Genome-Wide Characterization and Comparative Analyses of Simple Sequence Repeats among Four Miniature Pig Breeds
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals
2.3. Dna Extraction and Sequencing Based on Simple Sequence Repeat (SSR)-Enriched Library
2.4. Data Treatment and SSRs Scanning
2.5. Analysis of Polymorphic SSRs and Functional Annotation
2.6. Designing Primers and Experimental Validation
3. Results
3.1. Overview of SSRs and Repeat Units in the Pig Reference Genome
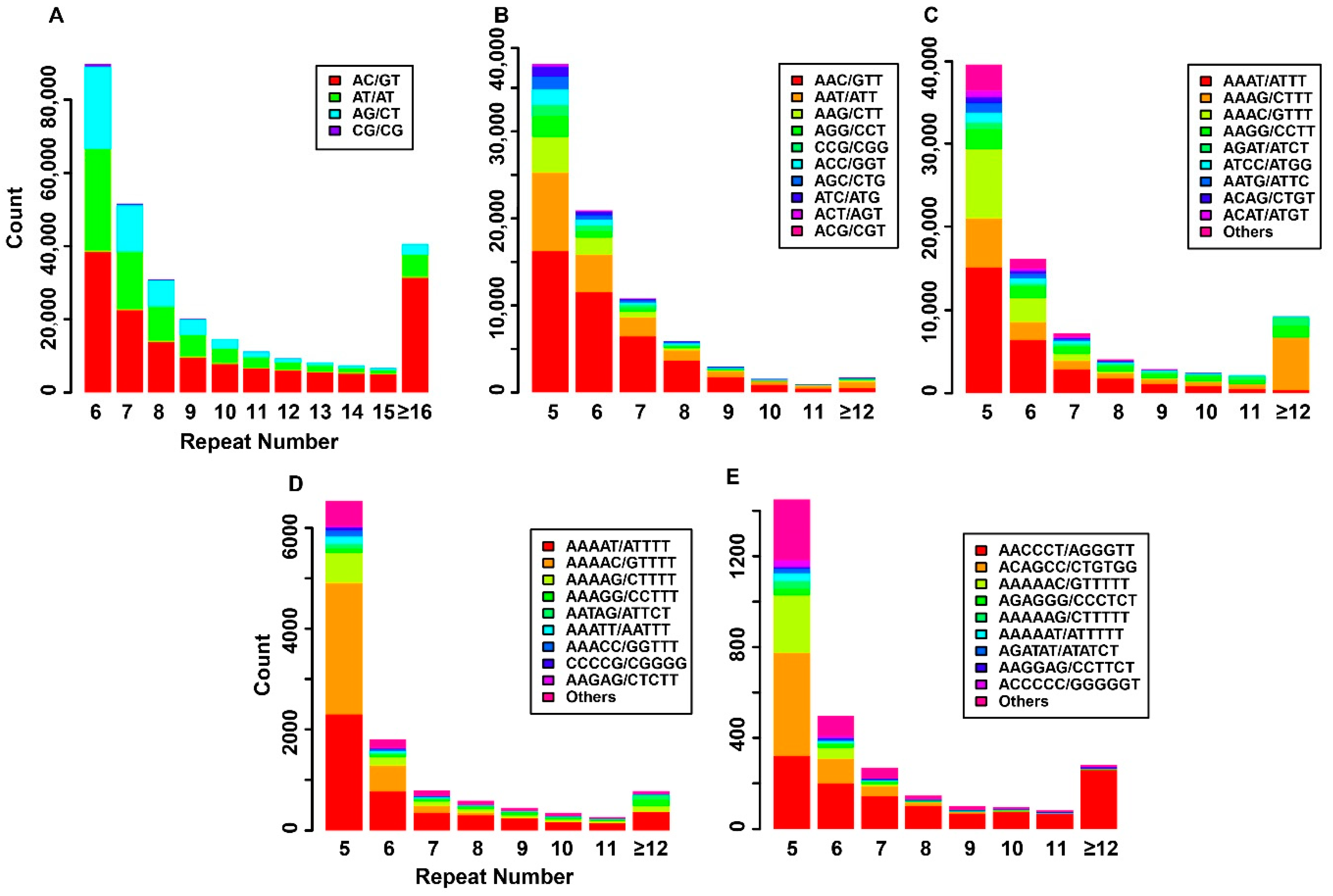
3.2. SRR Discovery from Four Miniature Pig Breeds
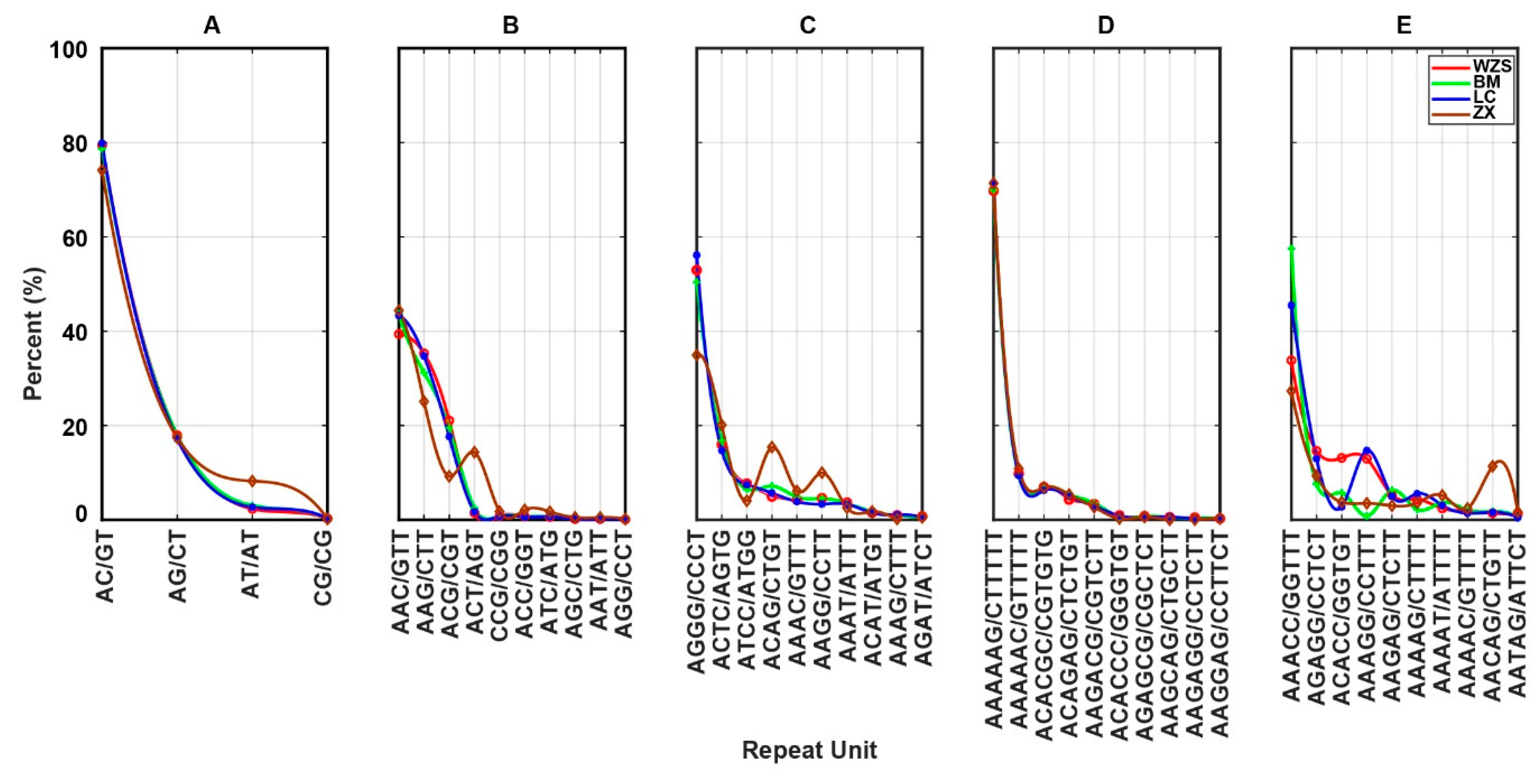
3.3. Frequency of Repeat Units
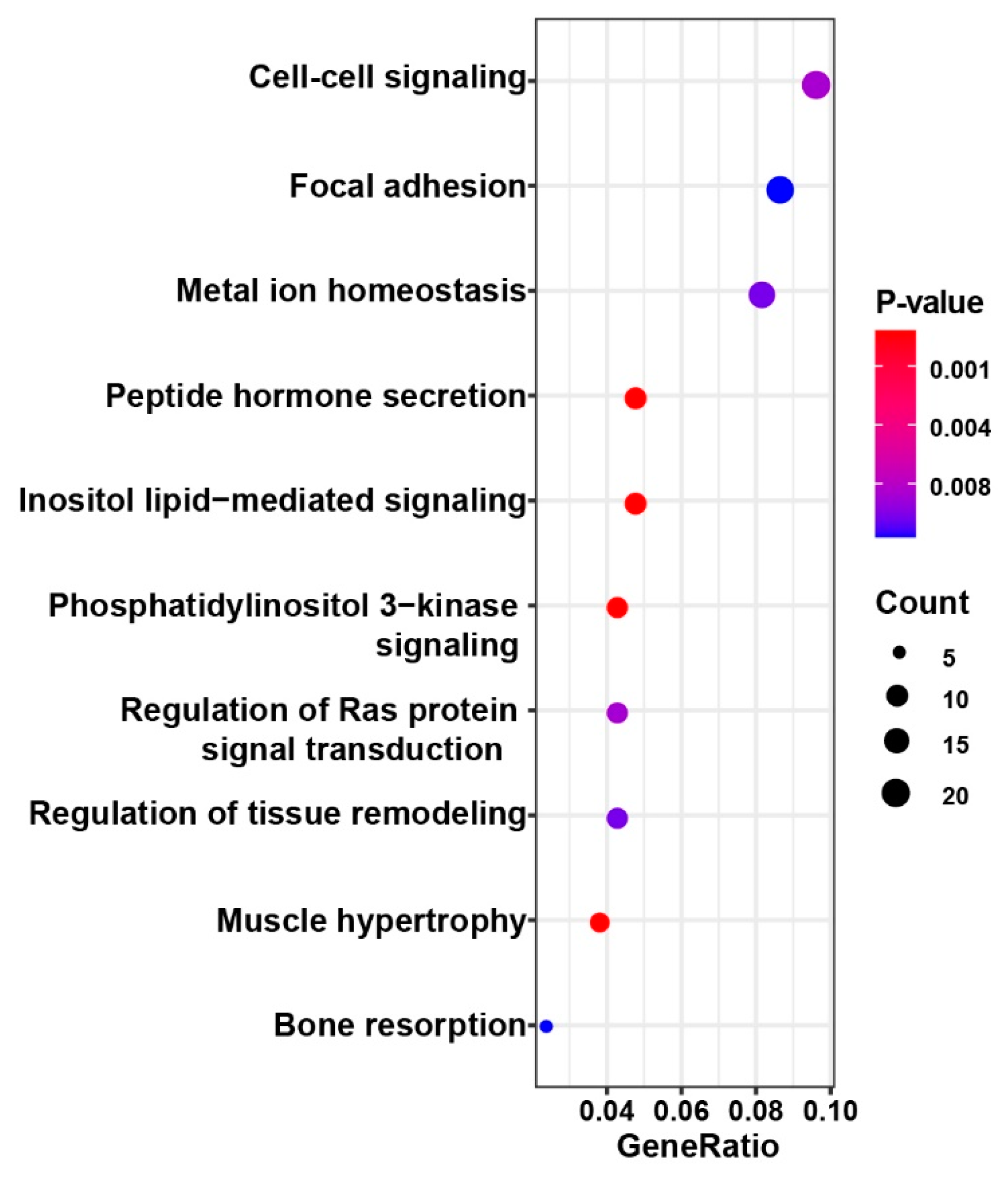
3.4. Polymorphic and Functional SSRs in Four Miniature Pig Breeds
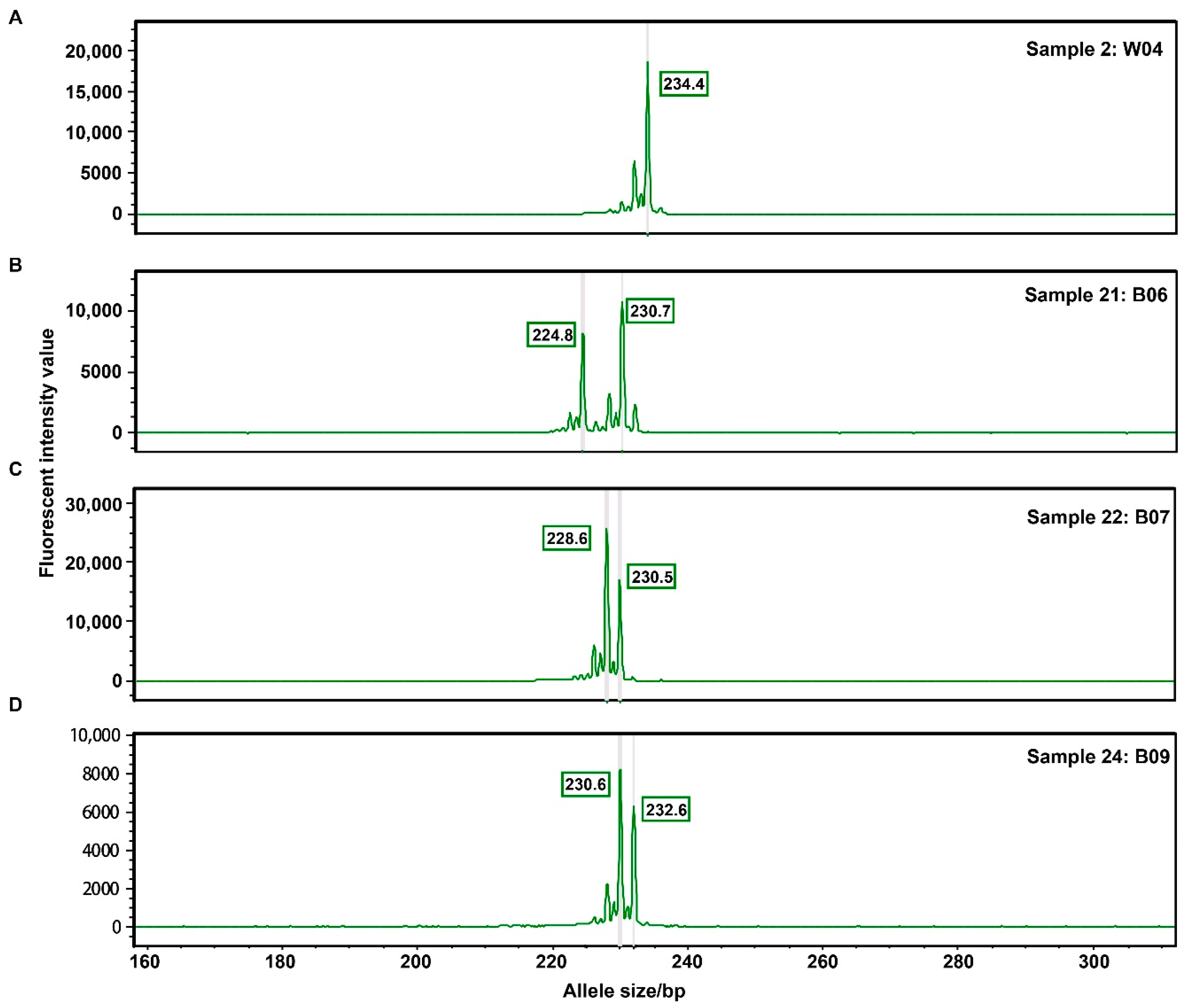
3.5. Experiment Validation Using Fluorescence Polymerase Chain Reaction (PCR) and Capillary Electrophoresis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Toth, G.; Gáspári, Z.; Jurka, J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef] [Green Version]
- Seyoum, M.; Du, X.M.; He, S.P.; Jia, Y.H.; Pan, Z.; Sun, J.L. Analysis of genetic diversity and population structure in upland cotton (Gossypium hirsutum L.) germplasm using simple sequence repeats. J. Genet. 2018, 97, 513–522. [Google Scholar] [CrossRef]
- Park, D.H.; Sa, K.J.; Lim, S.E.; Ma, S.J.; Lee, J.K. Genetic diversity and population structure of Perilla frutescens collected from Korea and China based on simple sequence repeats (SSRs). Genes Genom. 2019, 41, 1329–1340. [Google Scholar] [CrossRef]
- Chombe, D.; Bekele, E. Genetic diversity analysis of cultivated Korarima [Aframomum corrorima (Braun) P.C.M. Jansen] populations from southwestern Ethiopia using inter simple sequence repeats (ISSR) marker. J. Biol. Res. (Thessalon) 2018, 25, 1. [Google Scholar] [CrossRef] [Green Version]
- Silva, A.V.; Nascimento, A.L.; Vitoria, M.F.; Rabbani, A.R.; Soares, A.N.; Ledo, A.S. Diversity and genetic stability in banana genotypes in a breeding program using inter simple sequence repeats (ISSR) markers. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef] [PubMed]
- Rebala, K.; Rabtsava, A.A.; Kotova, S.A.; Kipen, V.N.; Zhurina, N.V.; Gandzha, A.I.; Tsybovsky, I.S. STR Profiling for Discrimination between Wild and Domestic Swine Specimens and between Main Breeds of Domestic Pigs Reared in Belarus. PLoS ONE 2016, 11, e0166563. [Google Scholar] [CrossRef] [PubMed]
