Association between Rumen Microbiota and Marbling Score in Korean Native Beef Cattle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Diet
2.2. DNA Extraction from Rumen Samples
2.3. 16S rRNA Gene Amplicon Sequencing
2.4. Taxonomic and Diversity Analyses
2.5. PICRUSt2 Functional Prediction
2.6. Identification of Taxonomic Drivers of PICRUSt2-Predicted Functions Using FishTaco
2.7. Statistical Analyses
3. Results
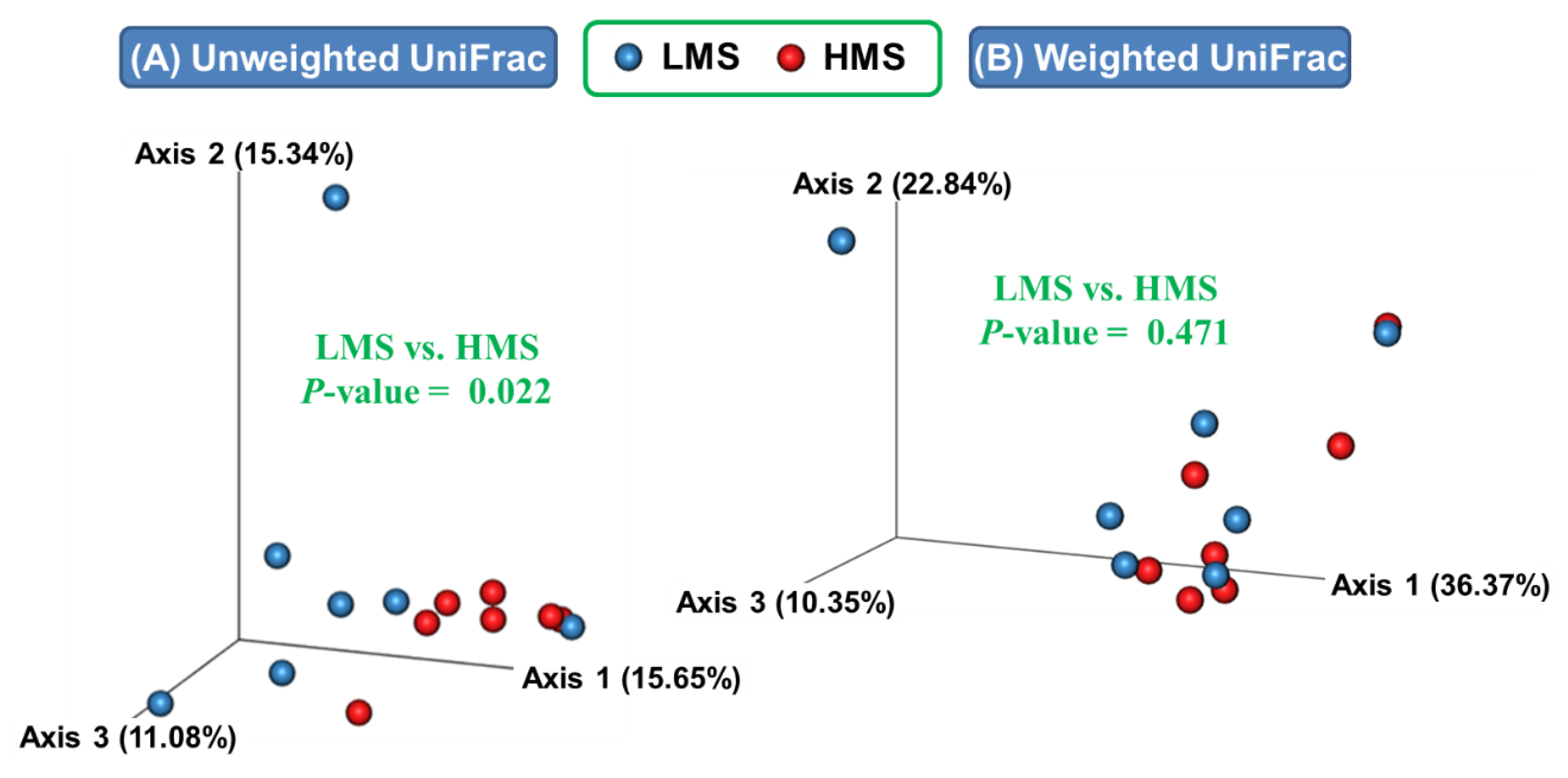
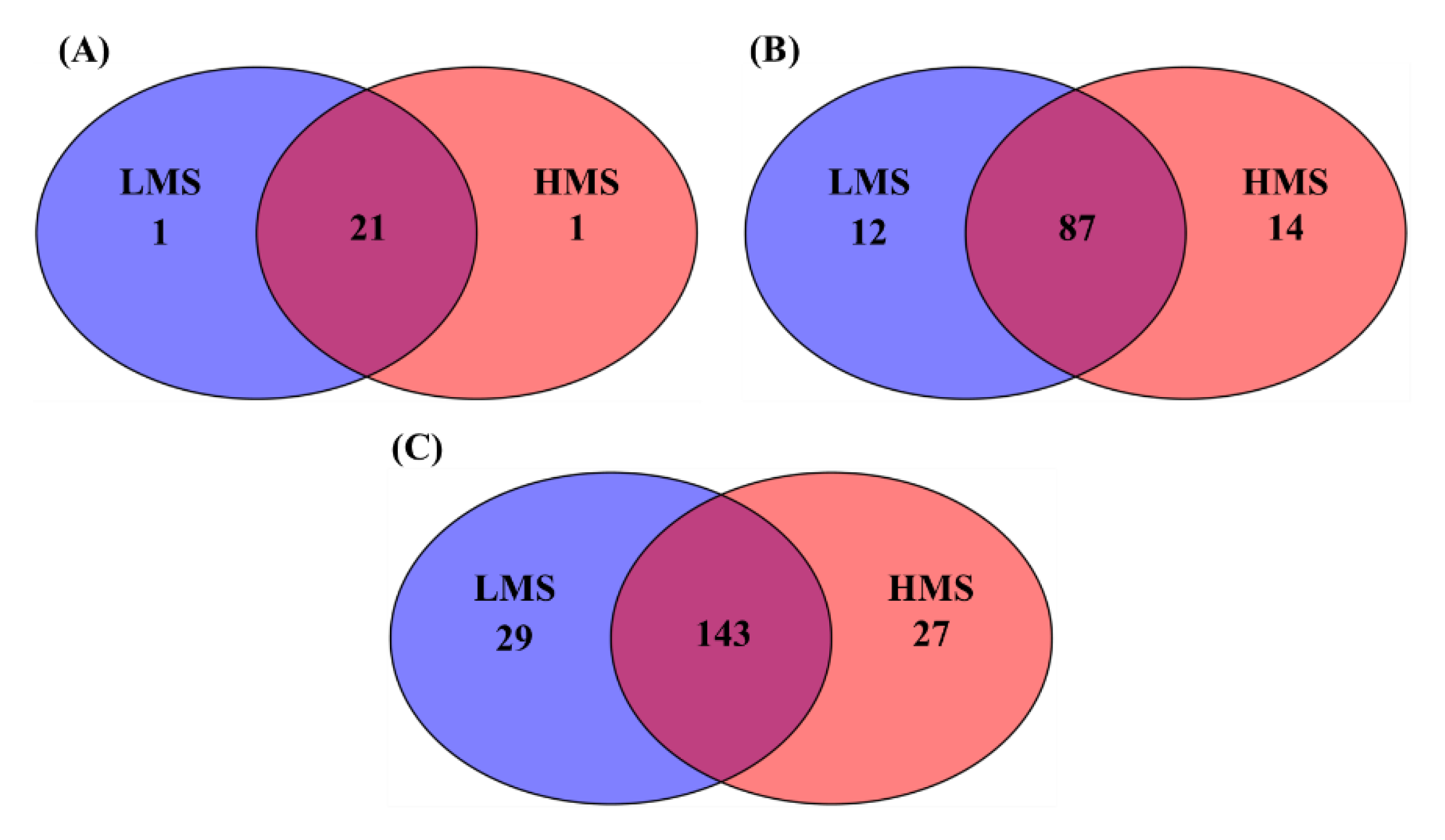
3.1. Alpha- and Beta-Diversity Analyses
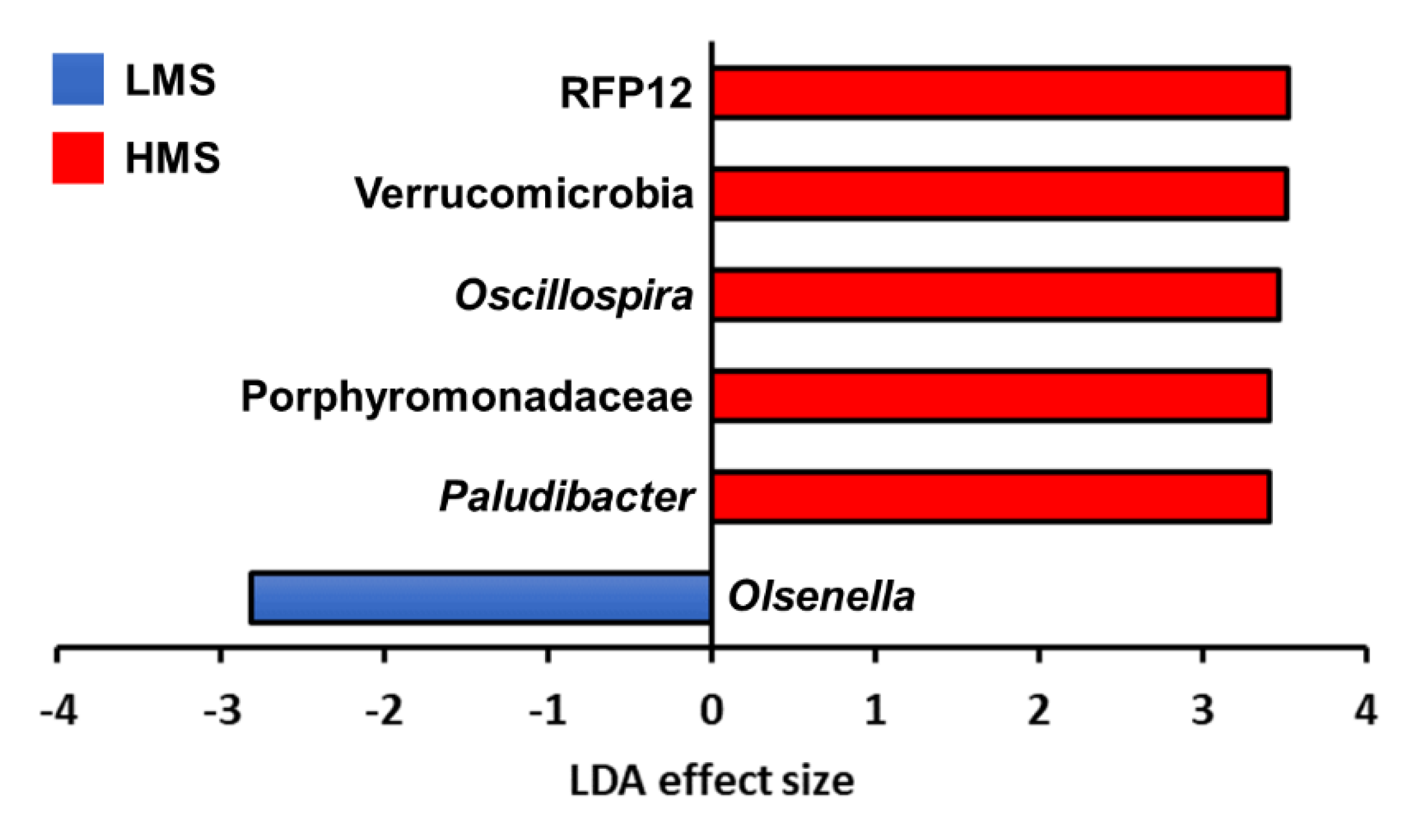
3.2. Taxonomic Analysis with Major Classified Taxa
3.3. Functional Prediction and Taxonomic Contributors to Lipid Metabolism
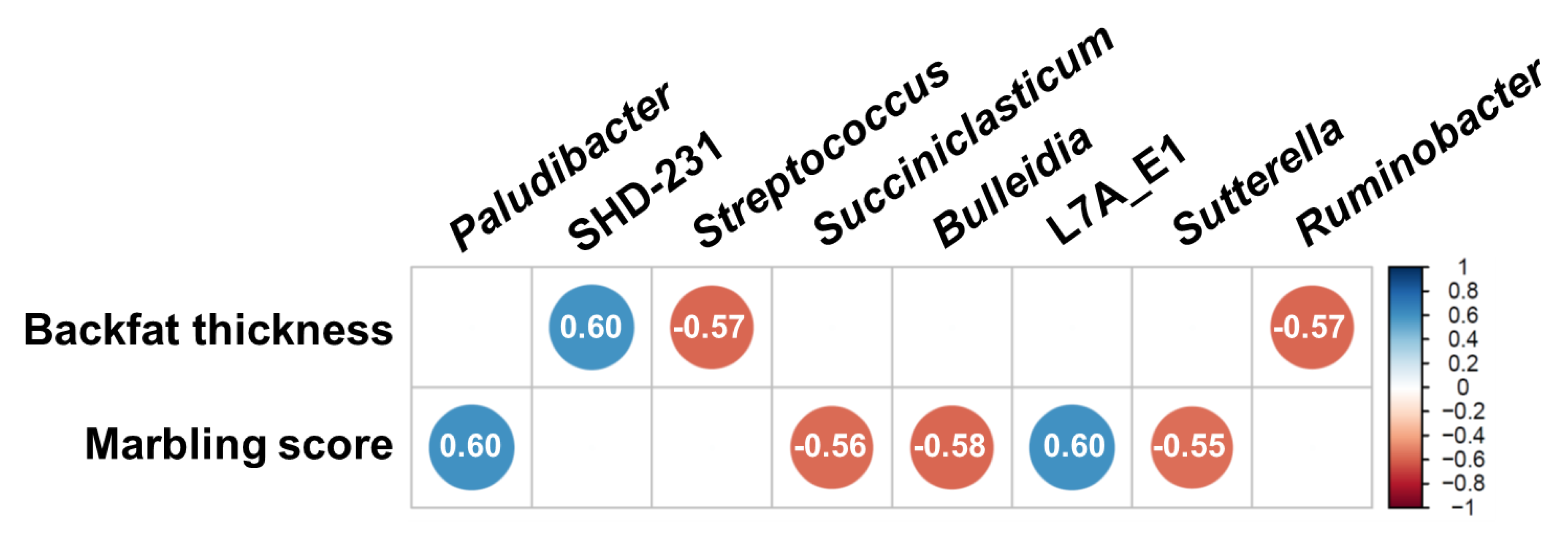
3.4. Correlation between Major Classified Genera and Meat Quality Measurements
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kim, W.H.; Kang, S.-N.; Arasu, M.V.; Chu, G.-M.; Kim, D.H.; Park, J.-H.; Oh, Y.K.; Choi, K.C. Profile of Hanwoo steer carcass characteristics, meat quality and fatty acid composition after feeding Italian ryegrass silage. Korean J. Food Sci. Anim. Resour. 2015, 35, 299. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smith, S.B. Marbling and its nutritional impact on risk factors for cardiovascular disease. Korean J. Food Sci. Anim. Resour. 2016, 36, 435. [Google Scholar] [CrossRef] [PubMed]
- Garmyn, A.; Hilton, G.; Mateescu, R.; Morgan, J.; Reecy, J.M.; Tait, R.G., Jr.; Beitz, D.C.; Duan, Q.; Schoonmaker, J.P.; Mayes, M.S. Estimation of relationships between mineral concentration and fatty acid composition of longissimus muscle and beef palatability traits. J. Anim. Sci. 2011, 89, 2849–2858. [Google Scholar] [CrossRef]
- Sasaki, S.; Yamada, T.; Sukegawa, S.; Miyake, T.; Fujita, T.; Morita, M.; Ohta, T.; Takahagi, Y.; Murakami, H.; Morimatsu, F. Association of a single nucleotide polymorphism in akirin 2 gene with marbling in Japanese Black beef cattle. BMC Res. Notes 2009, 2, 131. [Google Scholar] [CrossRef]
