Bayesian Analysis of the Association between Casein Complex Haplotype Variants and Milk Yield, Composition, and Curve Shape Parameters in Murciano-Granadina Goats
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Sample and Sample Selection Process
2.2. Milk Yield Standardization and Composition Analysis
2.3. Milk Production Records
2.4. Selection of Best-Fitting Milk Yield and Composition Curve Models and Curve Shape Parameters Calculation
2.5. Genotyping and Linkage Disequilibrium (LD)
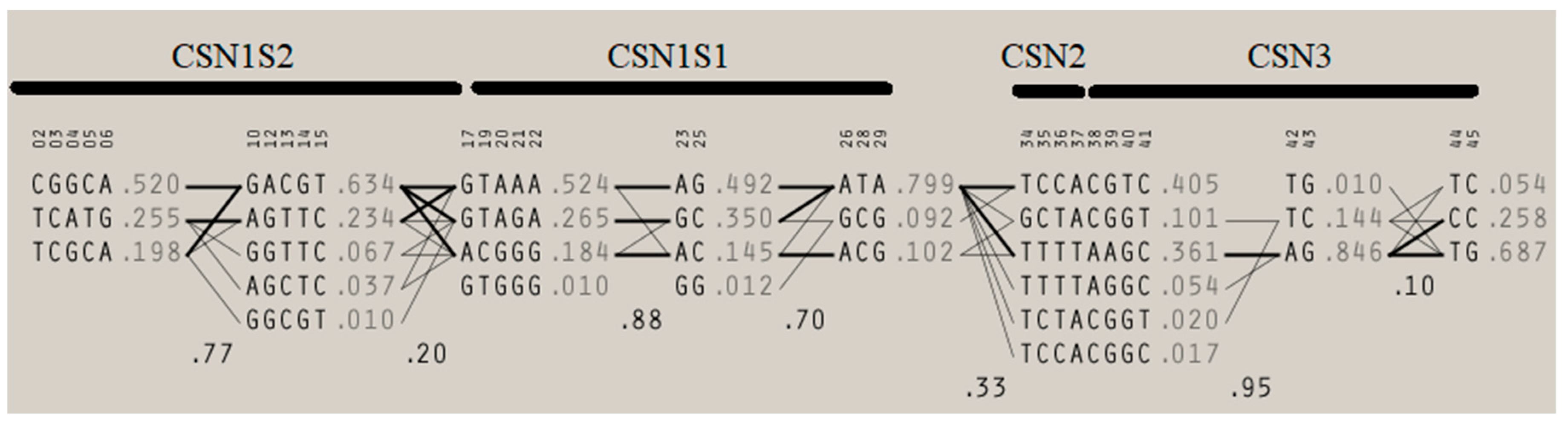
2.6. Haplotyping
2.7. Bayesian Analysis of Haplotype Factor
3. Results
3.1. Milk Yield and Composition Association with Potential Combinations of αS1- and αS2-Casein Loci Haplotypic Sequences
3.2. Milk Yield and Composition Association with Potential Combinations of αS1- and β-Casein Loci Haplotypic Sequences
3.3. Milk Yield and Composition Association with Potential Combinations of αS1- and κ-Casein Loci Haplotypic Sequences
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Caravaca, F.; Ares, J.L.; Carrizosa, J.; Urrutia, B.; Baena, F.; Jordana, J.; Badaoui, B.; Sànchez, A.; Angiolillo, A.; Amills, M. Effects of α s1-casein (CSN1S1) and κ-casein (CSN3) genotypes on milk coagulation properties in Murciano-Granadina goats. J. Dairy Res. 2011, 78, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Rychtářová, J.; Sztankoova, Z.; Kyselova, J.; Zink, V.; Štípková, M.; Vacek, M.; Štolc, L. Effect of DGAT1, BTN1A1, OLR1, and STAT1 genes on milk production and reproduction traits in the Czech Fleckvieh breed. Czech J. Anim. Sci 2014, 59, 45–53. [Google Scholar]
- Pizarro Inostroza, M.; Landi, V.; Navas González, F.; León Jurado, J.; Martínez Martínez, A.; Fernández Álvarez, J.; Delgado Bermejo, J. Non-parametric association analysis of additive and dominance effects of casein complex SNPs on milk content and quality in Murciano-Granadina goats. J. Anim. Breed. Genet. 2020, 137, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; Szymanowska, M.; Zwierzchowski, L.; Leroux, C. The impact of genetic polymorphisms on the protein composition of ruminant milks. Reprod. Nutr. Dev. 2002, 42, 433–459. [Google Scholar] [CrossRef]
- Pizarro Inostroza, M.G.; Landi, V.; Navas González, F.J.; León Jurado, J.M.; Martínez Martínez, M.d.A.; Fernández Álvarez, J.; Delgado Bermejo, J.V. Non-parametric analysis of casein complex genes epistasis and their effect on phenotypic expression of milk yield and composition in Murciano-Granadina goats. J. Dairy Sci. 2020, 103, 8274–8291. [Google Scholar] [CrossRef]
- Yahyaoui, M.H.; Coll, A.; Sanchez, A.; Folch, J.M. Genetic polymorphism of the caprine kappa casein gene. J. Dairy Res. 2001, 68, 209–216. [Google Scholar]
- Martin, C.; Morgavi, D.; Doreau, M. Methane mitigation in ruminants: From microbe to the farm scale. Animal 2010, 4, 351–365. [Google Scholar] [CrossRef] [Green Version]
- Boettcher, P.; Caroli, A.; Stella, A.; Chessa, S.; Budelli, E.; Canavesi, F.; Ghiroldi, S.; Pagnacco, G. Effects of casein haplotypes on milk production traits in Italian Holstein and Brown Swiss cattle. J. Dairy Sci. 2004, 87, 4311–4317. [Google Scholar] [CrossRef]
- Caroli, A.; Chiatti, F.; Chessa, S.; Rignanese, D.; Bolla, P.; Pagnacco, G. Focusing on the goat casein complex. J. Dairy Sci. 2006, 89, 3178–3187. [Google Scholar] [CrossRef] [Green Version]