- Lorenzini, R.; Fanelli, R.; Tancredi, F.; Siclari, A.; Garofalo, L. Matching STR and SNP genotyping to discriminate between wild boar, domestic pigs and their recent hybrids for forensic purposes. Sci. Rep. 2020, 10, 3188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandran, S.; Pukalenthy, B.; Adhimoolam, K.; Manickam, D.; Sampathrajan, V.; Chocklingam, V.; Eswaran, K.; Arunachalam, K.; Joikumar Meetei, L.; Rajasekaran, R.; et al. Marker-Assisted Selection to Pyramid the Opaque-2 (O2) and beta-Carotene (crtRB1) Genes in Maize. Front. Genet. 2019, 10, 859. [Google Scholar] [CrossRef] [Green Version]
- Ma, P.; Xu, H.; Xu, Y.; Song, L.; Liang, S.; Sheng, Y.; Han, G.; Zhang, X.; An, D. Characterization of a Powdery Mildew Resistance Gene in Wheat Breeding Line 10V-2 and Its Application in Marker-Assisted Selection. Plant. Dis. 2018, 102, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Leite, D.C.; Pinheiro, J.B.; Campos, J.B.; Di Mauro, A.O.; Uneda-Trevisoli, S.H. QTL mapping of soybean oil content for marker-assisted selection in plant breeding program. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, J.; Baczkiewicz, A.; Buczkowska, K.; Gorski, P.; Krawczyk, K.; Mizia, P.; Myszczynski, K.; Slipiko, M.; Szczecinska, M. The Increase of Simple Sequence Repeats during Diversification of Marchantiidae, An Early Land Plant Lineage, Leads to the First Known Expansion of Inverted Repeats in the Evolutionarily-Stable Structure of Liverwort Plastomes. Genes (Basel) 2020, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witkos, T.M.; Krzyzosiak, W.J.; Fiszer, A.; Koscianska, E. A potential role of extended simple sequence repeats in competing endogenous RNA crosstalk. RNA Biol. 2018, 15, 1399–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flickinger, R. Polymorphism of simple sequence repeats may quantitatively regulate gene transcription. Exp. Cell Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Joy, N.; Beevi, Y.P.M.; Soniya, E.V. A deeper view into the significance of simple sequence repeats in pre-miRNAs provides clues for its possible roles in determining the function of microRNAs. BMC Genet. 2018, 19, 29. [Google Scholar] [CrossRef] [PubMed]