- Kwon, A.; Srikanth, K.; Lee, E.; Kim, S.; Chung, H. Confirmation of genotypic effects for the bovine APM1 gene on marbling in Hanwoo cattle. J. Anim. Sci. Technol. 2016, 58, 15. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-C.; Lee, S.-H.; Lee, J.-W.; Kim, T.-H.; Choi, B.-H. Identification of single nucleotide polymorphism marker and association analysis of marbling score in Fas gene of Hanwoo. Asian-Australas. J. Anim. Sci. 2016, 29, 23. [Google Scholar] [CrossRef]
- Lee, M.R.; Tweed, J.K.; Moloney, A.P.; Scollan, N.D. The effects of fish oil supplementation on rumen metabolism and the biohydrogenation of unsaturated fatty acids in beef steers given diets containing sunflower oil. Anim. Sci. 2005, 80, 361–367. [Google Scholar] [CrossRef]
- Schmid, A.; Collomb, M.; Sieber, R.; Bee, G. Conjugated linoleic acid in meat and meat products: A review. Meat Sci. 2006, 73, 29–41. [Google Scholar] [CrossRef]
- Chilliard, Y.; Glasser, F.; Ferlay, A.; Bernard, L.; Rouel, J.; Doreau, M. Diet, rumen biohydrogenation and nutritional quality of cow and goat milk fat. Eur. J. Lipid Sci. Technol. 2007, 109, 828–855. [Google Scholar] [CrossRef]
- Noci, F.; French, P.; Monahan, F.; Moloney, A. The fatty acid composition of muscle fat and subcutaneous adipose tissue of grazing heifers supplemented with plant oil-enriched concentrates. J. Anim. Sci. 2007, 85, 1062–1073. [Google Scholar] [CrossRef]
- Crumb, D.; Vattem, D. Conjugated linoleic acid (CLA)-An overview. Int. J. Appl. Res. Nat. Prod. 2011, 4, 12–18. [Google Scholar]
- Gorocica-Buenfil, M.; Fluharty, F.; Bohn, T.; Schwartz, S.; Loerch, S. Effect of low vitamin A diets with high-moisture or dry corn on marbling and adipose tissue fatty acid composition of beef steers. J. Anim. Sci. 2007, 85, 3355–3366. [Google Scholar] [CrossRef] [PubMed]
- Pickworth, C.; Loerch, S.C.; Fluharty, F. Restriction of vitamin A and D in beef cattle finishing diets on feedlot performance and adipose accretion. J. Anim. Sci. 2012, 90, 1866–1878. [Google Scholar] [CrossRef] [PubMed]
- Myers, S.; Faulkner, D.B.; Ireland, F.; Berger, L.; Parrett, D. Production systems comparing early weaning to normal weaning with or without creep feeding for beef steers. J. Anim. Sci. 1999, 77, 300–310. [Google Scholar] [CrossRef]
