The Association of the Copy Number Variation of the MLLT10 Gene with Growth Traits of Chinese Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Selection and Measurement of Body Size Data
2.2. Sample Collected, Genomic DNA Extraction
2.3. Primer Design and Amplification Detection
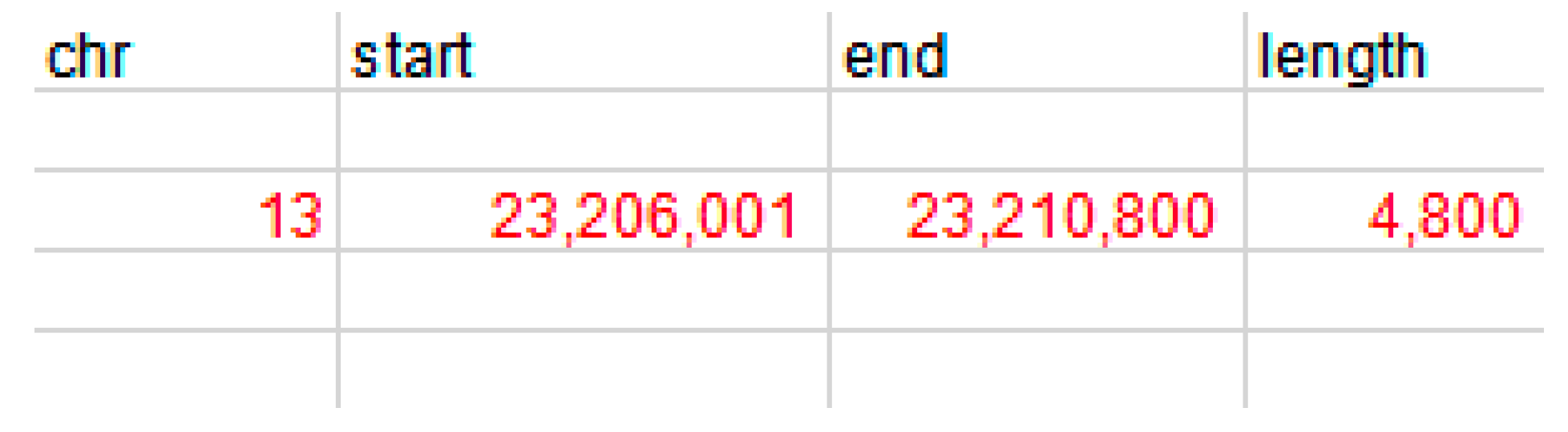
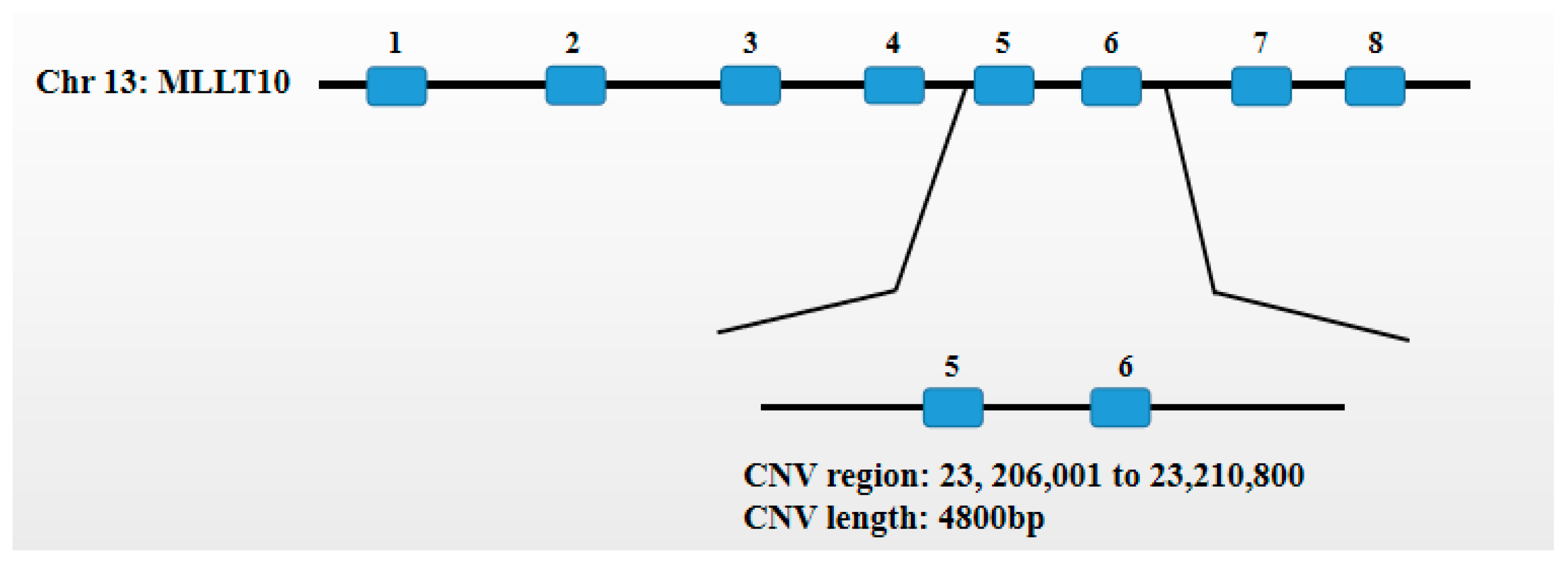
2.4. Detection of the MLLT10 Gene Copy Number
2.5. Data Analysis Processing
3. Results
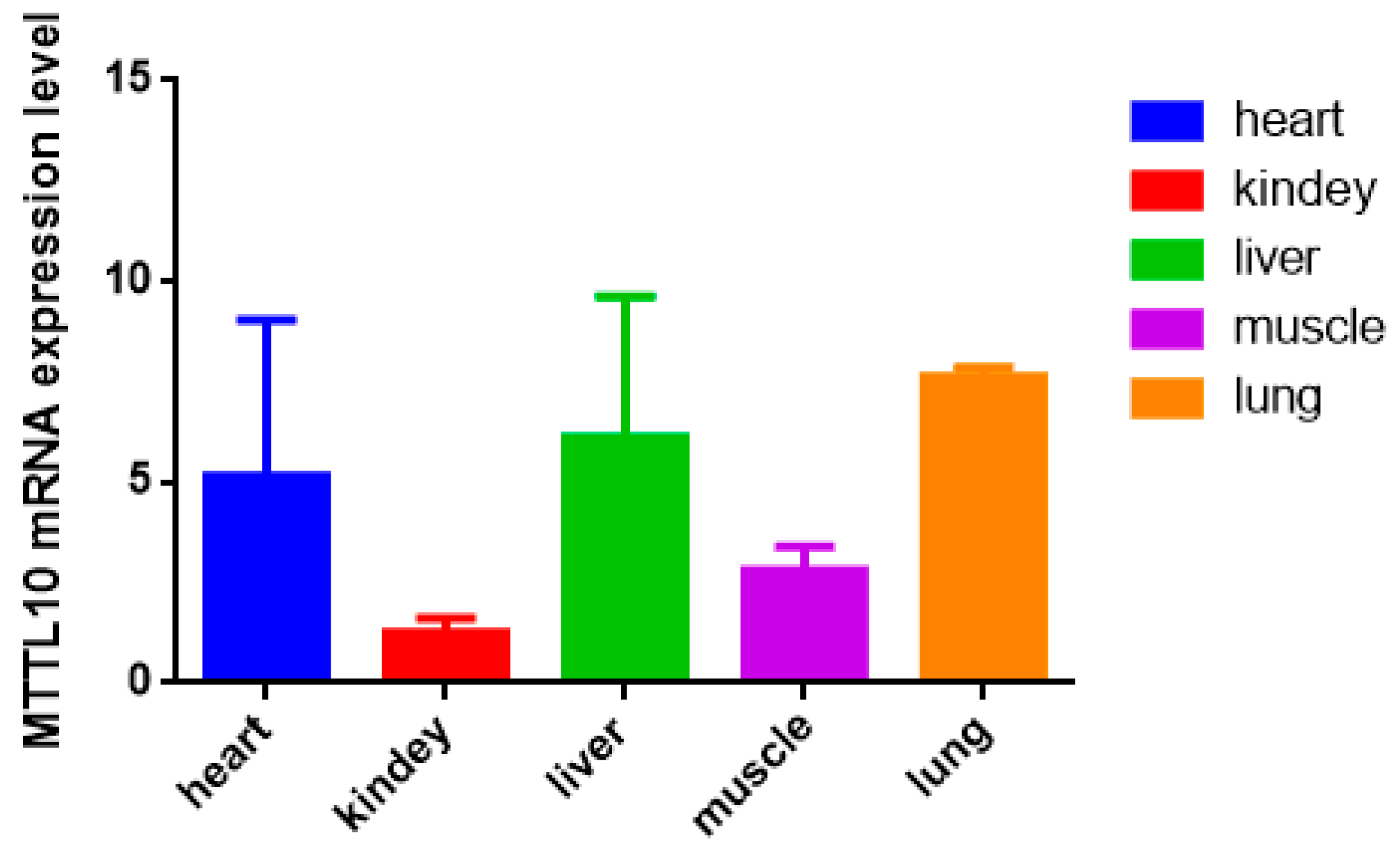
3.1. Expression Profile of the MLLT10 Gene
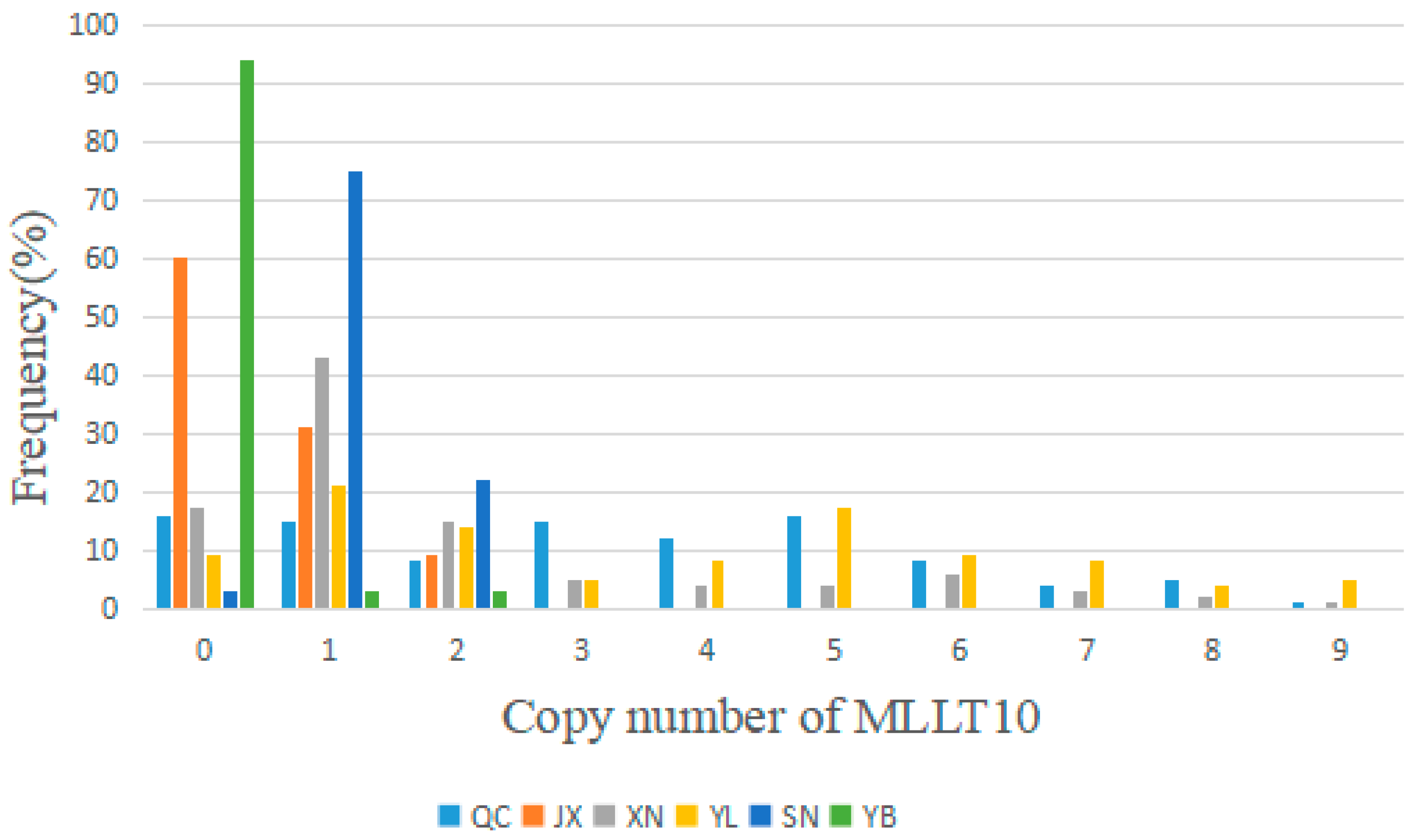
3.2. Distribution of the MLLT10 Gene Copy Number in the Experimental Sample Group
3.3. Correlation Analysis in the MLLT10 Gene CNVs and Growth Traits of Different Cattle Breeds
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Paul, S.; Jung-Woo, C.; Urmila, B.; Jennifer, M.; Yan, M.; Xiaoping, L.; Stephen, M. Whole genome resequencing of Black Angus and Holstein cattle for SNP and CNV discovery. BMC Genom. 2011, 12, 559. [Google Scholar] [CrossRef]
- Yujia, S.; Xianyong, L.; Chuzhao, L.; Chunlei, Z.; Hong, C. Haplotype combination of the bovine CFL2 gene sequence variants and association with growth traits in Qinchuan cattle. Gene 2015, 563, 136–141. [Google Scholar] [CrossRef]
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Lupski, J.R. Structural variation in the human genome and its role in disease. Ann. Rev. Med. 2010, 61, 437–455. [Google Scholar] [CrossRef]