- Sacchi, P.; Chessa, S.; Budelli, E.; Bolla, P.; Ceriotti, G.; Soglia, D.; Rasero, R.; Cauvin, E.; Caroli, A. Casein haplotype structure in five Italian goat breeds. J. Dairy Sci. 2005, 88, 1561–1568. [Google Scholar] [CrossRef] [Green Version]
- Atashi, H.; Salavati, M.; De Koster, J.; Ehrlich, J.; Crowe, M.; Opsomer, G.; Gplus, E.C.; Hostens, M. Genome-wide association for milk production and lactation curve parameters in Holstein dairy cows. J. Anim. Breed. Genet. 2020, 137, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.; Wang, D.; Zheng, X.; Zhang, Q.; Zhang, S.; Mrode, R.; Liu, J.-F. Eigen decomposition expedites longitudinal genome-wide association studies for milk production traits in Chinese Holstein. Genet. Sel. Evol. 2018, 50, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bionaz, M.; Loor, J.J. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol. Genom. 2007, 29, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Berget, I.; Martens, H.; Kohler, A.; Sjurseth, S.; Afseth, N.; Narum, B.; Ådnøy, T.; Lien, S. Caprine CSN1S1 haplotype effect on gene expression and milk composition measured by Fourier transform infrared spectroscopy. J. Dairy Sci. 2010, 93, 4340–4350. [Google Scholar] [CrossRef] [Green Version]
- Strucken, E.; Bortfeldt, R.; De Koning, D.; Brockmann, G. Genome-wide associations for investigating time-dependent genetic effects for milk production traits in dairy cattle. Anim. Genet. 2012, 43, 375–382. [Google Scholar] [CrossRef]
- Leroux, C.; Le Provost, F.; Petit, E.; Bernard, L.; Chilliard, Y.; Martin, P. Real-time RT-PCR and cDNA macroarray to study the impact of the genetic polymorphism at the alpha s1-casein locus on the expression of genes in the goat mammary gland during lactation. Reprod. Nutr. Dev. 2003, 43, 459–469. [Google Scholar] [CrossRef] [Green Version]
- López, S.; France, J.; Odongo, N.; McBride, R.; Kebreab, E.; AlZahal, O.; McBride, B.; Dijkstra, J. On the analysis of Canadian Holstein dairy cow lactation curves using standard growth functions. J. Dairy Sci. 2015, 98, 2701–2712. [Google Scholar] [CrossRef] [Green Version]
- Arnal, M.; Robert-Granié, C.; Larroque, H. Diversity of dairy goat lactation curves in France. J. Dairy Sci. 2018, 101, 11040–11051. [Google Scholar] [CrossRef] [Green Version]
- France, J.; Kebreab, E. Mathematical Modelling in Animal Nutrition; CABI: Wallingford, UK, 2008. [Google Scholar]
- Andonov, S.; Ødegård, J.; Boman, I.A.; Svendsen, M.; Holme, I.J.; Ådnøy, T.; Vukovic, V.; Klemetsdal, G. Validation of Test-Day Models for Genetic Evaluation of Dairy Goats in Norway. J. Dairy Sci. 2007, 90, 4863–4871. [Google Scholar] [CrossRef] [Green Version]
- Ferrell, J.E.; Tsai, T.Y.-C.; Yang, Q. Modeling the Cell Cycle: Why Do Certain Circuits Oscillate? Cell 2011, 144, 874–885. [Google Scholar] [CrossRef] [Green Version]
- Pizarro, G.; Landi, V.; León Jurado, J.M.; Navas González, F.J.; Delgado Bermejo, J.V. Non-parametric analysis of the effects of αS1-casein genotype and parturition nongenetic factors on milk yield and composition in Murciano-Granadina goats. Ital. J. Anim. Sci. 2019, 18, 1021–1034. [Google Scholar] [CrossRef] [Green Version]
- ICAR. Guidelines for Performance Recording in Dairy Sheep and Dairy Goats; International Committee for Animal Recording: Rome, Italy, 2018; p. 35. [Google Scholar]