- Myers, S.; Faulkner, D.; Ireland, F.; Parrett, D. Comparison of three weaning ages on cow-calf performance and steer carcass traits. J. Anim. Sci. 1999, 77, 323–329. [Google Scholar] [CrossRef]
- Sithyphone, K.; Yabe, M.; Horita, H.; Hayashi, K.; Fumita, T.; Shiotsuka, Y.; Etoh, T.; Ebara, F.; Samadmanivong, O.; Wegner, J. Comparison of feeding systems: Feed cost, palatability and environmental impact among hay-fattened beef, consistent grass-only-fed beef and conventional marbled beef in Wagyu (Japanese Black cattle). Anim. Sci. J. 2011, 82, 352–359. [Google Scholar] [CrossRef]
- Moriel, P.; Johnson, S.; Vendramini, J.; McCann, M.; Gerrard, D.; Mercadante, V.; Hersom, M.; Arthington, J. Effects of calf weaning age and subsequent management systems on growth performance and carcass characteristics of beef steers. J. Anim. Sci. 2014, 92, 3598–3609. [Google Scholar] [CrossRef]
- Moriel, P.; Johnson, S.; Vendramini, J.; Mercadante, V.; Hersom, M.; Arthington, J. Effects of calf weaning age and subsequent management system on growth and reproductive performance of beef heifers. J. Anim. Sci. 2014, 92, 3096–3107. [Google Scholar] [CrossRef]
- Smith, S.B.; Crouse, J.D. Relative contributions of acetate, lactate and glucose to lipogenesis in bovine intramuscular and subcutaneous adipose tissue. J. Nutr. 1984, 114, 792–800. [Google Scholar] [CrossRef]
- Griinari, J.; Corl, B.; Lacy, S.; Chouinard, P.; Nurmela, K.; Bauman, D. Conjugated linoleic acid is synthesized endogenously in lactating dairy cows by Δ9-desaturase. J. Nutr. 2000, 130, 2285–2291. [Google Scholar] [CrossRef]
- Polan, C.; McNeill, J.; Tove, S. Biohydrogenation of unsaturated fatty acids by rumen bacteria. J. Bacteriol. 1964, 88, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.J.; McKain, N.; Shingfield, K.J.; Devillard, E. Isomers of conjugated linoleic acids are synthesized via different mechanisms in ruminal digesta and bacteria. J. Lipid Res. 2007, 48, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Klieve, A.; Hennessy, D.; Ouwerkerk, D.; Forster, R.; Mackie, R.; Attwood, G. Establishing populations of Megasphaera elsdenii YE 34 and Butyrivibrio fibrisolvens YE 44 in the rumen of cattle fed high grain diets. J. Appl. Microbiol. 