- Fadista, J.; Thomsen, B.; Holm, L.E.; Bendixen, C. Copy number variation in the bovine genome. BMC Genom. 2010, 11, 284. [Google Scholar] [CrossRef]
- Estivill, X.; Armengol, L. Copy number variants and common disorders: Filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet. 2007, 3, e190. [Google Scholar] [CrossRef]
- Wang, X.; Nahashon, S.; Feaster, T.K.; Bohannon-Stewart, A.; Adefope, N. An initial map of chromosomal segmental copy number variations in the chicken. BMC Genom. 2010, 11, 351. [Google Scholar] [CrossRef]
- Richard, P.M.A.C.; Mark, S.F.; Tomas, W.F.; Shurnevia, S.; Hans, H.C.; Pete, K.; Richard, R.; Martien, A.M. Large scale variation in DNA copy number in chicken breeds. BMC Genom. 2013, 14, 398. [Google Scholar] [CrossRef]
- Fadista, J.; Nygaard, M.; Holm, L.E.; Thomsen, B.; Bendixen, C. A snapshot of CNVs in the pig genome. PLoS ONE 2008, 3, e3916. [Google Scholar] [CrossRef]
- Li, Y.; Mei, S.; Zhang, X.; Peng, X.; Liu, G.; Tao, H.; Wu, H.Y.; Jiang, S.; Xiong, Y.; Li, F. Identification of genome-wide copy number variations among diverse pig breeds by array CGH. BMC Genom. 2012, 13, 725. [Google Scholar] [CrossRef]
- Fontanesi, L.; Beretti, F.; Riggio, V.; Gómez, G.E.; Dall’Olio, S.; Davoli, R.; Russo, V.; Portolano, B. Copy number variation and missense mutations of the agouti signaling protein (ASIP) gene in goat breeds with different coat colors. Cytogenet. Genome Res. 2009, 126, 333–347. [Google Scholar] [CrossRef]