- Gymrek, M.; Willems, T.; Guilmatre, A.; Zeng, H.; Markus, B.; Georgiev, S.; Daly, M.J.; Price, A.L.; Pritchard, J.K.; Sharp, A.J.; et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet. 2016, 48, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Malausa, T.; Gilles, A.; Meglecz, E.; Blanquart, H.; Duthoy, S.; Costedoat, C.; Dubut, V.; Pech, N.; Castagnone-Sereno, P.; Delye, C.; et al. High-throughput microsatellite isolation through 454 GS-FLX Titanium pyrosequencing of enriched DNA libraries. Mol. Ecol. Resour. 2011, 11, 638–644. [Google Scholar] [CrossRef]
- Conyers, C.M.; Allnutt, T.R.; Hird, H.J.; Kaye, J.; Chisholm, J. Development of a microsatellite-based method for the differentiation of European wild boar (Sus scrofa scrofa) from domestic pig breeds (Sus scrofa domestica) in food. J. Agric. Food Chem. 2012, 60, 3341–3347. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Y.; Zhang, X.; Xu, X.; Zhao, S. Characterization of porcine simple sequence repeat variation on a population scale with genome resequencing data. Sci. Rep. 2017, 7, 2376. [Google Scholar] [CrossRef]
- Fang, X.; Mou, Y.; Huang, Z.; Li, Y.; Han, L.; Zhang, Y.; Feng, Y.; Chen, Y.; Jiang, X.; Zhao, W.; et al. The sequence and analysis of a Chinese pig genome. Gigascience 2012, 1, 16. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, M.; Sharma, N.; Gera, M.; Kim, N.; Sodhi, S.S.; Pulicherla, K.; Huynh, D.; Kim, D.C.; Zhang, J.; Kwon, T.; et al. The first comprehensive description of the expression profile of genes involved in differential body growth and the immune system of the Jeju Native Pig and miniature pig. Amino Acids 2019, 51, 495–511. [Google Scholar] [CrossRef]
- Schulze-Tanzil, G.; Silawal, S.; Hoyer, M. Anatomical feature of knee joint in Aachen minipig as a novel miniature pig line for experimental research in orthopaedics. Anat. Anz. 2020, 227, 151411. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.F.; Li, H.; Bai, G.; Zhang, Q.Z.; Gao, X.; Liu, T.; Wang, H.B. Establishment of Renal Failure Models by Laparoscopy in Bama Pigs Which Underwent Partial Nephrectomy and Radical Contralateral Nephrectomy. J. Vet. Res. 2019, 63, 447–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, J.; Zeng, H.; Zhang, M.; Wei, Q.; Wang, Y.; Yang, H.; Lu, Y.; Li, R.; Xiong, Q.; Zhang, L.; et al. OSBPL2-disrupted pigs recapitulate dual features of human hearing loss and hypercholesterolaemia. J. Genet. Genom. 2019, 46, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, Y.; Wang, M.; Guo, Y.; Liang, J.; Yang, X.; Qi, W.; Wu, Y.; Si, J.; Zhu, S.; et al. Development and Genome Sequencing of a Laboratory-Inbred Miniature Pig Facilitates Study of Human Diabetic Disease. iScience 2019, 19, 162–176. [Google Scholar] [CrossRef] [Green Version]