2003, 95, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Devillard, E.; McIntosh, F.M.; Newbold, C.J.; Wallace, R.J. Rumen ciliate protozoa contain high concentrations of conjugated linoleic acids and vaccenic acid, yet do not hydrogenate linoleic acid or desaturate stearic acid. Br. J. Nutr. 2006, 96, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Vahmani, P.; Mapiye, C.; Prieto, N.; Rolland, D.C.; McAllister, T.A.; Aalhus, J.L.; Dugan, M.E. The scope for manipulating the polyunsaturated fatty acid content of beef: A review. J. Anim. Sci. Biotechnol. 2015, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, M.; Ramos-Morales, E.; Wallace, R. The role of microbes in rumen lipolysis and biohydrogenation and their manipulation. Animal 2010, 4, 1008–1023. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Guan, L.L. Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl. Environ. Microbiol. 2017, 83, e00061-17. [Google Scholar] [CrossRef]
- Petri, R.M.; Mapiye, C.; Dugan, M.E.; McAllister, T.A. Subcutaneous adipose fatty acid profiles and related rumen bacterial populations of steers fed red clover or grass hay diets containing flax or sunflower-seed. PLoS ONE 2014, 9, e104167. [Google Scholar] [CrossRef]
- Petri, R.M.; Vahmani, P.; Yang, H.E.; Dugan, M.E.; McAllister, T.A. Changes in rumen microbial profiles and subcutaneous fat composition when feeding extruded flaxseed mixed with or before hay. Front. Microbiol. 2018, 9, 1055. [Google Scholar] [CrossRef]
- Reddy, K.E.; Jeong, J.; Lee, S.D.; Baek, Y.-C.; Oh, Y.; Kim, M.; So, K.M.; Kim, D.W.; Kim, J.H.; Park, S. Effect of different early weaning regimens for calves on adipogenic gene expression in Hanwoo loin at the fattening stage. Livest. Sci. 2017, 195, 87–98. [Google Scholar] [CrossRef]
- Jo, C.; Cho, S.; Chang, J.; Nam, K. Keys to production and processing of Hanwoo beef: A perspective of tradition and science. Anim. Front. 2012, 2, 32–38. [Google Scholar] [CrossRef]
