Mitogenome Diversity and Maternal Origins of Guangxi Buffalo Breeds
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sequencing
2.2. Reconstruction of Mitochondrial Genomes
2.3. Data Analysis
3. Results
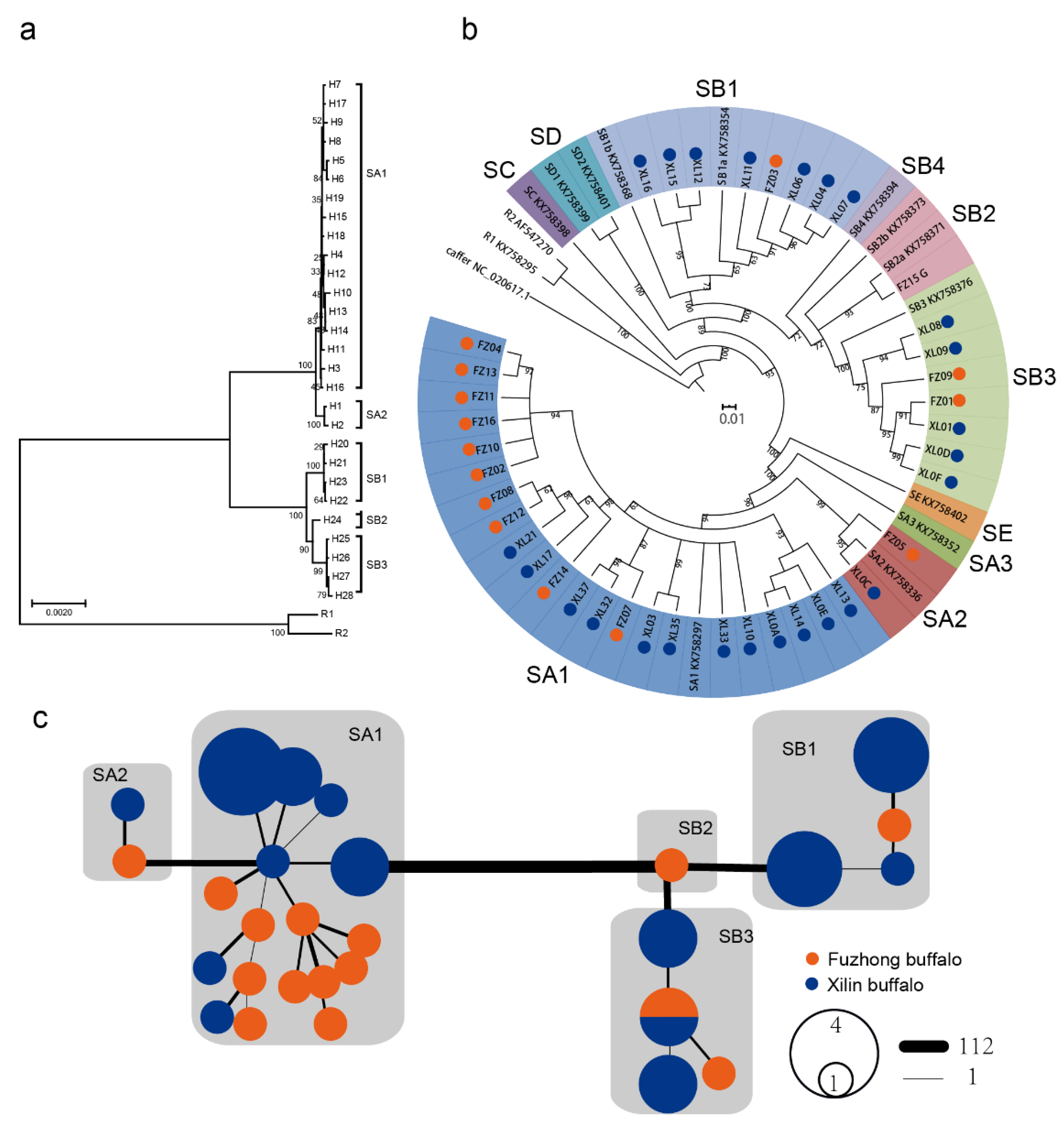
3.1. MtDNA Sequence Variation and Genetic Diversity
3.2. Population Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Michelizzi, V.N.; Dodson, M.V.; Pan, Z.; Amaral, M.E.J.; Michal, J.J.; Mclean, D.J.; Womack, J.E.; Jiang, Z. Water Buffalo Genome Science Comes of Age. Int. J. Biol. Sci. 2010, 6, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.; Ulbrich, F. Chromosomes of the Murrah buffalo and its crossbreds with the Asiatic swamp buffalo (Bubalus bubalis). Zeitschrift für Tierzüchtung und Züchtungsbiologie 1967, 84, 110–114. [Google Scholar] [CrossRef]
- Arnold, H.; Pette, D. Binding of Glycolytic Enzymes to Structure Proteins of the Muscle. Eur. J. Biochem. 1968, 6, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, Y.; Yindee, M.; Li, K.-Y.; Kuo, H.-Y.; Ju, Y.-T.; Ye, S.; Faruque, M.O.; Li, Q.; Wang, Y.; et al. Strong and stable geographic differentiation of swamp buffalo maternal and paternal lineages indicates domestication in the China/Indochina border region. Mol. Ecol. 2016, 25, 1530–1550. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Shen, J.; Achilli, A.; Chen, N.; Chen, Q.; Dang, R.; Zheng, Z.; Zhang, H.; Zhang, X.; Wang, S.; et al. Genomic analyses reveal distinct genetic architectures and selective pressures in buffaloes. GigaScience 2020, 9, giz166. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, S.; Chanthakhoun, V.; Dang, R.; Huang, Y.; Chen, H.; Lei, C. Multiple domestication of swamp buffalo in China and South East Asia. J. Anim. Breed. Genet. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nagarajan, M.; Sandhu, J.S.; Kumar, N.; Behl, V.; Nishanth, G. Mitochondrial DNA analyses of Indian water buffalo support a distinct genetic origin of river and swamp buffalo. Anim. Genet. 2007, 38, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.Z.; Zhang, W.; Chen, H.; Lu, F.; Ge, Q.L.; Liu, R.Y.; Dang, R.H.; Yao, Y.Y.; Yao, L.B.; Lu, Z.F. Two Maternal Lineages Revealed by Mitochondrial DNA D-loop Sequences in Chinese Native Water Buffaloes (Bubalus bubalis). Asian-Australas J. Anim. Sci. 2007, 20, 471–476. [Google Scholar] [CrossRef]
