55 |
| Uncharacterised Mollicutes | 0 | 0.36 | 65.05 |
| Pirellulaceae | 0.09 | 0.38 | 66.52 |
| Streptococcaceae | 0.82 | 0.58 | 67.94 |
| Uncharacterised Bradymonadales | 0.12 | 0.33 | 69.34 |
| Pasteurellaceae | 0.63 | 0.53 | 70.73 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowland, T.L.; Torok, V.A.; Low, W.Y.; Barton, M.D.; Plush, K.J.; Kirkwood, R.N. Faecal Microbiota Analysis of Piglets During Lactation. Animals 2020, 10, 762. https://doi.org/10.3390/ani10050762
Nowland TL, Torok VA, Low WY, Barton MD, Plush KJ, Kirkwood RN. Faecal Microbiota Analysis of Piglets During Lactation. Animals. 2020; 10(5):762. https://doi.org/10.3390/ani10050762
Chicago/Turabian StyleNowland, Tanya L., Valeria A. Torok, Wai Y. Low, Mary D. Barton, Kate J. Plush, and Roy N. Kirkwood. 2020. "Faecal Microbiota Analysis of Piglets During Lactation" Animals 10, no. 5: 762. https://doi.org/10.3390/ani10050762
APA StyleNowland, T. L., Torok, V. A., Low, W. Y., Barton, M. D., Plush, K. J., & Kirkwood, R. N. (2020). Faecal Microbiota Analysis of Piglets During Lactation. Animals, 10(5), 762. https://doi.org/10.3390/ani10050762