- IBM Corp. IBM SPSS Statistics for Windows, Version 25.0; IBM Corp: Armonk, NY, USA, 2017. [Google Scholar]
- Arora, J.S. Chapter 14-Practical Applications of Optimization. In Introduction to Optimum Design, 4th ed.; Arora, J.S., Ed.; Academic Press: Boston, MA, USA, 2017; pp. 601–680. [Google Scholar]
- IBM Corp. IBM SPSS Statistics Algorithms, Version 25.0; IBM Corp.: Armonk, NY, USA, 2017; p. 110. [Google Scholar]
- Durbin, J. Testing for serial correlation in least-squares regression when some of the regressors are lagged dependent variables. Econometrica 1970, 38, 410–421. [Google Scholar] [CrossRef]
- Val-Arreola, D.; Kebreab, E.; Dijkstra, J.; France, J. Study of the lactation curve in dairy cattle on farms in central Mexico. J. Dairy Sci. 2004, 87, 3789–3799. [Google Scholar] [CrossRef]
- Otwinowska-Mindur, A.; Ptak, E. Genetic analysis of lactation persistency in the Polish Holstein-Friesian cows. Anim. Sci. Pap. Rep. 2015, 33, 107–117. [Google Scholar]
- Yahuza, I. Yield-density equations and their application for agronomic research: A review. Int. J. Biosci. 2011, 1, 1–17. [Google Scholar]
- Miller, S.; Dykes, D.; Polesky, H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. Spec. Publ. 1988, 16, 1215. [Google Scholar] [CrossRef] [Green Version]
- Hubbard, T.; Barker, D.; Birney, E.; Cameron, G.; Chen, Y.; Clark, L.; Cox, T.; Cuff, J.; Curwen, V.; Down, T. The Ensembl genome database project. Nucleic Acids Res. Spec. Publ. 2002, 30, 38–41. [Google Scholar] [CrossRef] [Green Version]
- Hao, K.; Li, C.; Rosenow, C.; Wong, W.H. Detect and adjust for population stratification in population-based association study using genomic control markers: An application of Affymetrix Genechip® Human Mapping 10K array. Eur. J. Hum. Genet. 2004, 12, 1001–1006. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Dagnachew, B.S.; Thaller, G.; Lien, S.; Ådnøy, T. Casein SNP in Norwegian goats: Additive and dominance effects on milk composition and quality. Genet. Sel. Evol. 2011, 43, 31. [Google Scholar] [CrossRef] [Green Version]
- Glusman, G.; Cox, H.C.; Roach, J.C. Whole-genome haplotyping approaches and genomic medicine. Genome Med. 2014, 6, 73. [Google Scholar] [PubMed] [Green Version]
- Chen, Z.; Yao, Y.; Ma, P.; Wang, Q.; Pan, Y. Haplotype-based genome-wide association study identifies loci and candidate genes for milk yield in Holsteins. PLoS ONE 2018, 13, e0192695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernando, R.L.; Garrick, D. Bayesian methods applied to GWAS. In Genome-Wide Association Studies and Genomic Prediction. Methods in Molecular Biology (Methods and Protocols); Gondro, C., van der Werf, J., Hayes, B., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 1019, pp. 237–274. [Google Scholar]
- Navas, F.J.; Jordana, J.; León, J.M.; Arando, A.; Pizarro, G.; McLean, A.K.; Delgado, J.V. Measuring and modeling for the assessment of the genetic background behind cognitive processes in donkeys. Res. Vet. Sci. 2017, 113, 105–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navas González, F.J.; León Jurado, J.M.; Delgado Bermejo, J.V. Interpretación de las interacciones entre variables categóricas en modelos de regresión categórica para el metaanálisis de efectos fijos de modelos animales. In Proceedings of the XIX Simposio Iberoamericano Sobre Conservación y Utilización de Recursos Zoogenéticos, Riobamba, Ecuador, 22–26 October 2018. [Google Scholar]