- Lei, C.Z.; Zhang, W.; Chen, H.; Lu, F.; Liu, R.Y.; Yang, X.Y.; Zhang, H.C.; Liu, Z.G.; Yao, L.B.; Lu, Z.F.; et al. Independent maternal origin of Chinese swamp buffalo (Bubalus bubalis). Anim. Genet. 2007, 38, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, N.; Capodiferro, M.R.; Zhang, T.; Lancioni, H.; Zhang, H.; Miao, Y.; Chanthakhoun, V.; Wanapat, M.; Yindee, M.; et al. Whole Mitogenomes Reveal the History of Swamp Buffalo: Initially Shaped by Glacial Periods and Eventually Modelled by Domestication. Sci. Rep. 2017, 7, 4708. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Briggs, A.W.; Good, J.M.; Green, R.E.; Krause, J.; Maricic, T.; Stenzel, U.; Lalueza-Fox, C.; Rudan, P.; Brajković, D.; Kućan, Ž.; et al. Targeted Retrieval and Analysis of Five Neandertal mtDNA Genomes. Science 2009, 325, 318. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Rozas, R. DnaSP, DNA sequence polymorphism: An interactive program for estimating population genetics parameters from DNA sequence data. Comput. Appl. Biosci. Cabios 1995, 11, 621. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E. Pegas: An R package for population genetics with an integrated–modular approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.Z.; Zhang, C.M.; Weining, S.; Campana, M.G.; Bower, M.A.; Zhang, X.M.; Liu, L.; Lan, X.Y.; Chen, H. Genetic diversity of mitochondrial cytochrome b gene in Chinese native buffalo. Anim. Genet. 2011, 42, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.-P.; Li, R.; Xie, W.-M.; Xu, P.; Chang, T.-C.; Liu, L.; Cheng, F.; Zhang, R.-F.; Lan, X.-Y.; Chen, H. Phylogeography and domestication of Chinese swamp buffalo. PLoS ONE 2013, 8, e56552. [Google Scholar] [CrossRef] [PubMed]
| Breed | N | S | H | Haplogroup | Hd ± SE | Pi ± SE | k | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SA1 | SA2 | SB1 | SB2 | SB3 | |||||||
| Fuzhong | 15 | 156 | 15 | 10 | 1 | 1 | 1 | 2 | 1.000 ± 0.0006 | 0.00332 ± 0.00294 | 54.267 |
| Xilin | 25 | 153 | 14 | 12 | 1 | 7 | 0 | 5 | 0.947 ± 0.0005 | 0.00409 ± 0.00249 | 66.893 |
| Total | 40 | 164 | 28 | 22 | 2 | 8 | 1 | 7 | 0.978 ± 0.0001 | 0.00381 ± 0.00237 | 62.286 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, T.; Huang, G.; Sun, J.; Wang, Z.; Teng, S.; Cao, Y.; Hanif, Q.; Chen, N.; Lei, C.; Liao, Y. Mitogenome Diversity and Maternal Origins of Guangxi Buffalo Breeds. Animals 2020, 10, 547. https://doi.org/10.3390/ani10040547
Sun T, Huang G, Sun J, Wang Z, Teng S, Cao Y, Hanif Q, Chen N, Lei C, Liao Y. Mitogenome Diversity and Maternal Origins of Guangxi Buffalo Breeds. Animals. 2020; 10(4):547. https://doi.org/10.3390/ani10040547
Chicago/Turabian StyleSun, Ting, Guangyun Huang, Junli Sun, Zihao Wang, Shaohua Teng, Yanhong Cao, Quratulain Hanif, Ningbo Chen, Chuzhao Lei, and Yuying Liao. 2020. "Mitogenome Diversity and Maternal Origins of Guangxi Buffalo Breeds" Animals 10, no. 4: 547. https://doi.org/10.3390/ani10040547
APA StyleSun, T., Huang, G., Sun, J., Wang, Z., Teng, S., Cao, Y., Hanif, Q., Chen, N., Lei, C., & Liao, Y. (2020). Mitogenome Diversity and Maternal Origins of Guangxi Buffalo Breeds. Animals, 10(4), 547. https://doi.org/10.3390/ani10040547





