Comparative Transcriptome Analysis of Gill Tissue in Response to Hypoxia in Silver Sillago (Sillago sihama)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statements
2.2. Fish and Hypoxia Experiment
2.3. RNA Extraction and Illumina Library Preparation
2.4. Data Filtering, Reads Mapping and Differential Expression Analysis
2.5. Validation of DEGs by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
3. Results and Discussion
3.1. Illumina Sequencing Assembly
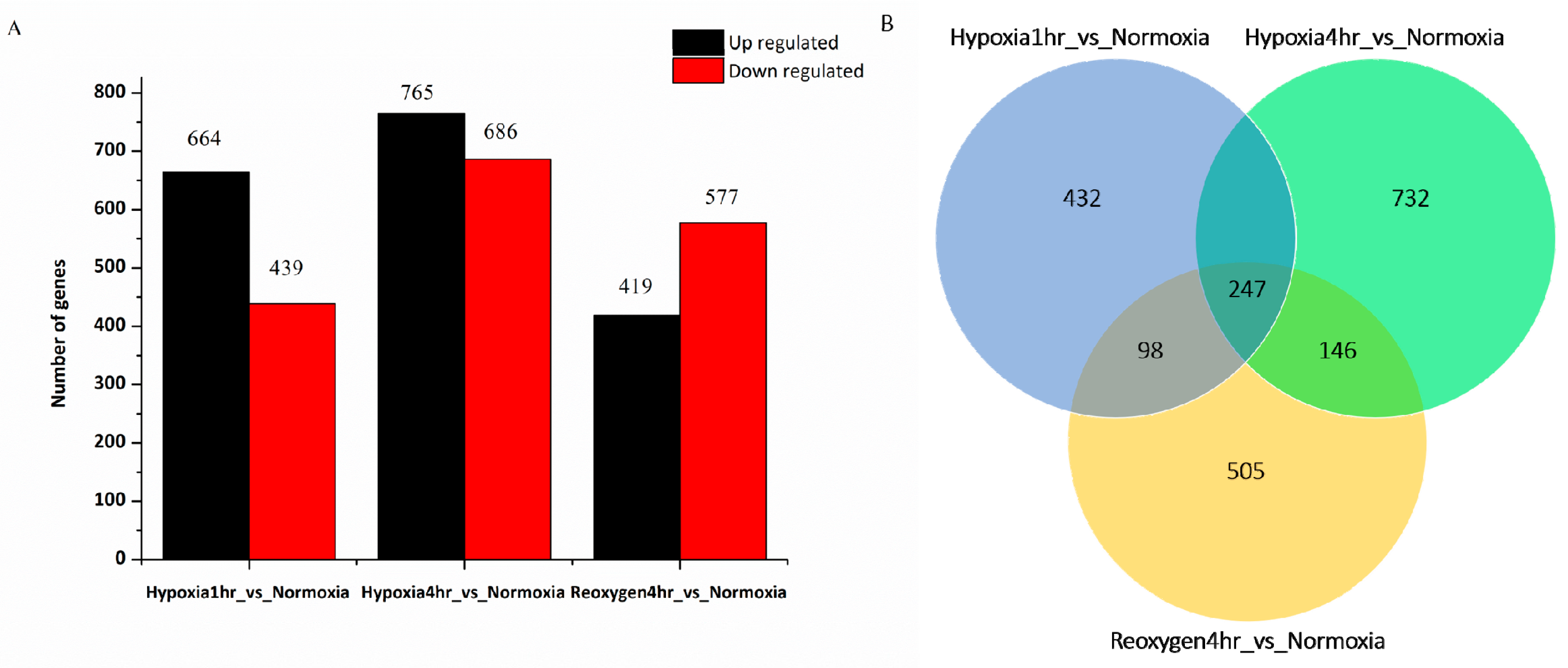
3.2. Differential Gene Expression Analysis
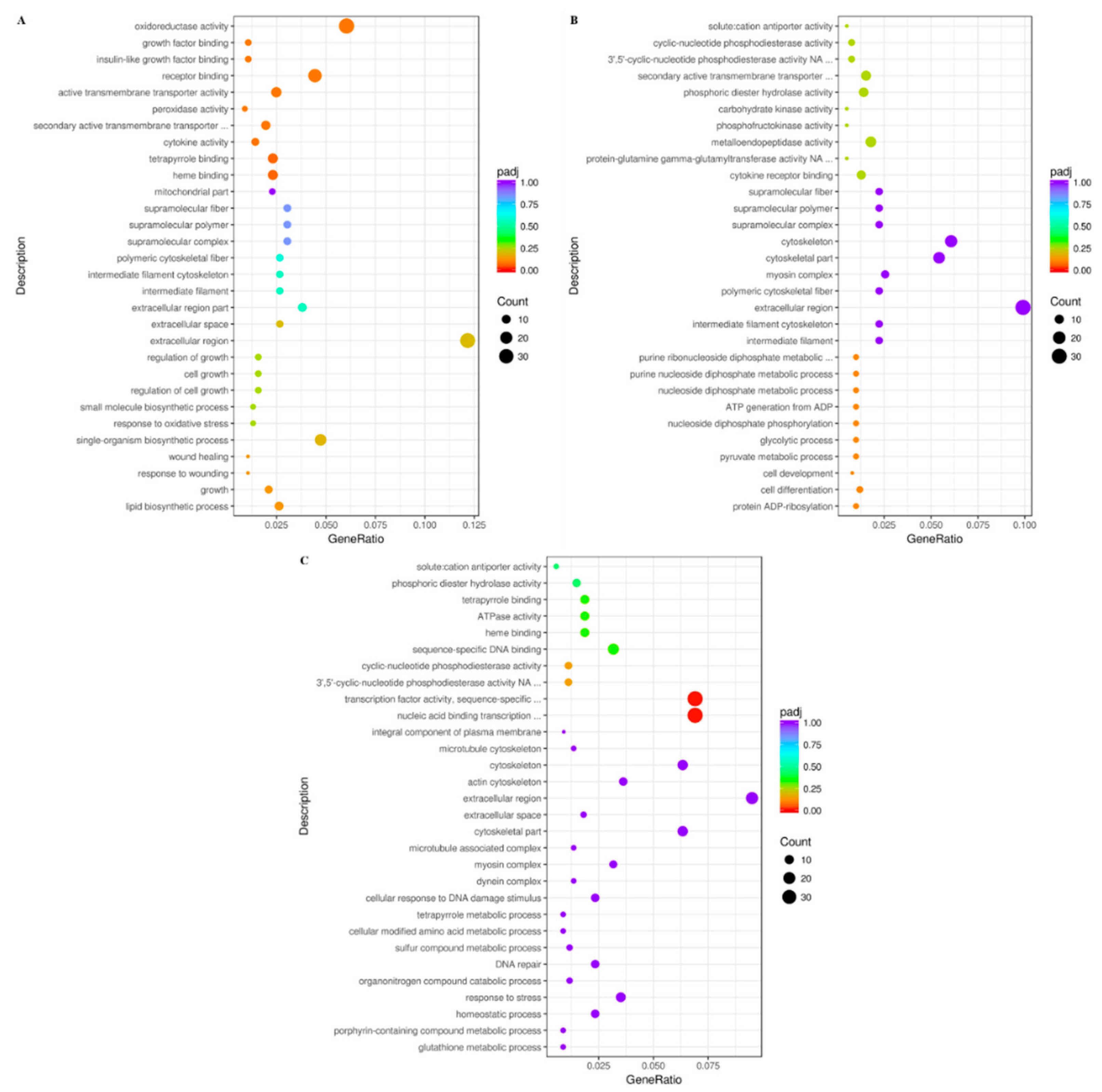
3.3. GO Term Enrichment Analysis
3.4. KEGG Pathway Enrichment Analysis
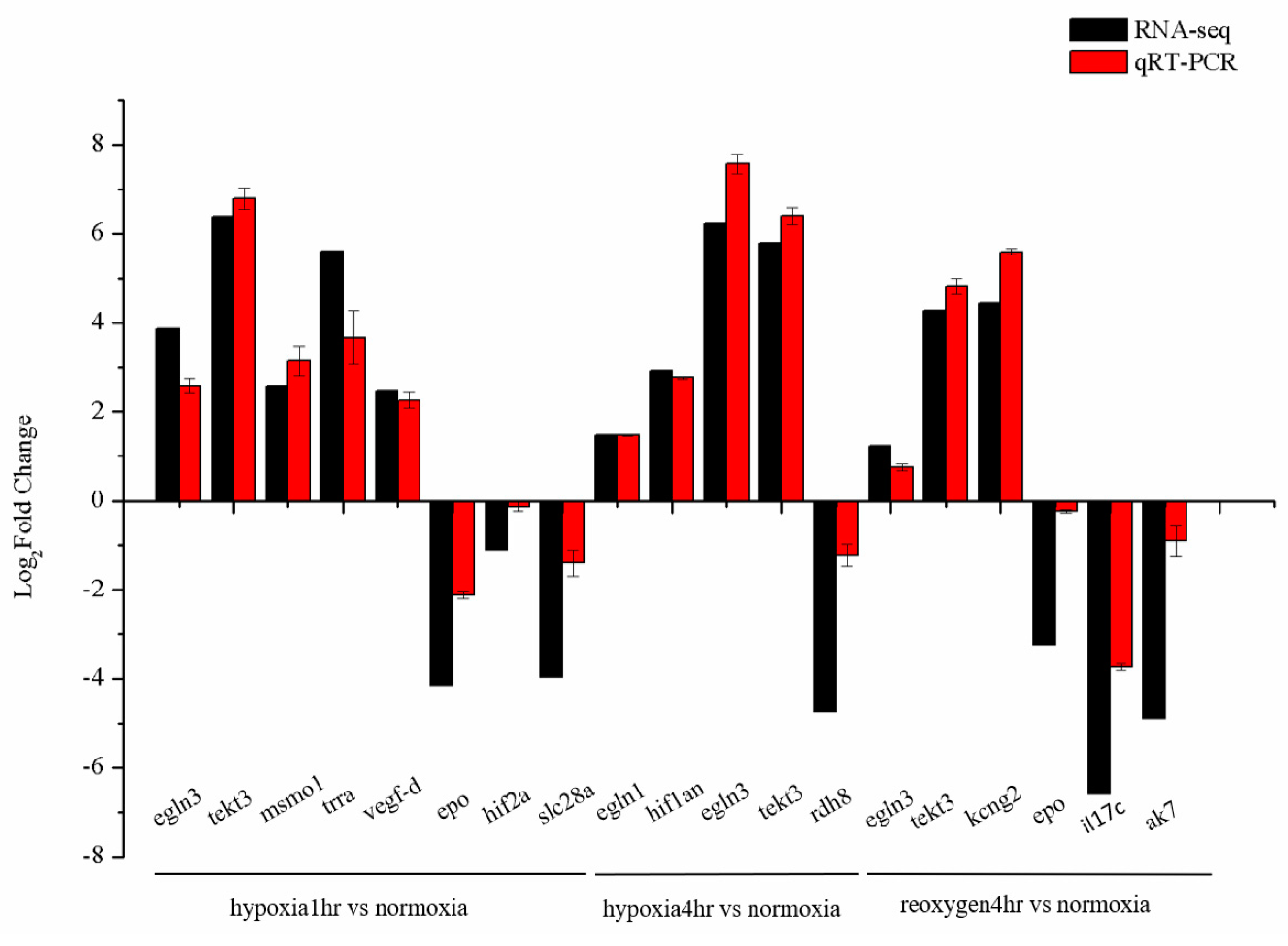
3.5. Validation of RNA-Seq Data with qRT-PCR
3.6. DEGs as Adaptive Response to Hypoxia
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rogers, N.J.; Urbina, M.A.; Reardon, E.E.; McKenzie, D.J.; Wilson, R.W. A new analysis of hypoxia tolerance in fishes using a database of critical oxygen level (Pcrit). Conserv. Physiol. 2016, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.G. Physiological, behavioral and biochemical adaptations of intertidal fishes to hypoxia. J. Exp. Biol. 2011, 214, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Turko, A.J.; Cooper, C.A.; Wright, P.A. Gill remodelling during terrestrial acclimation reduces aquatic respiratory function of the amphibious fish Kryptolebias marmoratus. J. Exp. Biol. 2012, 215, 3973–3980. [Google Scholar] [CrossRef] [PubMed]
- Mitrovic, D.; Dymowska, A.; Nilsson, G.E.; Perry, S.F. Physiological consequences of gill remodeling in goldfish (Carassius auratus) during exposure to long-term hypoxia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R224–R234. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Lehmann, D.W.; Harms, C.A.; Law, J.M. Acute hypoxia-reperfusion triggers immunocompromise in Nile Tilapia. J. Aquat. Anim. Health 2007, 19, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Onukwufor, J.O.; Wood, C.M. The osmorespiratory compromise in rainbow trout (Oncorhynchus mykiss): The effects of fish size, hypoxia, temperature and strenuous exercise on gill diffusive water fluxes and sodium net loss rates. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2018, 219–220, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.H.; Piermarini, P.M.; Choe, K.P. The multifunctional fish gill: Dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol. Rev. 2005, 85, 97–177. [Google Scholar] [CrossRef]