- Cleophas, T.J.; Zwinderman, A.H. Bayesian Analysis of Variance (Anova). In Modern Bayesian Statistics in Clinical Research; Cleophas, T.J., Zwinderman, A.H., Eds.; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar]
- Bayarri, M.J.; Berger, J.O.; Forte, A.; García-Donato, G. Criteria for Bayesian model choice with application to variable selection. Ann. Stat. 2012, 40, 1550–1577. [Google Scholar] [CrossRef] [Green Version]
- Scholtens, M.; Jiang, A.; Smith, A.; Littlejohn, M.; Lehnert, K.; Snell, R.; Lopez-Villalobos, N.; Garrick, D.; Blair, H. Genome-wide association studies of lactation yields of milk, fat, protein and somatic cell score in New Zealand dairy goats. J. Anim. Sci. Biotechnol. 2020, 11, 55. [Google Scholar] [CrossRef]
- Abdel-Shafy, H.; Bortfeldt, R.H.; Tetens, J.; Brockmann, G.A. Single nucleotide polymorphism and haplotype effects associated with somatic cell score in German Holstein cattle. Genet. Sel. Evol. GSE 2014, 46, 35. [Google Scholar] [CrossRef] [Green Version]
- Laramie, J.M.; Wilk, J.B.; DeStefano, A.L.; Myers, R.H. HaploBuild: An algorithm to construct non-contiguous associated haplotypes in family based genetic studies. Bioinformatics 2007, 23, 2190–2192. [Google Scholar] [CrossRef]
- Liu, F.; Schmidt, R.H.; Reif, J.C.; Jiang, Y. Selecting Closely-Linked SNPs Based on Local Epistatic Effects for Haplotype Construction Improves Power of Association Mapping. G3 Genes Genomes Genet. 2019, 9, 4115–4126. [Google Scholar] [CrossRef] [Green Version]
- Hayes, B.; Hagesæther, N.; Ådnøy, T.; Pellerud, G.; Berg, P.R.; Lien, S. Effects on production traits of haplotypes among casein genes in Norwegian goats and evidence for a site of preferential recombination. Genetics 2006, 174, 455–464. [Google Scholar] [PubMed] [Green Version]
- Perna, A.; Intaglietta, I.; Gambacorta, E.; Simonetti, A. The influence of casein haplotype on quality, coagulation, and yield traits of milk from Italian Holstein cows. J. Dairy Sci. 2016, 99, 3288–3294. [Google Scholar] [CrossRef] [Green Version]
- Criscione, A.; Tumino, S.; Avondo, M.; Marletta, D.; Bordonaro, S. Casein haplotype diversity in seven dairy goat breeds. Arch. Tierzucht. 2019, 62, 447–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Criscione, A.; Bordonaro, S.; Moltisanti, V.; Marletta, D. Differentiation of South Italian goat breeds in the focus of biodiversity conservation. Small Rumin. Res. 2016, 145, 12–19. [Google Scholar] [CrossRef]
- Mucha, S.; Mrode, R.; Coffey, M.; Kizilaslan, M.; Desire, S.; Conington, J. Genome-wide association study of conformation and milk yield in mixed-breed dairy goats. J. Dairy Sci. 2018, 101, 2213–2225. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Song, Y.; Hou, J.; Han, P.; Peng, J.; Zhang, L.; Wang, J.; Cao, B. Mutations in the MTHFR gene and their associations with milk production traits in dairy goats. Small Rumin. Res. 2015, 130, 76–80. [Google Scholar] [CrossRef]
- Dagnachew, B.S.; Ådnøy, T. Additive and dominance effects of casein haplotypes on milk composition and quality in Norwegian dairy goats. Small Rumin. Res. 2014, 122, 59–69. [Google Scholar] [CrossRef]