- Fontanesi, L.; Beretti, F.; Martelli, P.L.; Colombo, M.; Dall’Olio, S.; Occidente, M.; Portolano, B.; Casadio, R.; Matassino, D.; Russo, V. A first comparative map of copy number variations in the sheep genome. Genomics 2011, 97, 158–165. [Google Scholar] [CrossRef]
- George, E.; Derek, M. Bickhart. Copy number variation in the cattle genome. Funct. Integr. Genom. 2012, 12, 609–624. [Google Scholar] [CrossRef]
- Da Silva, J.M.; Giachetto, P.F.; da Silva, L.O.; Cintra, L.C.; Paiva, S.R.; Yamagishi, M.E.B.; Caetano, A.R. Genome-wide copy number variation (CNV) detection in Nelore cattle reveals highly frequent variants in genome regions harboring QTLs affecting production traits. BMC Genom. 2016, 17, 454. [Google Scholar] [CrossRef] [PubMed]
- Morerio, C.; Rapella, A.; Tassano, E.; Rosanda, C.; Panarello, C. MLL–MLLT10 fusion gene in pediatric acute megakaryoblastic leukemia. Leukemia Res. 2005, 29, 1223–1226. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.; Schnittger, S.; Klaus, M.; Kern, W.; Hiddemann, W.; Haferlach, T. AML with 11q23/MLL abnormalities as defined by the WHO classification: Incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood 2003, 102, 2395–2402. [Google Scholar] [CrossRef]
- Köchl, S.; Niederstätter, H.; Parson, W. DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time. Forensic DNA Typing Protocols 2005, 13–29. [Google Scholar] [CrossRef]