- Wu, C. Bin; Liu, Z.Y.; Li, F.G.; Chen, J.; Jiang, X.Y.; Zou, S.M. Gill remodeling in response to hypoxia and temperature occurs in the hypoxia sensitive blunt snout bream (Megalobrama amblycephala). Aquaculture 2017, 479, 479–486. [Google Scholar] [CrossRef]
- Sollid, J.; Nilsson, G.E. Plasticity of respiratory structures—Adaptive remodeling of fish gills induced by ambient oxygen and temperature. Respir. Physiol. Neurobiol. 2006, 154, 241–251. [Google Scholar] [CrossRef]
- KAGA, T. Phylogenetic systematics of the family Sillaginidae (Percomorpha: Order Perciformes). Zootaxa 2013, 3642, 001–105. [Google Scholar] [CrossRef]
- Tian, C.; Li, Z.; Dong, Z.; Huang, Y.; Du, T.; Chen, H.; Jiang, D.; Deng, S.; Zhang, Y.; Wanida, S.; et al. Transcriptome Analysis of Male and Female Mature Gonads of Silver Sillago (Sillago sihama). Genes 2019, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- FAO. The State of World Fisheries and Aquaculture 2018—Meeting the sustainable development goals; FAO: Rome, Italy, 2018; ISBN 9789251305621. [Google Scholar]
- Yousif, O.; Menon, K.K.; A-Fatah, A.-R. Spawning, Larval Rearing and Growth of the Silver Sillago in Abu Dhabi. World Aquac. 2015, 46, 53–55. [Google Scholar]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2017, 8, e1364. [Google Scholar] [CrossRef] [PubMed]
- Qi, D.; Chen, Q.; Zheng, Z.; Wu, R.; Xia, M.; Chao, Y. Transcriptome Analysis Provides Insights Into the Adaptive Responses to Hypoxia of a Schizothoracine Fish (Gymnocypris eckloni). Front. Physiol. 2018, 9, 1326. [Google Scholar] [CrossRef]
- Hoff, M.L.M.; Fabrizius, A.; Czech-Damal, N.U.; Folkow, L.P.; Burmester, T. Transcriptome analysis identifies key metabolic changes in the hooded seal (Cystophora cristata) brain in response to hypoxia and reoxygenation. PLoS ONE 2017, 12, 1–21. [Google Scholar] [CrossRef]
- Chen, B.X.; Yi, S.K.; Wang, W.F.; He, Y.; Huang, Y.; Gao, Z.X.; Liu, H.; Wang, W.M.; Wang, H.L. Transcriptome comparison reveals insights into muscle response to hypoxia in blunt snout bream (Megalobrama amblycephala). Gene 2017, 624, 6–13. [Google Scholar] [CrossRef]
- Geng, X.; Feng, J.; Liu, S.; Wang, Y.; Arias, C.; Liu, Z. Transcriptional regulation of hypoxia inducible factors alpha (HIF-α) and their inhibiting factor (FIH-1) of channel catfish (ictalurus punctatus) under hypoxia. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014, 169, 38–50. [Google Scholar] [CrossRef]