- Yu, Z.; Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 2004, 36, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Meth. 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 2019. [Google Scholar] [CrossRef]
- Manor, O.; Borenstein, E. Systematic characterization and analysis of the taxonomic drivers of functional shifts in the human microbiome. Cell Host Microbe 2017, 21, 254–267. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Loor, J.; Elolimy, A.; McCann, J. Dietary impacts on rumen microbiota in beef and dairy production. Anim. Front. 2016, 6, 22–29. [Google Scholar] [CrossRef]
- Shabat, S.K.B.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Miller, M.E.B.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef] [PubMed]
- Roehe, R.; Dewhurst, R.J.; Duthie, C.-A.; Rooke, J.A.; McKain, N.; Ross, D.W.; Hyslop, J.J.; Waterhouse, A.; Freeman, T.C.; Watson, M. Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PLoS Genet. 2016, 12, e1005846. [Google Scholar] [CrossRef]
- Basarab, J.; Price, M.; Aalhus, J.; Okine, E.; Snelling, W.; Lyle, K. Residual feed intake and body composition in young growing cattle. Can. J. Anim. Sci. 2003, 83, 189–204. [Google Scholar] [CrossRef]
- Nkrumah, J.; Basarab, J.; Price, M.; Okine, E.; Ammoura, A.; Guercio, S.; Hansen, C.; Li, C.; Benkel, B.; Murdoch, B. Different measures of energetic efficiency and their phenotypic relationships with growth, feed intake, and ultrasound and carcass merit in hybrid cattle. J. Anim. Sci. 2004, 82, 2451–2459. [Google Scholar] [CrossRef]
- Ahola, J.; Skow, T.; Hunt, C.; Hill, R. Relationship between residual feed intake and end product palatability in longissimus steaks from steers sired by Angus bulls divergent for intramuscular fat expected progeny difference. Prof. Anim. Sci. 2011, 27, 109–115. [Google Scholar] [CrossRef]
- Sainz, R.; Cruz, G.; Monteiro, R.; Rodriguez, J.; Monteiro, D.; Guidi, V.; Anaruma, R. Carcass composition and visceral organs are similar at harvest in low-and high-residual feed intake groups of Angus-Hereford steers. In Proceedings of the Proceedings-American Society of Animal Science Western Section, Logan, Utah, 21–23 June 2006; p. 401. [Google Scholar]