- Bickhart, D.M.; Hou, Y.; Schroeder, S.G.; Alkan, C.; Cardone, M.F.; Matukumalli, L.K.; Song, J.; Schnabel, R.D.; Ventura, M.; Taylor, J.F.; et al. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res. 2012, 22, 778–790. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, T.; Cai, H.; Zhou, Y.; Lan, X.; Zhang, C.; Lei, C.; Qi, X.; Chen, H. Associations of MYH3 gene copy number variations with transcriptional expression and growth traits in Chinese cattle. Gene 2014, 535, 106–111. [Google Scholar] [CrossRef]
- Chen, C.; Qiao, R.; Wei, R.; Guo, Y.; Ai, H.; Ma, J.; Ren, J.; Huang, L. A comprehensive survey of copy number variation in 18 diverse pig populations and identification of candidate copy number variable genes associated with complex traits. BMC Genom. 2012, 13, 733. [Google Scholar] [CrossRef]
- Sebat, J.; Lakshmi, B.; Troge, J.; Chi, M.; Navin, N.; Lucito, R.; Healy, J.; Hicks, J.; Ye, K.; Reiner, A.; et al. Large-scale copy number polymorphism in the human genome. Science 2004, 305, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Richard, R.; Shumpei, I.; Karen, R.F.; Lars, F.; George, H.P.; Daniel, A.; Heike, F.; Michael, H.S.; Andrew, R.C.; Wenwei, C.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444. [Google Scholar] [CrossRef]