- Liao, X.; Cheng, L.; Xu, P.; Lu, G.; Wachholtz, M.; Sun, X.; Chen, S. Transcriptome Analysis of Crucian Carp (Carassius auratus), an Important Aquaculture and Hypoxia-Tolerant Species. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef]
- Zhou, T.; Gui, L.; Liu, M.; Li, W.; Hu, P.; Duarte, D.F.C.; Niu, H.; Chen, L. Transcriptomic responses to low temperature stress in the Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 2019, 84, 1145–1156. [Google Scholar] [CrossRef]
- Long, Y.; Yan, J.; Song, G.; Li, X.; Li, X.; Li, Q.; Cui, Z. Transcriptional events co-regulated by hypoxia and cold stresses in Zebrafish larvae. BMC Genomics 2015, 16, 385. [Google Scholar] [CrossRef]
- Olsvik, P.A.; Vikeså, V.; Lie, K.K.; Hevrøy, E.M. Transcriptional responses to temperature and low oxygen stress in Atlantic salmon studied with next-generation sequencing technology. BMC Genomics 2013, 14, 817. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- ZHANG, N.; DU, W.; WANG, Z.; HUANG, Y.; DU, T.; DONG, Z. Screening of Reference Genes for Real-time PCR in Different Tissues from Sillago sihama. J. Guangdong Ocean Univ. 2018, 38. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Khansari, A.R.; Balasch, J.C.; Vallejos-Vidal, E.; Parra, D.; Reyes-López, F.E.; Tort, L. Comparative Immune- and Stress-Related Transcript Response Induced by Air Exposure and Vibrio anguillarum Bacterin in Rainbow Trout (Oncorhynchus mykiss) and Gilthead Seabream (Sparus aurata) Mucosal Surfaces. Front. Immunol. 2018, 9, 856. [Google Scholar] [CrossRef]
- Liu, Y.; Xin, Z.Z.; Zhang, D.Z.; Wang, Z.F.; Zhu, X.Y.; Tang, B.P.; Jiang, S.H.; Zhang, H. Bin; Zhou, C.L.; Chai, X.Y.; et al. Transcriptome analysis of yellow catfish (Pelteobagrus fulvidraco) liver challenged with polyriboinosinic polyribocytidylic acid (poly I:C). Fish Shellfish Immunol. 2017, 68, 395–403. [Google Scholar] [CrossRef]
- Jin, J.; Wang, Y.; Wu, Z.; Hergazy, A.; Lan, J.; Zhao, L.; Liu, X.; Chen, N.; Lin, L. Transcriptomic analysis of liver from grass carp (Ctenopharyngodon idellus) exposed to high environmental ammonia reveals the activation of antioxidant and apoptosis pathways. Fish Shellfish Immunol. 2017, 63, 444–451. [Google Scholar] [CrossRef]
- Xiao, W. The hypoxia signaling pathway and hypoxic adaptation in fishes. Sci. China Life Sci. 2015, 58, 148–155. [Google Scholar] [CrossRef]
