Genetic Diversity and Ancestral Study for Korean Native Pigs Using 60K SNP Chip
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Genotypes
2.2. Statistical Analysis
3. Results
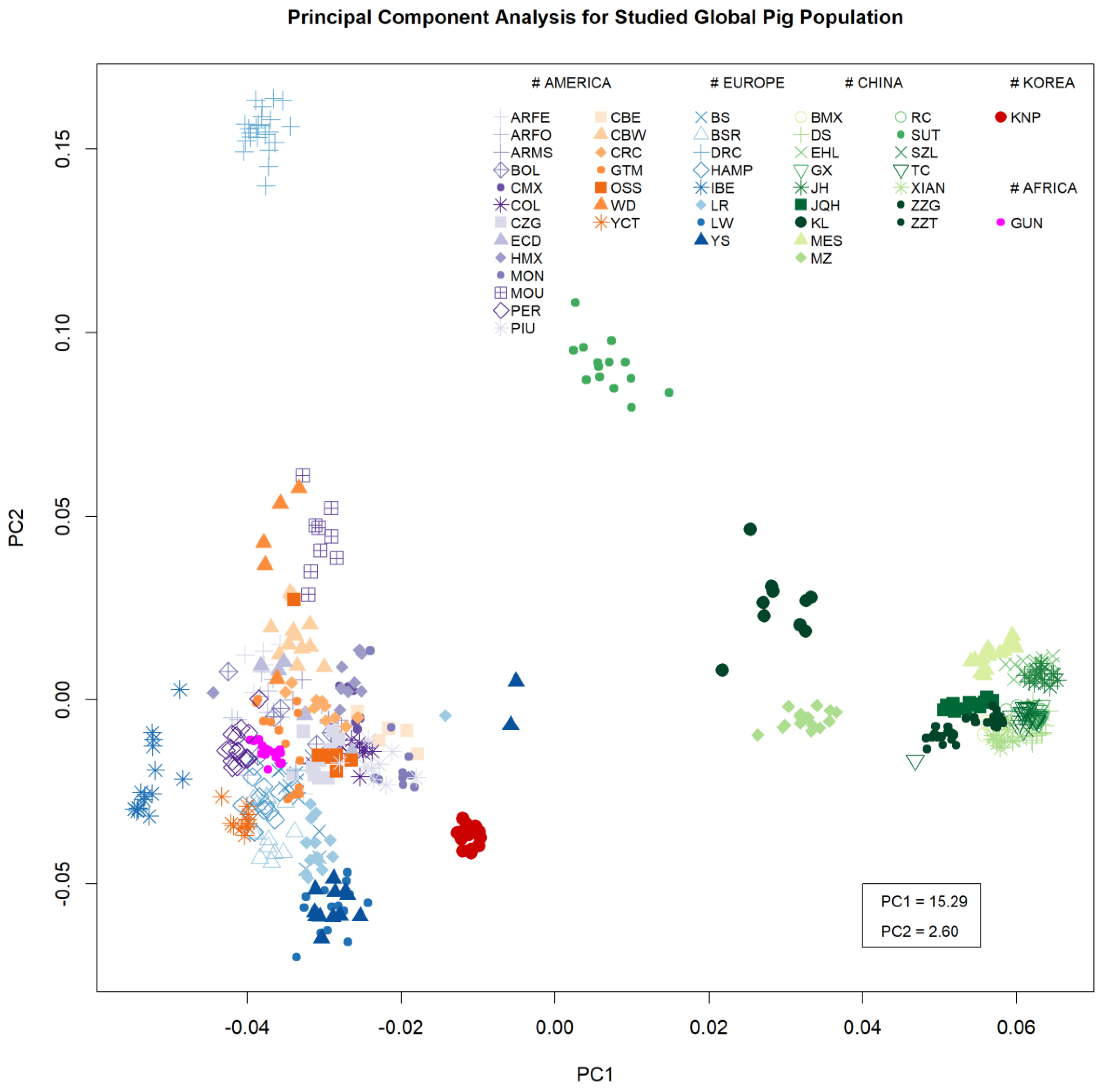
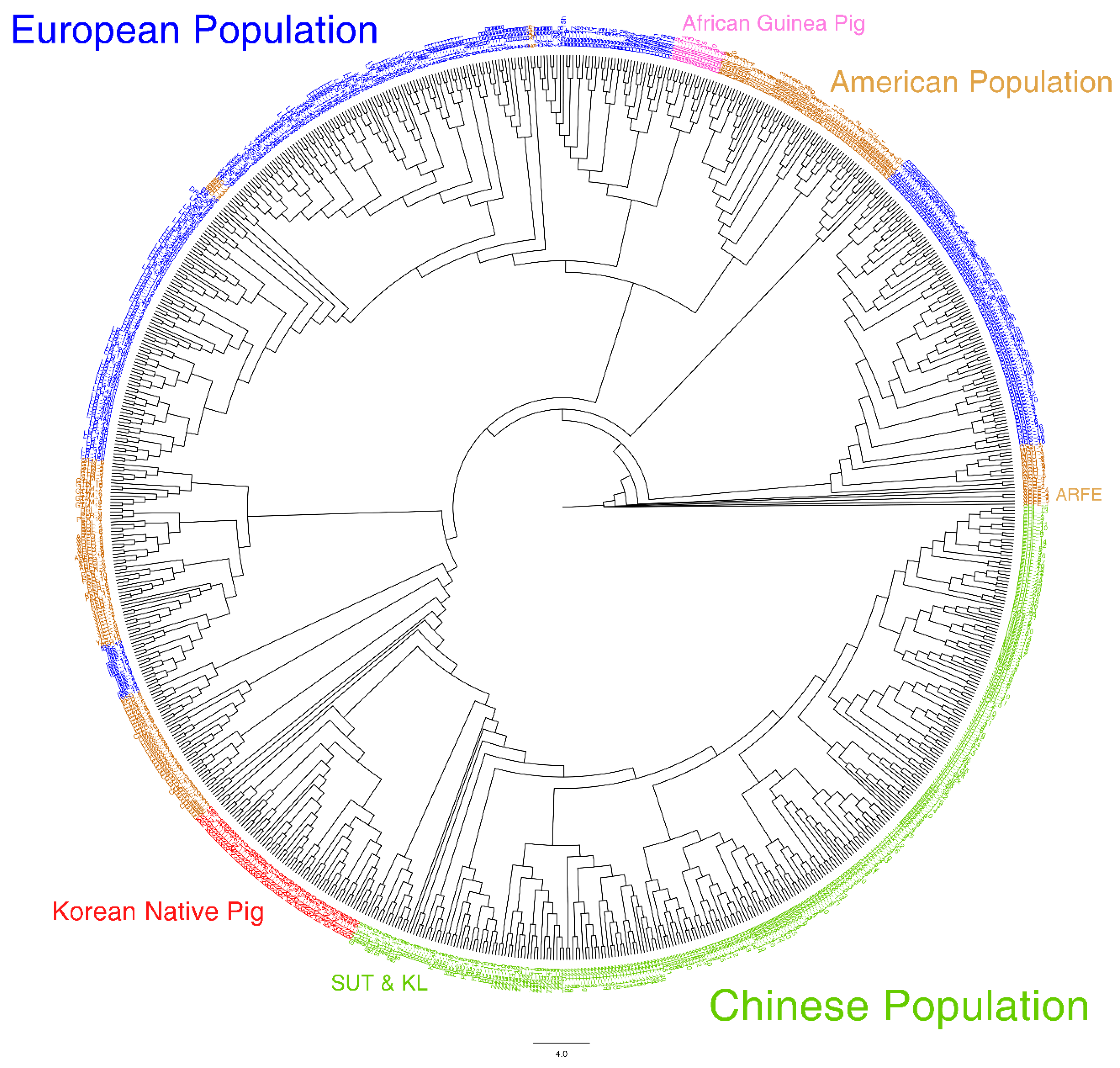
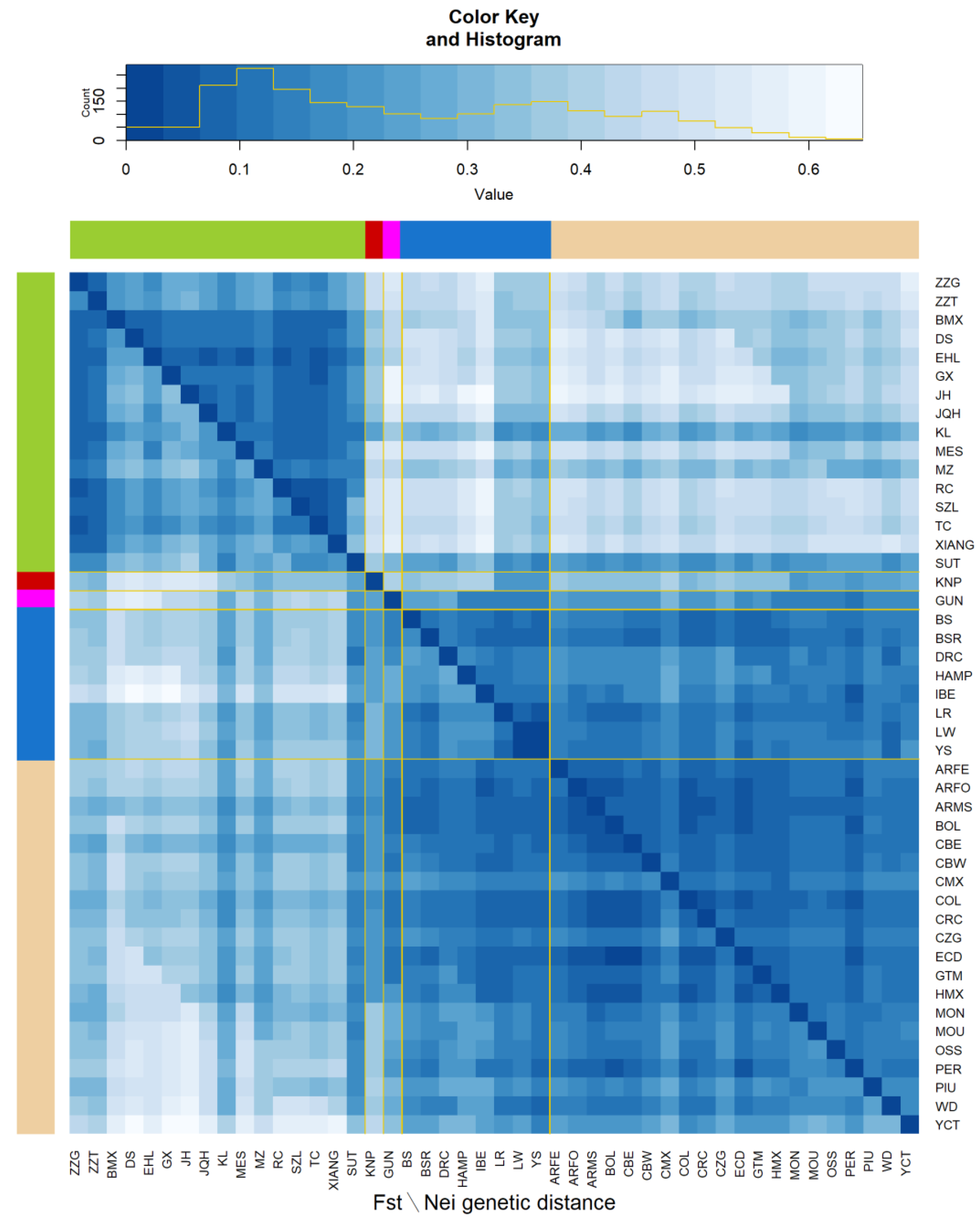
3.1. Breed Diversity
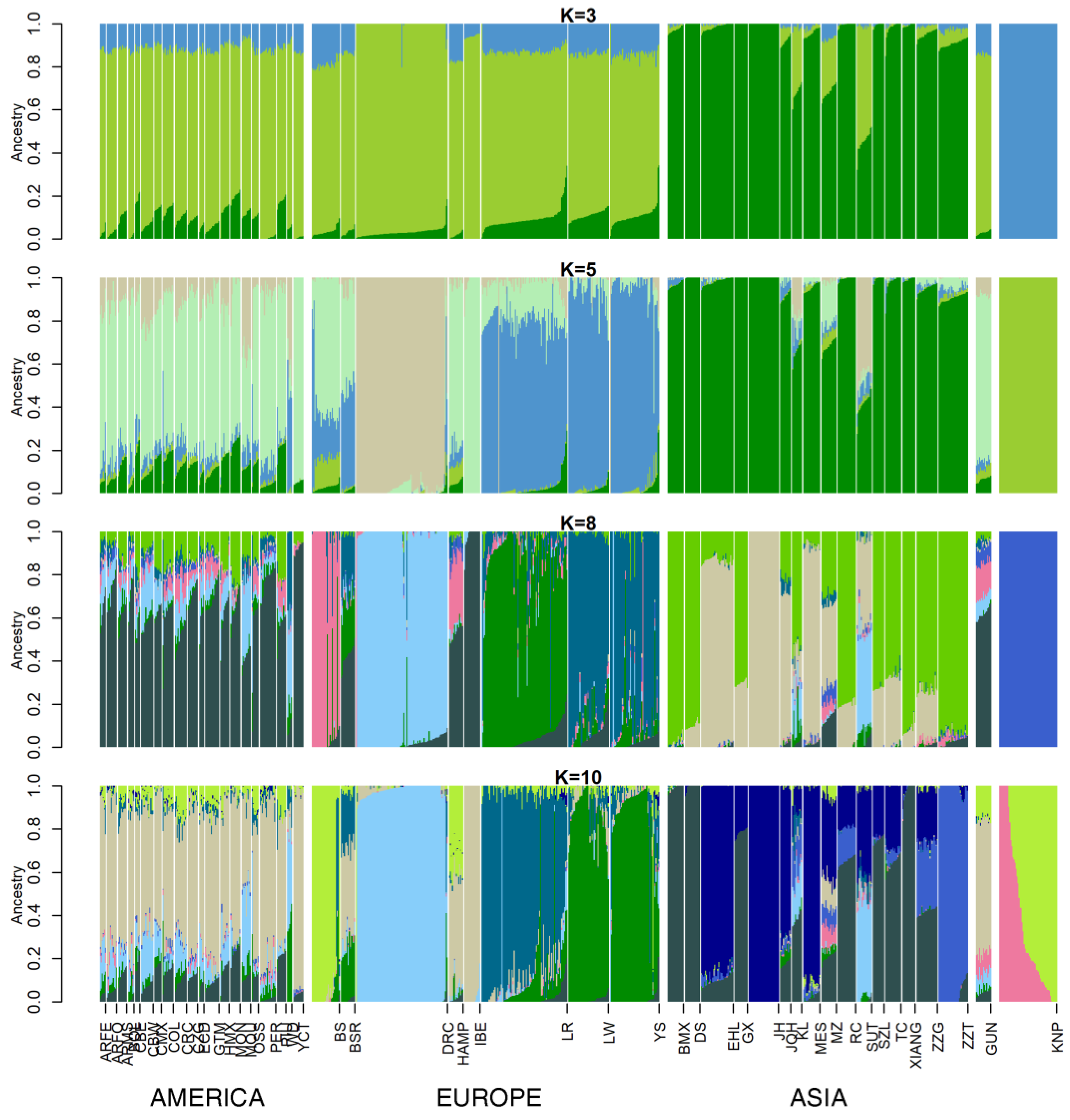
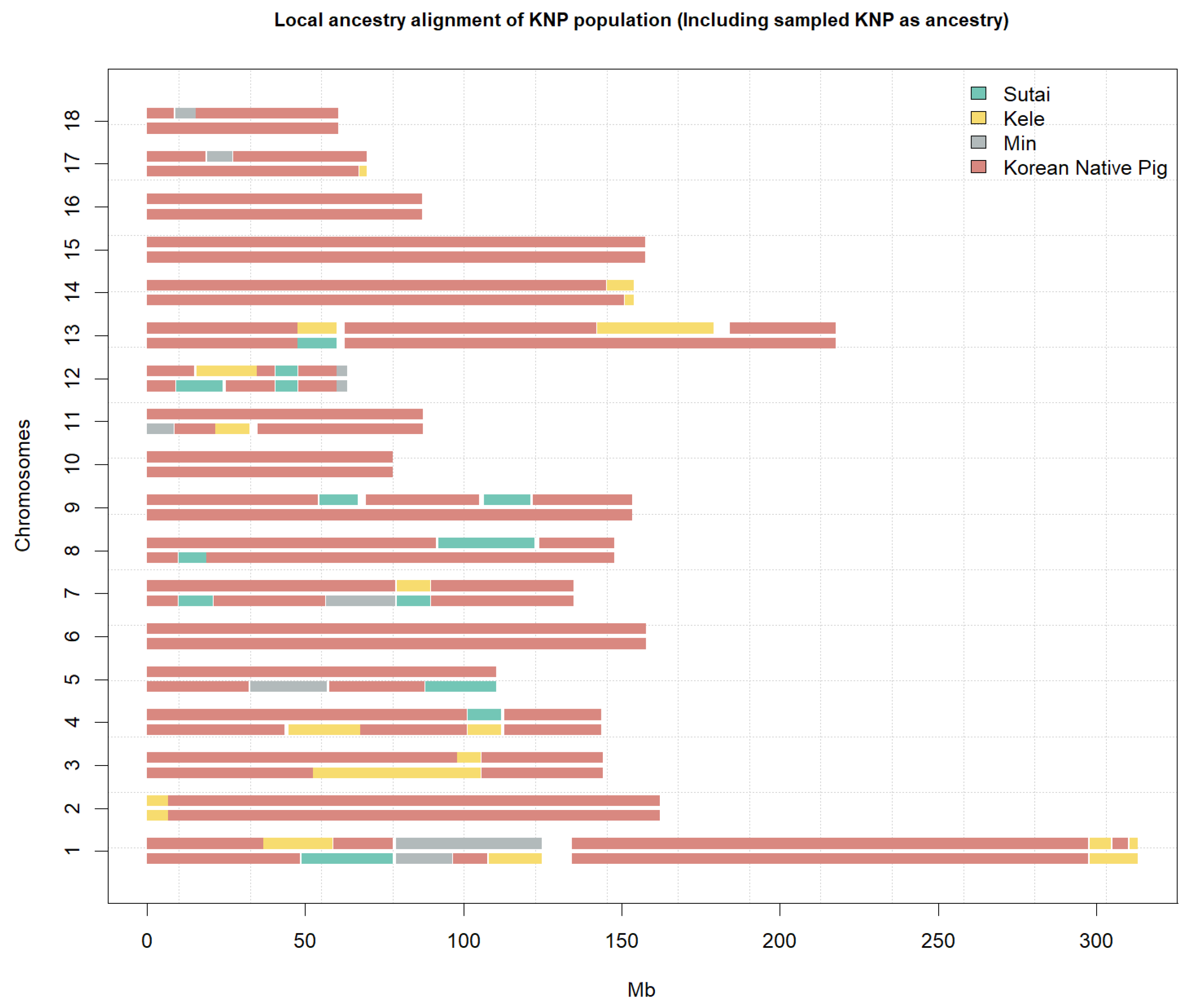