- Gigli, I.; Maizon, D.O.; Riggio, V.; Sardina, M.; Portolano, B. Casein haplotype variability in Sicilian dairy goat breeds. J. Dairy Sci. 2008, 91, 3687–3692. [Google Scholar] [CrossRef] [Green Version]
- Kucerova, J.; Matejicek, A.; Jandurová, O.; Sorensen, P.; Nemcova, E.; Stipkova, M.; Kott, T.; Bouska, J.; Frelich, J. Milk protein genes CSN1S1, CSN2, CSN3, LGB and their relation to genetic values of milk production parameters in Czech Fleckvieh. Czech J. Anim. Sci. 2006, 51, 241. [Google Scholar] [CrossRef] [Green Version]
- Grosclaude, F.; Ricordeau, G.; Martin, P.; Remeuf, F.; Vassal, L.; Bouillon, J. Du gène au fromage: Le polymorphisme de la caséine alphas1 caprine, ses effets, son évolution. INRA Prod. Anim. 1994, 7, 3–19. [Google Scholar] [CrossRef]
- Barbieri, M. Polymorphisme de la Caseine Alpha s1 Effets des Genotyppes sur des Performances Zootechiniques et Utilisation en Selection Caprine; Institut National Agronomique Paris-Grignon: Paris, France, 1995. [Google Scholar]
- Sanchez, A.; Angulo, C.; Amills, M.; Ares, J.; Serradilla, J. Effect of αS1-casein genotype on yield, composition and cheese making properties of milk in the Malagueña breed of goats. In Proceedings of the 6th World Congress on Genetics Applied to Livestock Production, Armidale, Australia, 11–16 January 1998; p. 242. [Google Scholar]
- Vásquez, J.; Novoa, C.; Carulla, J. Efecto del recuento de células somáticas sobre la aptitud quesera de la leche y la calidad fisicoquímica y sensorial del queso campesino. Rev. Med. Vet. Zoot. 2014, 61, 171–185. [Google Scholar] [CrossRef]
- Recio, I.; Pérez-Rodríguez, M.L.; Amigo, L.; Ramos, M. Study of the polymorphism of caprine milk caseins by capillary electrophoresis. J. Dairy Res. 1997, 64, 515–523. [Google Scholar] [CrossRef]
- Baltrėnaitė, L.; Liucvaikienė, K.; Makštutienė, N.; Morkūnienė, K.; Šalomskienė, L.; Miceikienė, I.; Stankevičius, R.; Kerzienė, S. Ožkų pieno baltymų genų įvairovės poveikis pieninėms savybėms. Vet. Med. Zoot. 2013, 62, 8–13. [Google Scholar]
- Chessa, S.; Budelli, E.; Chiatti, F.; Cito, A.; Bolla, P.; Caroli, A. Predominance of β-casein (CSN2) C allele in goat breeds reared in Italy. J. Dairy Sci. 2005, 88, 1878–1881. [Google Scholar] [PubMed]
- Vallas, M.; Kaart, T.; Värv, S.; Pärna, K.; Jõudu, I.; Viinalass, H.; Pärna, E. Composite β-κ-casein genotypes and their effect on composition and coagulation of milk from Estonian Holstein cows. J. Dairy Sci. 2012, 95, 6760–6769. [Google Scholar] [CrossRef] [PubMed]
- Noeparvar, P.; Morison, K.R. The effects of lactose on calcium phosphate precipitation. Chemeca 2018, 206, 1–8. [Google Scholar]
- Linderstrøm-Lang, K. Studies on casein. III. On the fractionation of casein. Compt. Rend. Trav. Lab. Carlsberg. Ser. Chim. 1929, 17, 1–116. [Google Scholar]
- Ercili-Cura, D.; Huppertz, T.; Kelly, A. Enzymatic modification of dairy product texture. In Modifying Food Texture; Elsevier: Amsterdam, The Netherlands, 2015; pp. 71–97. [Google Scholar]
- Cardona, S.J.C.; Cadavid, H.C.; Corrales, J.D.; Munilla, S.; Cantet, R.J.; Rogberg-Muñoz, A. Longitudinal data analysis of polymorphisms in the κ-casein and β-lactoglobulin genes shows differential effects along the trajectory of the lactation curve in tropical dairy goats. J. Dairy Sci. 2016, 99, 7299–7307. [Google Scholar] [CrossRef] [Green Version]