- Dobbins, S.E.; Broderick, P.; Melin, B.; Feychting, M.; Johansen, C.; Andersson, U.; Brännström, T.; Schramm, J.; Olver, B.; Lloyd, A.; et al. Common variation at 10p12. 31 near MLLT10 influences meningioma risk. Nat. Genet. 2011, 43, 825. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.J.; Pérusse, L.; Sarzynski, M.A. Genome-wide association studies suggest sex-specific loci associated with abdominal and visceral fat. Int. J. Obesity 2016, 40, 662. [Google Scholar] [CrossRef] [PubMed]
- Kejun, W.; Dewu, L.; Jules, H.-S.; Jie, C.; Chengkun, L.; Zhenfang, W.; Meiying, F.; Ning, L. Genome wide association analysis reveals new production trait genes in a male Duroc population. PLoS ONE 2015, 10, e0139207. [Google Scholar] [CrossRef]
| Primer | Sequence (5′–3′) | Product Length | |
|---|---|---|---|
| MLLT10-CNV | Forward primer | TCCAAGGACAAGAACCCTGC | 114 bp |
| Reverse primer | GCTCCTCTTAGGCCCTTGTC | ||
| BTF3 | Forward primer | AACCAGGAGAAACTCGCCAA | 166 bp |
| Reverse primer | TTCGGTGAAATGCCCTCTCG | ||
| MLLT10 (mRNA) | Forward primer | GGAAGTCTCTGCGCACACTA | 175 bp |
| Reverse primer | TGAAAAACTGCCTTGCTGGA | ||
| β-actin (mRNA) | Forward primer | GTCATCACCATCGGCAATGAG | 84 bp |
| Reverse primer | AATGCCGCAGGATTCCATG |
| Chinese Yellow Cattle Breeds | Growth Traits | CNV Types (Average Value ± Standard Error) | p Value | ||
|---|---|---|---|---|---|
| Deletion (n = 26) | Normal (n = 12) | Duplication (n = 56) | |||
| YL | Rump Length (cm) | 51.73 ± 4.285 a | 52.67 ± 3.367 a | 49.98 ± 3.952 b | 0.045 * |