3.2. Ancestry Analysis
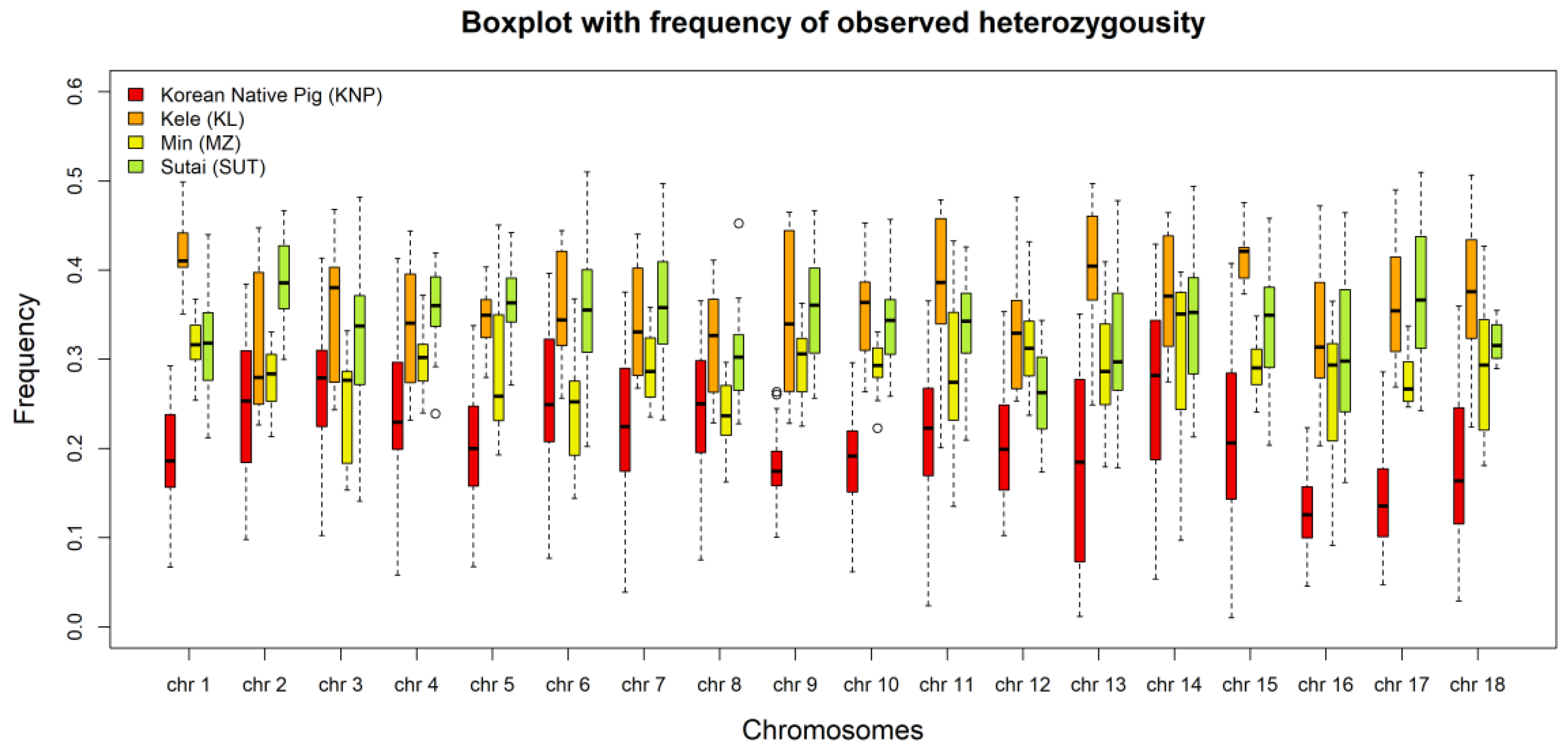
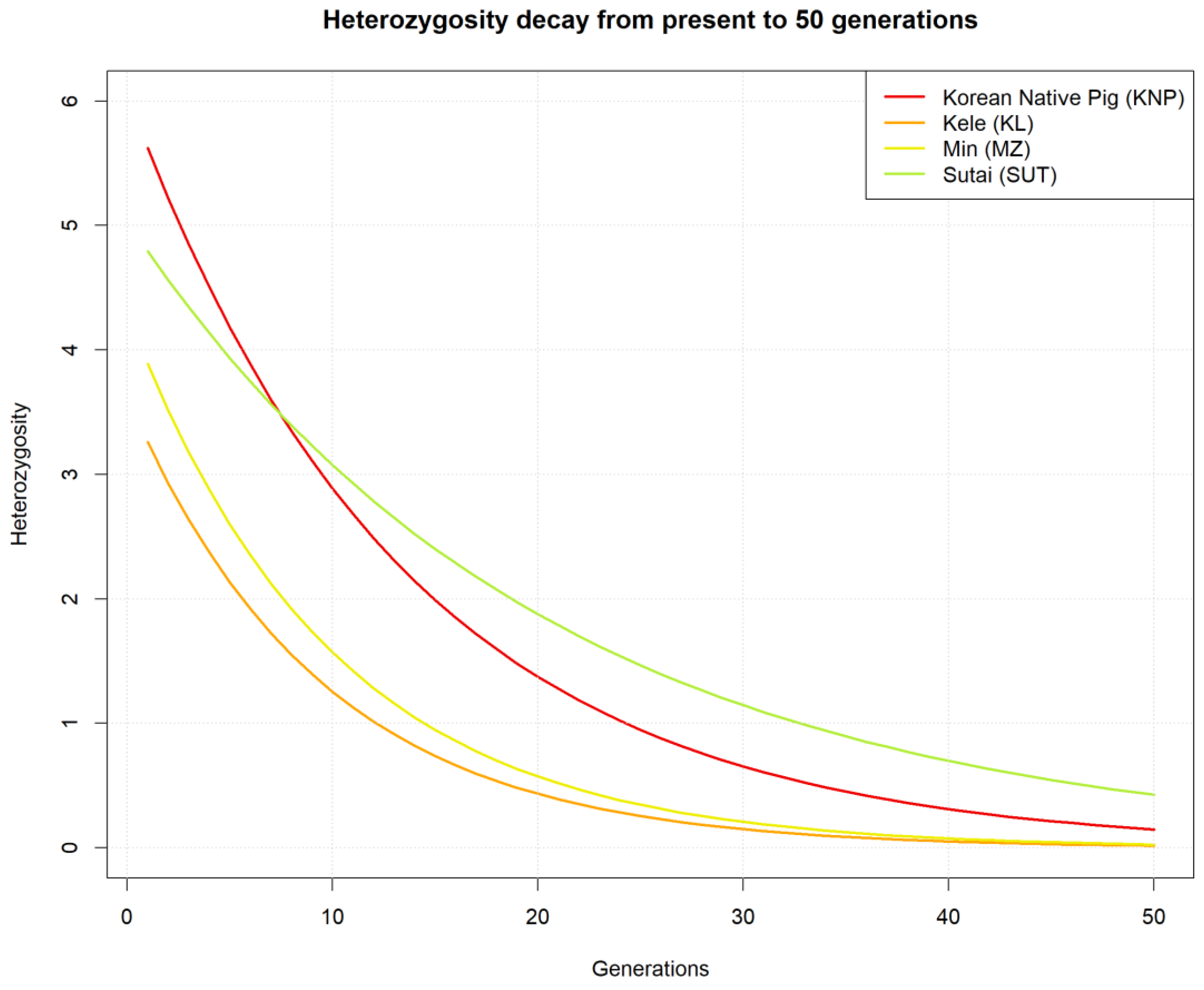
3.3. Genetic Diversity
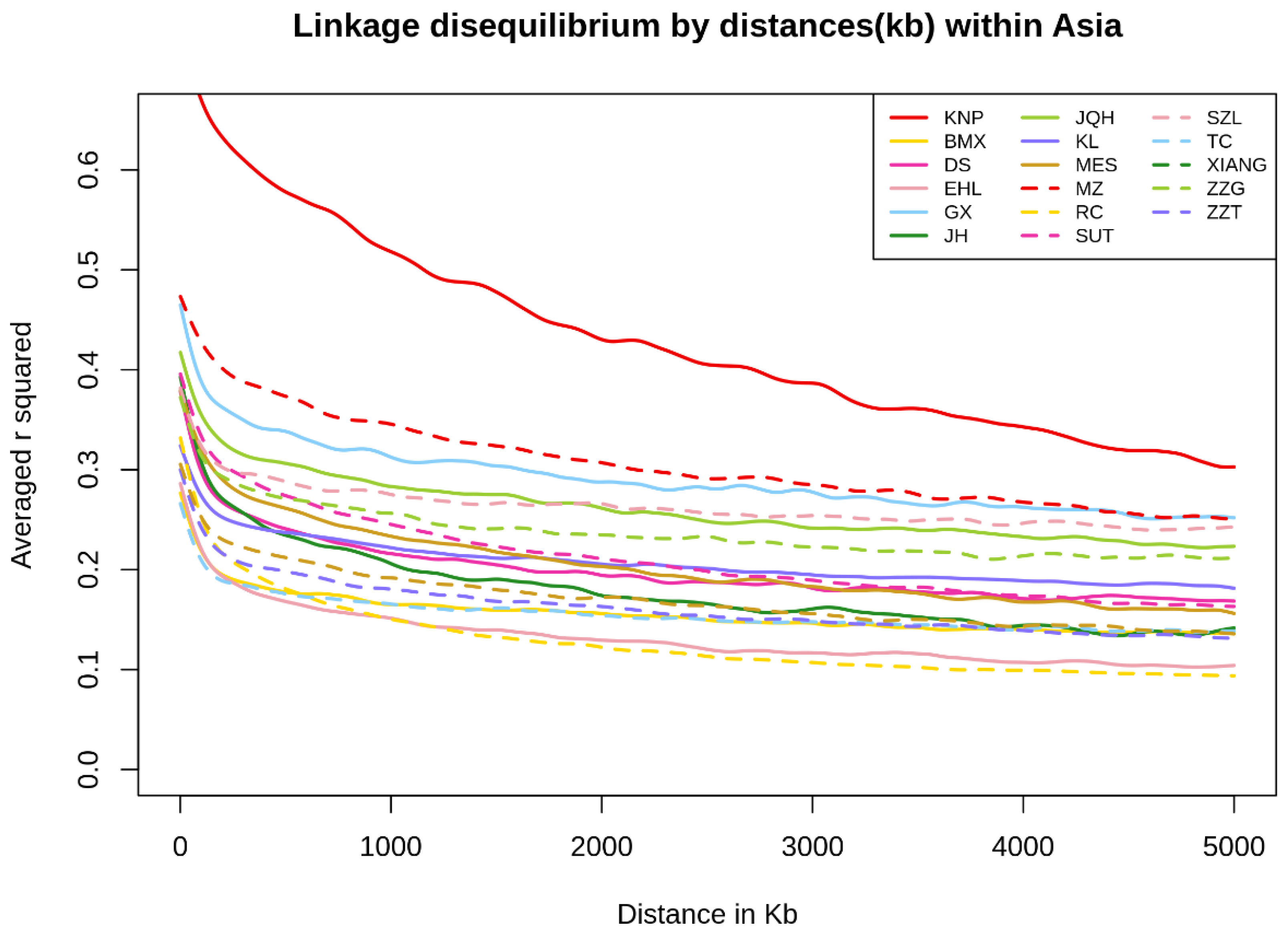
3.4. Linkage Disequilibrium and Observed Heterozygosity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Larson, G.; Dobney, K.M.; Albarella, U.; Fang, M.; Matisoo-Smith, E.; Robins, J.; Lowden, S.; Finlayson, H.; Brand, T.; Willerslev, E.; et