- Strucken, E.; De Koning, D.; Rahmatalla, S.; Brockmann, G.A. Lactation curve models for estimating gene effects over a timeline. J. Dairy Sci. 2011, 94, 442–449. [Google Scholar] [CrossRef] [Green Version]
- Caroli, A.; Chessa, S.; Erhardt, G. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef] [Green Version]
- Rupp, R.; Clément, V.; Piacere, A.; Robert-Granié, C.; Manfredi, E. Genetic parameters for milk somatic cell score and relationship with production and udder type traits in dairy Alpine and Saanen primiparous goats. J. Dairy Sci. 2011, 94, 3629–3634. [Google Scholar] [CrossRef] [Green Version]
- Alhussien, M.; Manjari, P.; Sheikh, A.; Seman, S.M.; Reddi, S.; Mohanty, A.; Mukherjee, J.; Dang, A. Immunological attributes of blood and milk neutrophils isolated from crossbred cows during different physiological conditions. Czech J. Anim. Sci. 2016, 61, 223–231. [Google Scholar] [CrossRef] [Green Version]
- Braunschweig, M.; Hagger, C.; Stranzinger, G.; Puhan, Z. Associations between casein haplotypes and milk production traits of Swiss Brown cattle. J. Dairy Sci. 2000, 83, 1387–1395. [Google Scholar] [CrossRef]
- Gambacorta, E.; Perna, A.; Cosentino, E. Attitudine alla caseificazione del latte di bovine Jersey in funzione del genotipo ai loci αs1, β, k-caseina. In Proceedings of the 18th World Buiatrics Congress, Bologna, Italy, 29 August–2 September; pp. 953–956.
- Gambacorta, E.; Cosentino, C.; Gambacorta, M.; Marsico, D.; Perna, A. Podolian breed cows: Casein genotype and cheesemaking attitude in pasta filata and in hard pressed curd cheese. In Proceedings of the 40th Simposio Internacional de Zootecnia From Genome to Proteome in Animal Science. Istituto Sperimentale Italiano “Lazzaro Spallanzani”, Lodi, Italy, 1 January 2005; pp. 153–162. [Google Scholar]
- Pereira, R.; Verneque, R.; Lopes, P.; Santana, M.; Lagrotta, M.; Torres, R.; Vercesi Filho, A.; Machado, M. Milk yield persistency in Brazilian Gyr cattle based on a random regression model. Genet. Mol. Res. 2012, 11, 1599–1609. [Google Scholar] [CrossRef] [PubMed]
- Kelm, S.; Freeman, A. Direct and Correlated Responses to Selection for Milk Yield: Results and Conclusions of Regional Project NC-2,“Improvement of Dairy Cattle through Breeding, with Emphasis on Selection” 1. J. Dairy Sci. 2000, 83, 2721–2732. [Google Scholar] [CrossRef]
- Muir, B.; Fatehi, J.; Schaeffer, L. Genetic relationships between persistency and reproductive performance in first-lactation Canadian Holsteins. J. Dairy Sci. 2004, 87, 3029–3037. [Google Scholar] [CrossRef] [Green Version]
- Strucken, E.M.; Laurenson, Y.C.; Brockmann, G.A. Go with the flow—biology and genetics of the lactation cycle. Front. Genet. 2015, 6, 118. [Google Scholar] [CrossRef] [Green Version]
- Watson, C.J.; Khaled, W.T. Mammary development in the embryo and adult: A journey of morphogenesis and commitment. Development 2008, 135, 995–1003. [Google Scholar] [CrossRef] [Green Version]
- McNally, S.; Martin, F. Molecular regulators of pubertal mammary gland development. Ann. Med. 2011, 43, 212–234. [Google Scholar]
- Wheeler, T.; Smolenski, G.; Harris, D.; Gupta, S.; Haigh, B.; Broadhurst, M.; Molenaar, A.; Stelwagen, K. Host-defence-related proteins in cows’ milk. Animal 2012, 6, 415–422. [Google Scholar]