| Hucklebone Width (cm) | 22.85 ± 2.572 a | 20.58 ± 2.712 b | 22.79 ± 2.708 a | 0.031 * | |
| XN | Deletion (n = 70) | Normal (n = 22) | Duplication (n = 13) | ||
| Cannon Bone Circumference (cm) | 19.19 ± 1.477 a | 19.23 ± 1.343 a | 20.38 ± 1.261 b | 0.022 * | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, P.; Zhang, Z.; Xu, J.; Qu, K.; Lyv, S.; Wang, X.; Cai, C.; Li, Z.; Wang, E.; Xie, J.; et al. The Association of the Copy Number Variation of the MLLT10 Gene with Growth Traits of Chinese Cattle. Animals 2020, 10, 250. https://doi.org/10.3390/ani10020250
Yang P, Zhang Z, Xu J, Qu K, Lyv S, Wang X, Cai C, Li Z, Wang E, Xie J, et al. The Association of the Copy Number Variation of the MLLT10 Gene with Growth Traits of Chinese Cattle. Animals. 2020; 10(2):250. https://doi.org/10.3390/ani10020250
Chicago/Turabian StyleYang, Peng, Zijing Zhang, Jiawei Xu, Kaixing Qu, Shijie Lyv, Xianwei Wang, Cuicui Cai, Zhiming Li, Eryao Wang, Jianliang Xie, and et al. 2020. "The Association of the Copy Number Variation of the MLLT10 Gene with Growth Traits of Chinese Cattle" Animals 10, no. 2: 250. https://doi.org/10.3390/ani10020250
APA StyleYang, P., Zhang, Z., Xu, J., Qu, K., Lyv, S., Wang, X., Cai, C., Li, Z., Wang, E., Xie, J., Ru, B., Xu, Z., Lei, C., Chen, H., Huang, B., & Huang, Y. (2020). The Association of the Copy Number Variation of the MLLT10 Gene with Growth Traits of Chinese Cattle. Animals, 10(2), 250. https://doi.org/10.3390/ani10020250