- Elolimy, A.A.; Abdelmegeid, M.K.; McCann, J.C.; Shike, D.W.; Loor, J.J. Residual feed intake in beef cattle and its association with carcass traits, ruminal solid-fraction bacteria, and epithelium gene expression. J. Anim. Sci. Biotechnol. 2018, 9, 67. [Google Scholar] [CrossRef]
- Kraatz, M.; Wallace, R.J.; Svensson, L. Olsenella umbonata sp. nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of Olsenella, Olsenella uli and Olsenella profusa. Int. J. Syst. Evol. Microbiol. 2011, 61, 795–803. [Google Scholar] [CrossRef]
- Ogata, T.; Makino, H.; Ishizuka, N.; Iwamoto, E.; Masaki, T.; Ikuta, K.; Kim, Y.-H.; Sato, S. Long-term high-grain diet altered the ruminal pH, fermentation, and composition and functions of the rumen bacterial community, leading to enhanced lactic acid production in Japanese Black beef cattle during fattening. PLoS ONE 2019, 14, e0225448. [Google Scholar] [CrossRef]
- Boone, D.; Castenholz, R.; Garrity, G.; Krieg, N. Bergey’s Manual of Systematic Bacteriology: Volume 4: The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes; Springer: New York, NY, USA, 2011; Volume 4. [Google Scholar]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.C.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef] [PubMed]
- Weimer, P.; Stevenson, D.; Mantovani, H.; Man, S. Host specificity of the ruminal bacterial community in the dairy cow following near-total exchange of ruminal contents. J. Dairy Sci. 2010, 93, 5902–5912. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.D.; Auffret, M.D.; Warr, A.; Wiser, A.H.; Press, M.O.; Langford, K.W.; Liachko, I.; Snelling, T.J.; Dewhurst, R.J.; Walker, A.W. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat. Commun. 2018, 9, 870. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Insights into the role of Erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, E.N.; Maclean, P.; Thomas, D.G.; Cave, N.J.; Young, W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ 2017, 5, e3019. [Google Scholar] [CrossRef] [PubMed]