- Chaneton, L.; Bonta, M.; Pol, M.; Tirante, L.; Bussmann, L. Milk lactoferrin in heifers: Influence of health status and stage of lactation. J. Dairy Sci. 2013, 96, 4977–4982. [Google Scholar] [CrossRef] [Green Version]
- Pizarro, M.I.; Landi, V.; Navas, F.G.; León, J.J.; Martínez, A.M.; Fernández, J.Á.; Delgado, J.B. Does the Acknowledgement of αS1-Casein Genotype Affect the Estimation of Genetic Parameters and Prediction of Breeding Values for Milk Yield and Composition Quality-Related Traits in Murciano-Granadina? Animals 2019, 9, 679. [Google Scholar] [CrossRef] [Green Version]
- Iraqi, M.; El-Labban, A.; Khalil, M. Estimation of breeding values and their accuracies using multivariates animal model analysis for growth traits in three local strains of chickens. Egypt. Poult. Sci. 2000, 20, 981–1002. [Google Scholar]
- Carillier-Jacquin, C.; Larroque, H.; Robert-Granié, C. Including α s1 casein gene information in genomic evaluations of French dairy goats. Genet. Sel. Evol. 2016, 48, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizarro Inostroza, M.G.; Landi, V.; Navas González, F.J.; León Jurado, J.M.; Delgado Bermejo, J.V.; Fernández Álvarez, J.; Martínez Martínez, M.d.A. Integrating Casein Complex SNPs Additive, Dominance and Epistatic Effects on Genetic Parameters and Breeding Values Estimation for Murciano-Granadina Goat Milk Yield and Components. Genes 2020, 11, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mucha, A.; Wierzbicki, H. Linear models for breeding values prediction in haplotype-assisted selection–An analysis of QTL-MAS Workshop 2011 Data. BMC Proc. 2012, 6 (Suppl. 2), S2–S11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Model | Parameters | Peak Yield | Persistency | Reference |
|---|---|---|---|---|
| Ali and Schaeffer | Milk yield (kg), protein, fat, dry matter and lactose (%) | [29] | ||
| Parabolic yield-density | Somatic cells count (SCC × 103 sc/mL) | [30] |
| Trait | Parameter | Groups | Sum of Squares | df | Mean Square | F | Sig. | Bayes Factor |
|---|---|---|---|---|---|---|---|---|
| Milk Yield | Peak | Between | 69,689.375 | 86.000 | 810.342 | 0.545 | 0.958 | 0.000 |
| Within | 22,301.205 | 15.000 | 1486.747 | |||||
| Persitence (b1) | Between | 1.822 | 86.000 | 0.021 | 0.608 | 0.921 | 0.000 | |
| Within | 0.523 | 15.000 | 0.035 | |||||
| Persitence (b2) | Between | 0.000 | 86.000 | 0.000 | 0.000 | |||
| Within | 0.000 | 15.000 | 0.000 | |||||
| Kg | Between | 18,562.178 | 86.000 | 215.839 | 17.316 | 0.001 | 120,000,000 | |
| Within | 37,643.494 | 3020.000 | 12.465 | |||||
| Fat | Peak | Between | 177,982.014 | 86.000 | 2069.558 | 0.818 | 0.729 | 0.000 |
| Within | 37,941.157 | 15.000 | 2529.410 | |||||
| Persitence (b1) | Between | 5.985 | 86.000 | 0.070 | 1.208 | 0.355 | 0.000 | |
| Within | 0.864 | 15.000 | 0.058 | |||||
| Persitence (b2) | Between | 0.000 | 86.000 | 0.000 | 0.998 | 0.539 | 0.000 | |
| Within | 0.000 | 15.000 | 0.000 | |||||
| % | Between | 1024.046 | 86.000 | 11.908 | 11.52 | 0.001 | 178,000,000 | |
| Within | 3120.806 | 3020.000 | 1.033 | |||||
| Protein | Peak | Between | 11,785.747 | 86.000 | 137.044 | 3.142 | 0.008 | 0.001 |
| Within | 654.156 | 15.000 | 43.610 | |||||
| Persitence (b1) | Between | 0.490 | 86.000 | 0.006 | 1.630 | 0.144 | 0.000 | |