- Cai, Z.; Yu, C.; Fu, D.; Pan, Y.; Huang, J.; Rong, Y.; Deng, L.; Chen, J.; Chen, M. Differential metabolic and hepatic transcriptome responses of two miniature pig breeds to high dietary cholesterol. Life Sci. 2020, 250, 117514. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, G.; Wu, Z.; Fan, Z.; Yang, M.; Wu, T.; Wang, J.; Zhang, C.; Wang, S. Tracking diphyodont development in miniature pigs in vitro and in vivo. Biol. Open 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Cao, H.; Wang, S.; Zheng, L.; Chen, Z.; Wen, X.; Zhang, S.; Xiang, Y.; Gao, Y. [Isolation and culture of adipose-derived mesenchymal stem cells from inbreed line miniature pig of Wuzhishan and their biological characteristics]. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2019, 44, 297–306. [Google Scholar] [PubMed]
- Zhiqiang, P.; Cun, S.; Ying, J.; Ningli, W.; Li, W. WZS-pig is a potential donor alternative in corneal xenotransplantation. Xenotransplantation 2007, 14, 603–611. [Google Scholar] [CrossRef]
- Lepais, O.; Bacles, C.F.E. Comparison of random and SSR-enriched shotgun pyrosequencing for microsatellite discovery and single multiplex PCR optimization in Acacia harpophylla F. Muell. Ex Benth. Mol. Ecol. Resour. 2011, 11, 711–724. [Google Scholar] [CrossRef]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Yang, L.; Peng, Z.; Sun, H.; Yue, X.; Lou, Y.; Dong, L.; Wang, L.; Gao, Z. Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci. Rep. 2015, 5, 8018. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: A J. Integr. Boil. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Joachim, A.; Ruttkowski, B.; Palmieri, N. Microsatellite Analysis of Geographically Close Isolates of Cystoisospora suis. Front. Vet. Sci. 2019, 6, 96. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, G.; Lin, F.; Jiang, B.; Dong, F.; Liu, H. Expression of the insulin-like growth factor system in skeletal muscle during embryonic and postnatal development in the first filial generation pigs from Erhualian and Yorkshire reciprocal crosses. Gen. Comp. Endocrinol. 2011, 173, 56–62. [Google Scholar] [CrossRef] [PubMed]
- King, K.; Moody, A.; Fisher, S.A.; Mirza, M.M.; Cuthbert, A.P.; Hampe, J.; Sutherland-Craggs, A.; Sanderson, J.; MacPherson, A.J.; Forbes, A.; et al. Genetic variation in the IGSF6 gene and lack of association with inflammatory bowel disease. Eur. J. Immunogenet. 2003, 30, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shi, W.; Cao, X. Multiplicity of BMP signaling in skeletal development. Ann. N. Y. Acad. Sci. 2007, 1116, 29–49. [Google Scholar] [CrossRef]