- Chen, N.; Huang, C.H.; Chen, B.X.; Liu, H.; Wang, W.M.; Gul, Y.; Wang, H.L. Alternative splicing transcription of Megalobrama amblycephala HIF prolyl hydroxylase PHD3 and up-regulation of PHD3 by HIF-1α. Biochem. Biophys. Res. Commun. 2016, 469, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Sahlmann, C.; Sutherland, B.J.G.; Kortner, T.M.; Koop, B.F.; Krogdahl, Å.; Bakke, A.M. Early response of gene expression in the distal intestine of Atlantic salmon (Salmo salar L.) during the development of soybean meal induced enteritis. Fish Shellfish Immunol. 2013, 34, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Zhao, J.; Li, T.; Tafalla, C.; Zhang, Q.; Wang, X.; Gong, X.; Shen, Z.; Li, A. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. salmonicida. J. Proteomics 2015, 122, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Krücken, J.; Schroetel, R.M.U.; Müller, I.U.; Saïdani, N.; Marinovski, P.; Benten, W.P.M.; Stamm, O.; Wunderlich, F. Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene 2004, 341, 291–304. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, W.; Gao, C.; Ji, B.; Chi, X.; Zheng, W.; Wang, H.L. Dysregulation of GTPase IMAP family members in hepatocellular cancer. Mol. Med. Rep. 2016, 14, 4119–4123. [Google Scholar] [CrossRef]
- Råbergh, C.M.I.; Vrolijk, N.H.; Lipsky, M.M.; Chen, T.T. Differential expression of two CYP1A genes in rainbow trout (Oncorhynchys mykiss). Toxicol. Appl. Pharmacol. 2000, 165, 195–205. [Google Scholar] [CrossRef]
- Leiva, K.; Werner, N.; Sepúlveda, D.; Barahona, S.; Baeza, M.; Cifuentes, V.; Alcaíno, J. Identification and functional characterization of the CYP51 gene from the yeast Xanthophyllomyces dendrorhous that is involved in ergosterol biosynthesis. BMC Microbiol. 2015, 15, 89. [Google Scholar] [CrossRef]
- Liu, J.; Tian, Y.; Ding, Y.; Heng, D.; Xu, K.; Liu, W.; Zhang, C. Role of CYP51 in the regulation of T3 and FSH-induced steroidogenesis in female mice. Endocrinology 2017, 158, 3974–3987. [Google Scholar] [CrossRef]
- Morrison, A.M.S.; Goldstone, J.V.; Lamb, D.C.; Kubota, A.; Lemaire, B.; Stegeman, J.J. Identification, modeling and ligand affinity of early deuterostome CYP51s, and functional characterization of recombinant zebrafish sterol 14α-demethylase. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 1825–1836. [Google Scholar] [CrossRef]
- Weber, G.J.; Choe, S.E.; Dooley, K.A.; Paffett-Lugassy, N.N.; Zhou, Y.; Zon, L.I. Mutant-specific gene programs in the zebrafish. Blood 2005, 106, 521–530. [Google Scholar] [CrossRef]