al. Worldwide Phylogeography of Wild Boar Reveals Multiple Centers of Pig Domestication. Science 2005, 307, 1618–1621. [Google Scholar] [CrossRef]
- Kariyawasam, K.; Tsai, M. Access to genetic resources and benefit sharing: Implications of Nagoya Protocol on providers and users. J. World Intellect. Prop. 2018, 21, 289–305. [Google Scholar] [CrossRef]
- Chung, H.Y.; Ko, M.S. Characteristics and Application of Native Pigs in Jeju, South Korea. Soc. Cheju Stu. 2006, 1, 95–119. [Google Scholar]
- Seo, S.Y.; Park, K.M.; Seo, H.W.; Cho, S.H.; Park, B.Y.; Cho, I.C.; Sung, P.N. Pork Quality Characteristics of Different Parts from Crossbred (Jeju Native Pig × Landrace) Pigs. J. Agric. Life Sci. 2015, 49, 211–223. [Google Scholar] [CrossRef]
- Kim, D.W.; Kim, K.H.; Hong, J.K.; Cho, K.H.; Sa, S.J.; Park, J.C.; Choi, S.H. Comparison of Carcass Characteristics, Meat Quality, Amino Acids Contents, and Fatty Acid Profiles of Korea Native Pig by Gender. Reprod. Dev. Biol. 2013, 37, 129–134. [Google Scholar] [CrossRef]
- Seo, S.Y.; Park, K.; Cho, S.H.; Kang, G.; Hoa, V.B.; Kang, S.M.; Park, B.Y.; Cho, I.C.; Seong, P.N. Carcass Traits and Retail Cuts Characteristics of Crossbred (Jeju native pig × Landrace) Pigs. Ann. Anim. Resour. Sci. 2014, 25, 29–37. [Google Scholar] [CrossRef]
- Hong, J.K. The History of Korean Native Pig. Breeding Technology of Korean Native Pig; NIAS press: RDA, Seonghwan, Korea, 2013; Available online: https://www.nias.go.kr:3443/front/soboarddown.do?cmCode=M090814151125016&boardSeqNum=787&fileSeqNum=813 (accessed on 1 March 2020).