- Pitta, D.; Indugu, N.; Vecchiarelli, B.; Rico, D.; Harvatine, K.J. Alterations in ruminal bacterial populations at induction and recovery from diet-induced milk fat depression in dairy cows. J. Dairy Sci. 2018, 101, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Maia, M.R.; Chaudhary, L.C.; Figueres, L.; Wallace, R.J. Metabolism of polyunsaturated fatty acids and their toxicity to the microflora of the rumen. Antonie van Leeuwenhoek 2007, 91, 303–314. [Google Scholar] [CrossRef]
- Chen, X.; Liu, X.; Du, Y.; Wang, B.; Zhao, N.; Geng, Z. Green forage and fattening duration differentially modulate cecal microbiome of Wanxi white geese. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Mackie, R.I.; Aminov, R.I.; Hu, W.; Klieve, A.V.; Ouwerkerk, D.; Sundset, M.A.; Kamagata, Y. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl. Environ. Microbiol. 2003, 69, 6808–6815. [Google Scholar] [CrossRef]
- Hua, C.; Tian, J.; Tian, P.; Cong, R.; Luo, Y.; Geng, Y.; Tao, S.; Ni, Y.; Zhao, R. Feeding a high concentration diet induces unhealthy alterations in the composition and metabolism of ruminal microbiota and host response in a goat model. Front. Microbiol. 2017, 8, 138. [Google Scholar] [CrossRef]
- McKain, N.; Shingfield, K.J.; Wallace, R.J. Metabolism of conjugated linoleic acids and 18: 1 fatty acids by ruminal bacteria: Products and mechanisms. Microbiology 2010, 156, 579–588. [Google Scholar] [CrossRef] [PubMed]


| Evaluations | Marbling Score Groups | SEM 3 | p-Values | |||
|---|---|---|---|---|---|---|
| LMS 1 | HMS 2 | MS | Diet * | MS × Diet | ||
| Live weight (kg) | 600.29 | 665.86 | 17.26 | 0.2066 | 0.9777 | 0.8048 |
| Carcass weight (kg) | 331.12 | 375.76 | 11.53 | 0.2702 | 0.9761 | 0.9299 |
| Carcass percent (%) | 55.16 | 56.23 | 0.52 | 0.8267 | 0.9032 | 0.3424 |
| Lean meat weight (kg) | 233.50 | 259.92 | 8.03 | 0.3703 | 0.9746 | 0.8277 |
| Fat weight (kg) | 55.99 | 68.94 | 3.13 | 0.1851 | 0.9441 | 0.8723 |
| Bone weight (kg) | 43.74 | 47.79 | 1.14 | 0.2617 | 0.9773 | 0.9273 |
| Backfat thickness (mm) | 10.29 | 9.57 | 0.64 | 0.8693 | 0.6736 | 0.9717 |
| Sirloin area (cm2) | 90.71 | 100.14 | 2.90 | 0.2255 | 0.8071 | 0.2504 |
| Meat yield index | 71.69 | 69.44 | 1.34 | 0.5246 | 0.7485 | 0.5262 |
| Meat color | 4.57 | 4.29 | 0.14 | 0.2969 | 0.1826 | 0.6559 |