- Qian, L.; Qi, S.; Cao, F.; Zhang, J.; Li, C.; Song, M.; Wang, C. Effects of penthiopyrad on the development and behaviour of zebrafish in early-life stages. Chemosphere 2019, 214, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Estrada, D.F. Specificity of the Redox Complex between Cytochrome P450 24A1 and adrenodoxin relies on carbon-25 hydroxylation of vitamin-D Substrate. Drug Metab. Dispos. 2019, 47, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Chun, R.F.; Blatter, E.; Elliott, S.; Fitz-gibbon, S.; Rieger, S.; Sagasti, A.; Adams, J.S.; Hewison, M. Cloning of a functional 25-hydroxy vitamin D-1 α -hydroxylase in zebra fish (Danio rerio). Cell Biochem. Funct. 2014, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, Y.; Lu, J.W.; Huo, X.; Gong, Z. Liver-specific androgen receptor knockout attenuates early liver tumor development in zebrafish. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Thomas, P. Effects of hypoxia exposure on hepatic cytochrome P450 1A (CYP1A) expression in atlantic croaker: Molecular mechanisms of CYP1A down-regulation. PLoS ONE 2012, 7, e40825. [Google Scholar] [CrossRef] [PubMed]
- Alak, G.; Yeltekin, A.Ç.; Tas, I.H.; Ucar, A.; Parlak, V.; Topal, A.; Kocaman, E.M.; Atamanalp, M. Investigation of 8-OHdG, CYP1A, HSP70 and transcriptional analyses of antioxidant defence system in liver tissues of rainbow trout exposed to eprinomectin. Fish Shellfish Immunol. 2017, 65, 136–144. [Google Scholar] [CrossRef]
- Leveelahti, L.; Leskinen, P.; Leder, E.H.; Waser, W.; Nikinmaa, M. Responses of threespine stickleback (Gasterosteus aculeatus, L) transcriptome to hypoxia. Comp. Biochem. Physiol. Part D Genomics Proteomics 2011, 6, 370–381. [Google Scholar] [CrossRef]
- Zhang, X. Bin; Zeng, Y.M.; Chen, X.Y.; Zhang, Y.X.; Ding, J.Z.; Xue, C. Decreased expression of hepatic cytochrome P450 1A2 (CYP1A2) in a chronic intermittent hypoxia mouse model. J. Thorac. Dis. 2018, 10, 825–834. [Google Scholar] [CrossRef]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-modified cancer cell metabolism. Front. Cell Dev. Biol. 2019, 7, 1–15. [Google Scholar] [CrossRef]
- Strange, R.C.; Spiteri, M.A.; Ramachandran, S.; Fryer, A.A. Glutathione-S-transferase family of enzymes. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2001, 482, 21–26. [Google Scholar] [CrossRef]
- Sun, S.; Xuan, F.; Fu, H.; Ge, X.; Zhu, J.; Qiao, H.; Jin, S.; Zhang, Y. Comparative proteomic study of the response to hypoxia in the muscle of oriental river prawn (Macrobrachium nipponense). J. Proteomics 2016, 138, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, G.; Chen, J.; Mao, J.; Liang, F.; Yin, S.; Tang, Z.; Zhao, C.; Chen, S. Modulated expression and enzymatic activities of darkbarbel catfish, pelteobagrus vachelli for oxidative stress induced by acute hypoxia and reoxygenation. Chemosphere 2016, 151, 271–279. [Google Scholar]