- Nanda, N.; Bao, M.; Lin, H.; Clauser, K.; Komuves, L.; Quertermous, T.; Conley, P.B.; Phillips, D.R.; Hart, M.J. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J. Biol. Chem. 2005, 280, 24680–24689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.F.; Yan, Y.Q.; Liu, D.; Pang, Y.S.; Wu, J.; Li, S.F.; Tong, H.L. Platelet endothelial aggregation receptor-1 (PEAR1) is involved in C2C12 myoblast differentiation. Exp. Cell Res. 2018, 366, 199–204. [Google Scholar] [CrossRef]
- Kazarian, E.; Son, H.; Sapao, P.; Li, W.; Zhang, Z.; Strauss, J.F.; Teves, M.E. SPAG17 Is Required for Male Germ Cell Differentiation and Fertility. Int. J. Mol. Sci. 2018, 19, 1252. [Google Scholar] [CrossRef] [Green Version]
- Levine, A.J.; Brivanlou, A.H. GDF3, a BMP inhibitor, regulates cell fate in stem cells and early embryos. Development 2005, 133, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Just, F.; Reyer, H.; Murani, E.; Ponsuksili, S.; Oster, M.; Wimmers, K. Genetic variants of major genes contributing to phosphate and calcium homeostasis and their association with serum parameters in pigs. J. Appl. Genet. 2018, 59, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Fan, H.; Cinar, M.U.; Phatsara, C.; Tesfaye, D.; Tholen, E.; Looft, C.; Schellander, K. Molecular mechanism underlying the differential MYF6 expression in postnatal skeletal muscle of Duroc and Pietrain breeds. Gene 2011, 486, 8–14. [Google Scholar] [CrossRef]
- Henriquez-Rodriguez, E.; Bosch, L.; Tor, M.; Pena, R.N.; Estany, J. The effect of SCD and LEPR genetic polymorphisms on fat content and composition is maintained throughout fattening in Duroc pigs. Meat Sci. 2016, 121, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Yan, R.; Wu, C.; Wang, H.; Yang, G.; Zhong, Y.; Liu, Y.; Wan, L.; Tang, A. Spermatogenesis-associated 48 is essential for spermatogenesis in mice. Andrologia 2018, 50, e13027. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.M.; Liu, L.C.; Ho, C.T.; Lin, Y.C.; Way, T.D. Pterostilbene Enhances TRAIL-Induced Apoptosis through the Induction of Death Receptors and Downregulation of Cell Survival Proteins in TRAIL-Resistance Triple Negative Breast Cancer Cells. J. Agric. Food Chem. 2017, 65, 11179–11191. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, N.S.; Jenkins, N.A.; Gilbert, D.J.; Copeland, N.G.; Reddi, A.H.; Lee, S.J. Growth/differentiation factor-10: A new member of the transforming growth factor-beta superfamily related to bone morphogenetic protein-3. Growth Factors 1995, 12, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Avvaru, A.K.; Saxena, S.; Sowpati, D.T.; Mishra, R.K. MSDB: A Comprehensive Database of Simple Sequence Repeats. Genome Biol. Evol. 2017, 9, 1797–1802. [Google Scholar] [CrossRef] [Green Version]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Avvaru, A.K.; Sowpati, D.T.; Mishra, R.K. PERF: An exhaustive algorithm for ultra-fast and efficient identification of microsatellites from large DNA sequences. Bioinformatics 2018, 34, 943–948. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.T.; Zhang, Y.J.; Qiao, L.; Chen, B. Comparative analyses of simple sequence repeats (SSRs) in 23 mosquito species genomes: Identification, characterization and distribution (Diptera: Culicidae). Insect Sci. 2018, 26, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Qin, M.; Yang, L.; Song, Z.; Luo, L.; Bao, H.; Ma, Z.; Zhou, Z.; Xu, J. A genome-wide analysis of simple sequence repeats in Apis cerana and its development as polymorphism markers. Gene 2017, 599, 53–59. [Google Scholar] [CrossRef]
- Mladineo, I.; Trumbic, Z.; Radonic, I.; Vrbatovic, A.; Hrabar, J.; Buselic, I. Anisakis simplex complex: Ecological significance of recombinant genotypes in an allopatric area of the Adriatic Sea inferred by genome-derived simple sequence repeats. Int. J. Parasitol. 2017, 47, 215–223. [Google Scholar] [CrossRef]
- Sharma, P.C.; Grover, A.; Kahl, G. Mining microsatellites in eukaryotic genomes. Trends Biotechnol. 2007, 25, 490–498. [Google Scholar] [CrossRef]
- Murat, C.; Riccioni, C.; Belfiori, B.; Cichocki, N.; Labbe, J.; Morin, E.; Tisserant, E.; Paolocci, F.; Rubini, A.; Martin, F. Distribution and localization of microsatellites in the Perigord black truffle genome and identification of new molecular markers. Fungal Genet. Biol. 2011, 48, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Xu, H.; Song, J.; Xu, J.; Zhu, Y.; Chen, S. Genome-wide analysis of simple sequence repeats in the model medicinal mushroom Ganoderma lucidum. Gene 2013, 512, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, L.; Xu, Y.; Chen, C.; Rong, T.; Ali, F.; Zhou, S.; Wu, F.; Liu, Y.; Wang, J.; et