| Fat color | 3.14 | 3.29 | 0.11 | 0.5188 | 0.6885 | 0.4281 |
| Marbling score ** | 3.14 B | 7.43 A | 0.62 | <0.0001 | 0.3742 | 0.0618 |
| Sirloin crude fat (%) | 28.84 | 35.07 | 1.69 | 0.2585 | 0.9403 | 0.8442 |
| Renal fat (%) | 16.22 | 14.26 | 1.01 | 0.4523 | 0.9268 | 0.8684 |
| Cholesterol (mg/dL) | 120.17 | 127.57 | 8.94 | 0.3508 | 0.0884 | 0.2079 |
| Triglyceride (mg/dL) | 22.83 | 21.57 | 2.11 | 0.9478 | 0.9257 | 0.6944 |
| High-density lipoprotein (mg/dL) | 87.00 | 106.86 | 5.70 | 0.1908 | 0.2682 | 0.3136 |
| Low-density lipoprotein (mg/dL) | 30.50 | 33.00 | 2.79 | 0.4966 | 0.2349 | 0.5109 |
| Total Lipid (mg/dL) | 293.83 | 306.43 | 20.75 | 0.3753 | 0.0826 | 0.4457 |
| Glucose (mg/dL) | 96.67 | 112.57 | 7.50 | 0.7138 | 0.4471 | 0.9402 |
| Insulin (μU/ml) | 13.27 | 6.77 | 2.22 | 0.2038 | 0.8244 | 0.3795 |
| Measurements | Marbling Score Groups | SEM 3 | p-Value | |
|---|---|---|---|---|
| LMS 1 | HMS 2 | |||
| Observed ASVs | 855.57 b | 1039.14 a | 49.74 | 0.0614 |
| Chao1 estimates | 860.06 b | 1045.96 a | 50.39 | 0.0615 |
| Evenness | 0.80 | 0.82 | 0.01 | 0.1959 |
| Faith’s Phylogenetic diversity | 64.80 | 62.14 | 3.05 | 0.6818 |
| Shannon’s index | 7.76 | 8.24 | 0.15 | 0.1070 |
| Simpson’s index | 0.984 | 0.989 | 0.002 | 0.2979 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Park, T.; Jeong, J.Y.; Baek, Y.; Lee, H.-J. Association between Rumen Microbiota and Marbling Score in Korean Native Beef Cattle. Animals 2020, 10, 712. https://doi.org/10.3390/ani10040712
Kim M, Park T, Jeong JY, Baek Y, Lee H-J. Association between Rumen Microbiota and Marbling Score in Korean Native Beef Cattle. Animals. 2020; 10(4):712. https://doi.org/10.3390/ani10040712
Chicago/Turabian StyleKim, Minseok, Tansol Park, Jin Young Jeong, Youlchang Baek, and Hyun-Jeong Lee. 2020. "Association between Rumen Microbiota and Marbling Score in Korean Native Beef Cattle" Animals 10, no. 4: 712. https://doi.org/10.3390/ani10040712
APA StyleKim, M., Park, T., Jeong, J. Y., Baek, Y., & Lee, H.-J. (2020). Association between Rumen Microbiota and Marbling Score in Korean Native Beef Cattle. Animals, 10(4), 712. https://doi.org/10.3390/ani10040712