- Wu, F.; Zheng, Y.; Gao, J.; Chen, S.; Wang, Z. Induction of oxidative stress and the transcription of genes related to apoptosis in rare minnow (Gobiocypris rarus) larvae with aroclor 1254 exposure. Ecotoxicol. Environ. Saf. 2014, 110, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Olsvik, P.A.; Nordtug, T.; Altin, D.; Lie, K.K.; Overrein, I.; Hansen, B.H. Transcriptional effects on glutathione S-transferases in first feeding Atlantic cod (Gadus morhua) larvae exposed to crude oil. Chemosphere 2010, 79, 905–913. [Google Scholar] [CrossRef]
- Yang, M.; Chen, P.; Liu, J.; Zhu, S.; Kroemer, G.; Klionsky, D.J.; Lotze, M.T.; Zeh, H.J.; Kang, R.; Tang, D. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci. Adv. 2019, 5, eaaw2238. [Google Scholar] [CrossRef]
- Luo, W.; Hu, H.; Chang, R.; Zhong, J.; Knabel, M.; O’Meally, R.; Cole, R.N.; Pandey, A.; Semenza, G.L. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 2011, 145, 732–744. [Google Scholar] [CrossRef]
- Zhong, W.; Guo, J.; Cui, L.; Chionh, Y.H.; Li, K.; El Sahili, A.; Cai, Q.; Yuan, M.; Michels, P.A.M.; Fothergill-Gilmore, L.A.; et al. Pyruvate kinase regulates the pentose-phosphate pathway in response to hypoxia in Mycobacterium tuberculosis. J. Mol. Biol. 2019, 431, 3690–3705. [Google Scholar] [CrossRef]
- Ifeanyi, O.E. Hypoxia and enzyme metabolism: A Review. 2014, 5, 40–43. [Google Scholar]
- Xu, Y.; Li, F.; Lv, L.; Li, T.; Zhou, X.; Deng, C.-X.; Guan, K.-L.; Lei, Q.-Y.; Xiong, Y. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. 2014, 74, 3630–3642. [Google Scholar] [CrossRef]
- Nitzan, T.; Kokou, F.; Doron-Faigenboim, A.; Slosman, T.; Biran, J.; Mizrahi, I.; Zak, T.; Benet, A.; Cnaani, A. Transcriptome analysis reveals common and differential response to low temperature exposure between tolerant and sensitive blue tilapia (Oreochromis aureus). Front. Genet. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Martínez, M.L.; Landry, C.; Boehm, R.; Manning, S.; Cheek, A.O.; Rees, B.B. Effects of long-term hypoxia on enzymes of carbohydrate metabolism in the gulf killifish, Fundulus grandis. J. Exp. Biol. 2006, 209, 3851–3861. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, V.I.; Bahnjukova, T.V.; Storey, K.B. Effect of hypoxia on the activity and binding of glycolytic and associated enzymes in sea scorpion tissues. Brazilian J. Med. Biol. Res. 1998, 31, 1059–1067. [Google Scholar] [CrossRef]