al. Development and characterization of simple sequence repeat markers providing genome-wide coverage and high resolution in maize. DNA Res. 2013, 20, 497–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Zhao, J.; Liu, M.; Liu, P.; Dai, L.; Zhao, Z. Genome-Wide Characterization of Simple Sequence Repeat (SSR) Loci in Chinese Jujube and Jujube SSR Primer Transferability. PLoS ONE 2015, 10, e0127812. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Song, P.; Koo, D.H.; Guo, L.; Li, Y.; Sun, S.; Weng, Y.; Yang, L. Genome wide characterization of simple sequence repeats in watermelon genome and their application in comparative mapping and genetic diversity analysis. BMC Genom. 2016, 17, 557. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yang, S.; Chen, Y.; Zhang, S.; Zhao, Q.; Li, M.; Gao, Y.; Yang, L.; Bennetzen, J.L. Comparative genome-wide characterization leading to simple sequence repeat marker development for Nicotiana. BMC Genom. 2018, 19, 500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Speed, T.P. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012, 40, e72. [Google Scholar] [CrossRef] [Green Version]
- Cole, R.K. An autosomal dwarfism in the domestic fowl. Poult. Sci. 2000, 79, 1507–1516. [Google Scholar] [CrossRef] [Green Version]
- Kader, A.; Li, Y.; Dong, K.; Irwin, D.M.; Zhao, Q.; He, X.; Liu, J.; Pu, Y.; Gorkhali, N.A.; Liu, X.; et al. Population Variation Reveals Independent Selection toward Small Body Size in Chinese Debao Pony. Genome Biol. Evol. 2015, 8, 42–50. [Google Scholar] [CrossRef] [Green Version]

| Items | WZS 1 | BM 1 | LC 1 | ZX 1 |
|---|---|---|---|---|
| Raw reads | 17,083,436 | 10,335,212 | 10,466,408 | 22,759,784 |
| High quality reads | 15,473,282 | 9,660,700 | 9,255,072 | 20,279,502 |
| Total length of high quality reads (base pair, bp) | 3,549,570,313 | 2,367,772,949 | 2,010,410,274 | 4,756,156,243 |
| Combined sequences | 6,658,339 | 4,369,729 | 3,885,823 | 9,043,156 |
| Total length of combined reads (bp) | 2,103,692,216 | 1,435,035,633 | 1,177,952,198 | 2,948,109,147 |
| Dinucleotide repeats (Di-SSRs) | 4,127,838 | 1,843,882 | 2,216,458 | 951,588 |
| Trinucleotide repeat (Tri-SSRs) | 586,364 | 264,912 | 316,662 | 101,743 |
| Tetranucleotide repeat (Tetra-SSRs) | 234,811 | 125,631 | 134,298 | 122,506 |
| Pentanucleotide repeat (Penta-SSRs) | 33,676 | 23,977 | 18,932 | 46,849 |
| Hexanucleotide repeat (Hexa-SSRs) | 17,693 | 7088 | 9198 | 2382 |