| Within | 0.052 | 15.000 | 0.003 | |||||
| Persitence (b2) | Between | 0.000 | 86.000 | 0.000 | 0.000 | |||
| Within | 0.000 | 15.000 | 0.000 | |||||
| % | Between | 253.694 | 86.000 | 2.950 | 16.951 | 0.001 | 311,000,000 | |
| Within | 525.565 | 3020.000 | 0.174 | |||||
| Dry Matter | Peak | Between | 243,045.970 | 86.000 | 2826.12 | 1.089 | 0.452 | 0.000 |
| Within | 38,915.155 | 15.000 | 2594.34 | |||||
| Persitence (b1) | Between | 8.013 | 86.000 | 0.093 | 1.330 | 0.274 | 0.000 | |
| Within | 1.051 | 15.000 | 0.070 | |||||
| Persitence (b2) | Between | 0.000 | 86.000 | 0.000 | 0.661 | 0.882 | 0.000 | |
| Within | 0.000 | 15.000 | 0.000 | |||||
| % | Between | 1823.530 | 86.000 | 21.204 | 13.804 | 0.001 | 382,000,000 | |
| Within | 4638.983 | 3020.000 | 1.536 | |||||
| Lactose | Peak | Between | 3683.784 | 86.000 | 42.835 | 1.238 | 0.334 | 0.000 |
| Within | 518.983 | 15.000 | 34.599 | |||||
| Persitence (b1) | Between | 0.164 | 86.000 | 0.002 | 0.906 | 0.634 | 0.000 | |
| Within | 0.032 | 15.000 | 0.002 | |||||
| Persitence (b2) | Between | 0.000 | 86.000 | 0.000 | 0.000 | |||
| Within | 0.000 | 15.000 | 0.000 | |||||
| % | Between | 90.587 | 86.000 | 1.053 | 14.50 | 0.001 | 78,800,000 | |
| Within | 219.336 | 3020.000 | 0.073 | |||||
| Somatic cell counts | Peak | Between | 201,479,119.002 | 86.000 | 2,342,780.45 | 0.374 | 0.998 | 0.000 |
| Within | 93,871,863.089 | 15.000 | 6,258,124.21 | |||||
| Persitence (b1) | Between | 9,286,148,083,739 | 86.000 | 107,978,466 | 0.569 | 0.945 | 0.000 | |
| Within | 2,847,041,628,535 | 15.000 | 189,802,775 | |||||
| ×103 sc/mL | Between | 1,735,786,503.91 | 86.000 | 20,183,564 | 22.388 | 0.001 | 457,000,000 | |
| Within | 2,722,657,222.52 | 3020.000 | 901,542.13 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inostroza, M.G.P.; González, F.J.N.; Landi, V.; Jurado, J.M.L.; Bermejo, J.V.D.; Fernández Álvarez, J.; Martínez Martínez, M.d.A. Bayesian Analysis of the Association between Casein Complex Haplotype Variants and Milk Yield, Composition, and Curve Shape Parameters in Murciano-Granadina Goats. Animals 2020, 10, 1845. https://doi.org/10.3390/ani10101845
Inostroza MGP, González FJN, Landi V, Jurado JML, Bermejo JVD, Fernández Álvarez J, Martínez Martínez MdA. Bayesian Analysis of the Association between Casein Complex Haplotype Variants and Milk Yield, Composition, and Curve Shape Parameters in Murciano-Granadina Goats. Animals. 2020; 10(10):1845. https://doi.org/10.3390/ani10101845
Chicago/Turabian StyleInostroza, María Gabriela Pizarro, Francisco Javier Navas González, Vincenzo Landi, Jose Manuel León Jurado, Juan Vicente Delgado Bermejo, Javier Fernández Álvarez, and María del Amparo Martínez Martínez. 2020. "Bayesian Analysis of the Association between Casein Complex Haplotype Variants and Milk Yield, Composition, and Curve Shape Parameters in Murciano-Granadina Goats" Animals 10, no. 10: 1845. https://doi.org/10.3390/ani10101845
APA StyleInostroza, M. G. P., González, F. J. N., Landi, V., Jurado, J. M. L., Bermejo, J. V. D., Fernández Álvarez, J., & Martínez Martínez, M. d. A. (2020). Bayesian Analysis of the Association between Casein Complex Haplotype Variants and Milk Yield, Composition, and Curve Shape Parameters in Murciano-Granadina Goats. Animals, 10(10), 1845. https://doi.org/10.3390/ani10101845