- Cota-Ruiz, K.; Leyva-Carrillo, L.; Peregrino-Uriarte, A.B.; Valenzuela-Soto, E.M.; Gollas-Galván, T.; Gómez-Jiménez, S.; Hernández, J.; Yepiz-Plascencia, G. Role of HIF-1 on phosphofructokinase and fructose 1, 6-bisphosphatase expression during hypoxia in the white shrimp Litopenaeus vannamei. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2016, 198, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Aliparasti, M.R.; Alipour, M.R.; Almasi, S.; Feizi, H. Effect of ghrelin on aldolase gene expression in the heart of chronic hypoxic rat. Int. J. Endocrinol. Metab. 2012, 10, 553–557. [Google Scholar] [CrossRef] [PubMed]
| Group | Raw Reads | Clean Reads | Clean Bases (G) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|
| Hypoxia1hr. | ||||||
| G_HI_1 | 59,525,298 | 57,377,574 | 8.61 | 97.00 | 92.30 | 47.58 |
| G_HI_2 | 45,730,892 | 43,973,742 | 6.60 | 96.24 | 90.38 | 46.97 |
| G_HI_3 | 48,080,268 | 44,777,822 | 6.72 | 96.87 | 92.04 | 46.98 |
| Hypoxia4hr. | ||||||
| G_HT_1 | 48,635,558 | 46,533,572 | 6.98 | 96.32 | 90.58 | 47.03 |
| G_HT_2 | 49,539,404 | 47,922,836 | 7.19 | 96.94 | 92.18 | 47.52 |
| G_HT_3 | 53,859,006 | 51,597,942 | 7.74 | 96.91 | 92.17 | 46.87 |
| Reoxygen4hr. | ||||||
| G_RO_1 | 47,007,630 | 44,659,448 | 6.70 | 96.59 | 91.37 | 46.63 |
| G_RO_2 | 46,031,644 | 43,741,284 | 6.56 | 97.16 | 92.57 | 46.78 |
| G_RO_3 | 52,813,132 | 49,769,382 | 7.47 | 96.58 | 91.37 | 47.32 |
| Normoxia | ||||||
| G_NO_1 | 42,655,494 | 40,115,434 | 6.02 | 97.23 | 92.8 | 47.31 |
| G_NO_2 | 43,073,570 | 41,127,670 | 6.17 | 96.8 | 91.79 | 48.49 |
| G_NO_3 | 44,331,564 | 41,995,130 | 6.30 | 96.75 | 91.75 | 47.69 |
| Gene Name | Log2 (Fold Change) | Description | ||
|---|---|---|---|---|
| Hypoxia1h | Hypoxia4h | Reoxygen4h | ||
| gimap4 | 6.39 | 6.45 | 6.89 | GTPase IMAP family member 4 |
| tekt3 | 6.39 | 5.79 | 4.27 | Tektin-3 |
| hisat | 5.10 | 4.38 | 3.12 | Histidine N-acetyltransferase |
| kcnq2 | 4.23 | 4.27 | 4.44 | Potassium voltage-gated channel subfamily KQT r 2 |
| egln3 | 3.86 | 6.21 | −1.16 | Hypoxia-inducible factor prolyl hydroxylase |
| aqp9 | 3.19 | 2.83 | 3.57 | Aquaporin-9 |
| slc12a3 | 3.12 | 1.67 | −1.68 | Solute carrier family 12 member 3 |
| igfbp1 | 1.90 | 1.80 | −1.17 | Insulin-like growth factor-binding protein 1 |
| pde4 | 1.36 | 1.89 | −1.26 | cAMP-specific phosphodiesterase 4 |
| pck1 | 1.32 | 1.05 | −1.06 | Phosphoenolpyruvate carboxykinase (GTP) |
| bmp10 | 1.57 | 2.19 | −1.88 | bone morphogenetic protein 10 |
| trim16 | 1.17 | 2.05 | −1.31 | Tripartite motif-containing protein 16 |