- Edea, Z.; Kim, S.W.; Lee, K.T.; Kim, T.H.; Kim, K.S. Genetic Structure of and Evidence for Admixture between Western and Korean Native Pig Breeds Revealed by Single Nucleotide Polymorphisms. Asian-Australas. J. Anim. Sci. 2014, 27, 1263–1269. [Google Scholar] [CrossRef]
- Choi, J.W.; Chung, W.H.; Lee, K.T.; Cho, E.S.; Lee, S.W.; Choi, B.H.; Lee, S.H.; Lim, W.J.; Lim, D.; Lee, Y.G.; et al. Whole-genome resequencing analyses of five pig breeds, including Korean wild and native, and three European origin breeds. Curr. Neuropharmacol. 2015, 22, 259–267. [Google Scholar] [CrossRef]
- Kim, K.S.; Yeo, J.S.; Kim, J.W. Assessment of genetic diversity of Korean native pig (Sus scrofa) using AFLP markers. Genes Genet. Syst. 2002, 77, 361–368. [Google Scholar] [CrossRef]
- Kim, T.H.; Kim, K.S.; Choi, B.H.; Yoon, D.H.; Jang, G.W.; Lee, K.T.; Chung, H.Y.; Lee, H.Y.; Park, H.S.; Lee, J.W. Genetic structure of pig breeds from Korea and China using microsatellite loci analysis. J. Anim. Sci. 2005, 83, 2255–2263. [Google Scholar] [CrossRef]
- Burgos-Paz, W.; Souza, C.A.; Megens, H.J.; Ramayo-Caldas, Y.; Melo, M.; Lemús-Flores, C.; Caal, E.; Soto, H.W.; Martinez, R.; A Álvarez, L.; et al. Porcine colonization of the Americas: A 60k SNP story. Heredity 2012, 110, 321–330. [Google Scholar] [CrossRef]
- Ai, H.; Huang, L.; Ren, J. Genetic diversity, linkage disequilibrium and selection signatures in Chinese and Western pigs revealed by genome-wide SNP markers. PLoS ONE 2013, 8, e56001. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Shannon, L.M.; Boyko, R.H.; Castelhano, M.; Corey, E.; Hayward, J.J.; McLean, C.; White, M.E.; Said, M.A.; Anita, B.A.; Bondjengo, N.I.; et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc. Natl. Acad. Sci. USA 2015, 112, 13639–13644. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Levine, D.; Shen, J.; Gogarten, S.; Laurie, C.; Weir, B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012, 28, 3326–3328. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Lee, T.H.; Guo, H.; Wang, X.; Kim, C.; Paterson, A.H. SNPhylo: A pipeline to construct a phylogenetic tree from huge SNP data. BMC Genom. 2014, 15, 162. [Google Scholar] [CrossRef]
- Brisbin, A.; Bryc, K.; Byrnes, J.; Zakharia, F.; Omberg, L.; Degenhardt, J.; Reynolds, A.; Ostrer, H.; Mezey, J.G.; Bustamante, C.D. PCAdmix: Principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum. Biol. 2012, 84, 343–364. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012, 8, 1002967. [Google Scholar] [CrossRef]
- Pembleton, L.W.; Cogan, N.O.I.; Forster, J.W. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef]
- Cho, I.C.; Han, S.H.; Fang, M.; Lee, S.S.; Ko, M.S.; Lee, H.; Lim, H.T.; Yoo, C.K.; Lee, J.H.; Jeon, J.T. The robust phylogeny of Korean wild boar (Sus scrofa coreanus) using partial D-loop sequence of mtDNA. Mol. Cells 2009, 28, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Gorbach, D.; Rothschild, M. The Pig Genome Project Has Plenty to Squeal about. Cytogenet. Genome Res. 2011, 134, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.; Crooijmans, R.P.M.A.; Affara, N.A.; Amaral, A.; Archibald, A.L.; Beever, J.E.; Bendixen, C.; Churcher, C.; Clark, R.; Dehais, P.; et al. Design of a High Density SNP Genotyping Assay in the Pig Using SNPs Identified and Characterized by Next Generation Sequencing Technology. PLoS ONE 2009, 4, e6524. [Google Scholar] [CrossRef] [PubMed]
- Kerstens, H.H.; Kollers, S.; Kommadath, A.; Del Rosario, M.; Dibbits, B.W.; Kinders, S.M.; Crooijmans, R.P.M.A.; Groenen, M. Mining for single nucleotide polymorphisms in pig genome sequence data. BMC Genom. 2009, 10, 4. [Google Scholar] [CrossRef]
- Lachance, J.; Tishkoff, S.A. SNP ascertainment bias in population genetic analyses: Why it is important, and how to correct it. BioEssays 2013, 35, 780–786. [Google Scholar] [CrossRef]
- Yang, S.L.; Wang, Z.G.; Liu, B.; Zhang, G.X.; Zhao, S.; Yu, M.; Fan, B.; Li, M.H.; Xiong, T.A. Genetic variation and relationships of eighteen Chinese indigenous pig breeds. Genet. Sel. Evol. 2003, 35, 657. [Google Scholar] [CrossRef]
| Population | Abbr. | Number of Samples | Continent (Country) | FIS | HO | HS |
|---|---|---|---|---|---|---|
| Guinea Hog | GUN | 15 | Africa | −0.21561 | 0.24466 | 0.19339 |