| kcnk1 | 1.07 | 1.52 | −1.32 | Potassium channel subfamily K member 1 |
| cxcr4 | 1.19 | 1.07 | −1.69 | C-X-C chemokine receptor type 4 |
| p4hb | −6.47 | −3.99 | −6.60 | Protein disulfide-isomerase |
| tx_B | −4.21 | −4.59 | −2.75 | Tx beta-subunit |
| glipr2 | −4.15 | −1.58 | −3.73 | Golgi-associated plant pathogenesis-related protein 1 |
| gimap7 | −4.08 | −4.95 | −3.16 | GTPase IMAP family member 7 |
| endod1 | −3.57 | −3.61 | −4.53 | Endonuclease domain-containing 1 protein |
| Pathway ID | Pathway Term | Gene Name |
|---|---|---|
| Hypoxia1hr_vs_Normoxia | ||
| dre00100 | Steroid biosynthesis | meso1, cyp51, sqle, nsdhl, tm7sf2, fdft1, dhcr24, dhcr7, lss, ebp |
| dre01230 | Biosynthesis of amino acids | pk, aldo, phgdh, pfk, aco, glnA, pgam, cps1, ass1, tktA, tktB, idh1 idh2 |
| Hypoxia4hr_vs_Normoxia | ||
| dre00100 | Steroid biosynthesis | meso1, tm7sf2, sqle, dhcr24, lss, nsdhl, cyp51, fdft1, cyp24a1 |
| Reoxygen4hr_vs_Normoxia | ||
| dre00480 | Glutathione metabolism | gclc, gsto1, gsr, LOC108897969, gss, gclm, gst, g6pd, LOC104939687, LOC104921330 |
| dre00980 | Metabolism of xenobiotics by cytochrome P450 | gsto1, LOC108897969, cbr1, gst, cyp1a1, LOC104939687, ugt1a1 |
| dre04216 | Ferroptosis | gclc, fth1, gss, gclm, tf, hmox1, tfrc |
| dre00982 | Drug metabolism - cytochrome P450 | gsto1, LOC108897969, gst, LOC104939687, ugt1a1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saetan, W.; Tian, C.; Yu, J.; Lin, X.; He, F.; Huang, Y.; Shi, H.; Zhang, Y.; Li, G. Comparative Transcriptome Analysis of Gill Tissue in Response to Hypoxia in Silver Sillago (Sillago sihama). Animals 2020, 10, 628. https://doi.org/10.3390/ani10040628
Saetan W, Tian C, Yu J, Lin X, He F, Huang Y, Shi H, Zhang Y, Li G. Comparative Transcriptome Analysis of Gill Tissue in Response to Hypoxia in Silver Sillago (Sillago sihama). Animals. 2020; 10(4):628. https://doi.org/10.3390/ani10040628
Chicago/Turabian StyleSaetan, Wanida, Changxu Tian, Jiawang Yu, Xinghua Lin, Feixiang He, Yang Huang, Hongjuan Shi, Yulei Zhang, and Guangli Li. 2020. "Comparative Transcriptome Analysis of Gill Tissue in Response to Hypoxia in Silver Sillago (Sillago sihama)" Animals 10, no. 4: 628. https://doi.org/10.3390/ani10040628
APA StyleSaetan, W., Tian, C., Yu, J., Lin, X., He, F., Huang, Y., Shi, H., Zhang, Y., & Li, G. (2020). Comparative Transcriptome Analysis of Gill Tissue in Response to Hypoxia in Silver Sillago (Sillago sihama). Animals, 10(4), 628. https://doi.org/10.3390/ani10040628