| Bamaxiang | BMX | 16 | China | −0.18743 | 0.21812 | 0.17437 |
| Dongshan | DS | 15 | China | −0.22474 | 0.19643 | 0.15194 |
| Erhualian | EHL | 32 | China | −0.15704 | 0.19786 | 0.16120 |
| Ganxi | GX | 13 | China | −0.24343 | 0.17913 | 0.13651 |
| Jinhua | JH | 30 | China | −0.19250 | 0.15328 | 0.12247 |
| Jiangquhai | JQH | 11 | China | −0.20828 | 0.22652 | 0.17738 |
| Kele | KL | 10 | China | −0.22069 | 0.36251 | 0.28308 |
| Meishan | MES | 17 | China | −0.14848 | 0.17325 | 0.14384 |
| Min | MZ | 15 | China | −0.24263 | 0.28690 | 0.21950 |
| Rongchang | RC | 18 | China | −0.17866 | 0.20472 | 0.16476 |
| Sutai | SUT | 15 | China | −0.21308 | 0.33809 | 0.26723 |
| Shaziling | SZL | 11 | China | −0.23253 | 0.21988 | 0.16979 |
| Tongcheng | TC | 16 | China | −0.16408 | 0.22455 | 0.18138 |
| Xiang Pig | XIANG | 13 | China | −0.19884 | 0.19039 | 0.14982 |
| Tibetan Gansu | ZZG | 21 | China | −0.18170 | 0.22380 | 0.17871 |
| Tibetan Tibet | ZZT | 29 | China | −0.19512 | 0.24058 | 0.19119 |
| Korean Native Pig | KNP | 57 | Korea | −0.23283 | 0.21350 | 0.16689 |
| Berkshire | BS | 28 | Europe | −0.19111 | 0.30093 | 0.24058 |
| Bisaro | BSR | 14 | Europe | −0.20780 | 0.32414 | 0.25618 |
| Duroc | DRC | 91 | Europe | −0.17946 | 0.28352 | 0.22941 |
| Hampshire | HAMP | 14 | Europe | −0.17519 | 0.24523 | 0.19857 |
| Iberian | IBE | 16 | Europe | −0.14276 | 0.19132 | 0.15988 |
| Landrace | LR | 85 | Europe | −0.20920 | 0.34154 | 0.27423 |
| Large White | LW | 40 | Europe | −0.19803 | 0.32323 | 0.26175 |
| Yorkshire | YS | 49 | Europe | −0.22586 | 0.36596 | 0.28915 |
| Cuba Eastern Creole | CBE | 5 | Central America | −0.19443 | 0.36501 | 0.29197 |
| Cuba Western Creole | CBW | 12 | Central America | −0.20699 | 0.33331 | 0.26401 |
| Costa Rica Creole | CRC | 12 | Central America | −0.15263 | 0.27830 | 0.23168 |
| Guatemala Creole | GTM | 14 | Central America | −0.18924 | 0.29959 | 0.24041 |
| Ossabaw | OSS | 7 | North America | −0.18876 | 0.29628 | 0.23614 |
| White Duroc | WD | 5 | North America | −0.23209 | 0.35621 | 0.27499 |
| Yucatan | YCT | 10 | North America | −0.19945 | 0.21675 | 0.17234 |
| Argentina Feral | ARFE | 6 | South America | −0.16744 | 0.28001 | 0.22843 |
| Argentina Formosa Semi Feral | ARFO | 10 | South America | −0.16979 | 0.29542 | 0.24152 |
| Argentina Misiones Creole | ARMS | 9 | South America | −0.19796 | 0.34920 | 0.27903 |
| Bolivia Creole | BOL | 6 | South America | −0.14305 | 0.26133 | 0.21780 |
| Colombia Creole | COL | 11 | South America | −0.18893 | 0.33441 | 0.26967 |
| Colombia Zungo | CZG | 10 | South America | −0.22178 | 0.31298 | 0.24301 |
| Ecuador Creole | ECD | 5 | South America | −0.17857 | 0.32963 | 0.26671 |
| Mexico Cuino | CMX | 7 | South America | −0.26775 | 0.32025 | 0.24199 |
| Mexico Hairless | HMX | 9 | South America | −0.17319 | 0.30347 | 0.24695 |
| Peru Creole | PER | 16 | South America | −0.18019 | 0.28900 | 0.23336 |
| Brazil Monteiro | MON | 10 | South America | −0.14992 | 0.27048 | 0.22507 |
| Brazil Moura | MOU | 9 | South America | −0.21217 | 0.32382 | 0.25486 |
| Brazil Piau | PIU | 9 | South America | −0.21873 | 0.28493 | 0.22212 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.H.; Seo, D.W.; Cho, E.S.; Choi, B.H.; Kim, Y.M.; Hong, J.K.; Han, H.D.; Jung, Y.B.; Kim, D.J.; Choi, T.J.; et al. Genetic Diversity and Ancestral Study for Korean Native Pigs Using 60K SNP Chip. Animals 2020, 10, 760. https://doi.org/10.3390/ani10050760
Lee SH, Seo DW, Cho ES, Choi BH, Kim YM, Hong JK, Han HD, Jung YB, Kim DJ, Choi TJ, et al. Genetic Diversity and Ancestral Study for Korean Native Pigs Using 60K SNP Chip. Animals. 2020; 10(5):760. https://doi.org/10.3390/ani10050760
Chicago/Turabian StyleLee, Soo Hyun, Dong Won Seo, Eun Seok Cho, Bong Hwan Choi, Yong Min Kim, Joon Ki Hong, Hyo Dong Han, Yeon Bok Jung, Dong Jun Kim, Tae Jeong Choi, and et al. 2020. "Genetic Diversity and Ancestral Study for Korean Native Pigs Using 60K SNP Chip" Animals 10, no. 5: 760. https://doi.org/10.3390/ani10050760
APA StyleLee, S. H., Seo, D. W., Cho, E. S., Choi, B. H., Kim, Y. M., Hong, J. K., Han, H. D., Jung, Y. B., Kim, D. J., Choi, T. J., & Lee, S. H. (2020). Genetic Diversity and Ancestral Study for Korean Native Pigs Using 60K SNP Chip. Animals, 10(5), 760. https://doi.org/10.3390/ani10050760