| Items 1 | WZS | BM | LC | ZX |
|---|---|---|---|---|
| SSR-containing sequences | 3,804,507 | 1,849,907 | 2,061,668 | 1,448,491 |
| Total clusters | 377,558 | 398,960 | 433,945 | 453,677 |
| SSLP = 1 | 317,538 | 328,074 | 369,977 | 411,277 |
| SSLP = 2 | 37,841 | 44,188 | 41,098 | 30,024 |
| SSLP = 3 | 13,861 | 16,865 | 14,879 | 8542 |
| SSLP = 4 | 5420 | 6341 | 5210 | 2650 |
| SSLP = 5 | 1840 | 2221 | 1779 | 747 |
| SSLP = 6 | 567 | 704 | 525 | 202 |
| SSLP = 7 | 206 | 257 | 182 | 75 |
| SSLP = 8 | 88 | 103 | 101 | 39 |
| SSLP = 9 | 60 | 61 | 50 | 29 |
| SSLP ≥ 10 | 137 | 146 | 144 | 92 |
| Genomic Coordinate of SSR Flanking Sequence 1 | Repeat Sequence | Gene | Functional Region 2 | Gene Function |
|---|---|---|---|---|
| 1:137690845_137690967 | (GCG)6, (GCG)8, | IGF1R | 5′ UTR | Muscle development [41] |
| 3:23673114_23673194 | (GT)13, (CA)15 | IGSF6 | 5′ UTR | Inflammatory disease [42] |
| 3:73527603_73527759 | (AC)9, (GT)13, (GT)15, (GT)17, (GT)18, (GT)20 | BMP10 | 3′ UTR | Skeletal development [43] |
| 4:93179209_93179533 | (TG)15, (CA)16, (CA)17, (TG)18, (CA)22 | PEAR1 | 3′ UTR | Platelet activation [44], myoblast differentiation [45] |
| 4:102696359_102696552 | (CA)22, (GT)23 | SPAG17 | CDS | Sperm motility [46] |
| 5:62835807_62836251 | (AC)12, (TG)13, (AC)14, (AC)16, (GT)17 | GDF3 | CDS | BMP inhibiting [47] |
| 5:66028468_66028506 | (GT)15, (GT)16, (TG)17, (GT)18 | FGF23 | 5′ UTR | Bone remodeling [48] |
| 5:100760758_100761162 | (GT)14, (AC)18 | MYF6 | 3′ UTR | Muscle development [49] |
| 6:146979743_146980171 | (CA)18, (TG)19, (CA)20, (CA)22, (CA)23 | LEPROT | CDS | Fat content [50] |
| 9:136235771_136236041 | (AC)17, (AC)19 | SPATA48 | CDS | Spermatogenesis [51] |
| 13:111022378_111022649 | (GT)15, (GT)16 | TNFSF10 | 3′ UTR | Cell apoptosis [52] |
| 14:88520086_88520470 | (CA)15, (GT)18, (CA)21, (GT)22, (TG)23, (TG)24, (TG)25, (TG)26, (TG)28 | GDF10 | 3′ UTR | TGFβ superfamily [53] |
| Loci 1 | Primer Sequences (5′–3′) 2 | Repeat Sequence 2 | Allele Size/bp 2 | NO. of Alleles 3 |
|---|---|---|---|---|
| chr1: 272,578,714-272,578,954 | F: TACACACCACGCGTGTACCT | (AC)12, (AC)14, (AC)15, (AC)16, (AC)17 | 207~275 | 5 |
| R: ACAACGGTTGCTGCTTTTTC | ||||
| chr11:70,376,652-70,376,765 | F: CCAGCTTTCCAGTTTCGTGT | (GT)14, (GT)15, GT)16, (GT)17, (GT)18, (GT)19 | 77~149 | 6 |
| R: AGATTGTGAGTGCGCAGATG | ||||
| chr18:1,858,964-1,859,153 | F: CTCCAAGGTCAAAAGCCAAA | (AC)13, (AC)14, (AC)15, (AC)16, (AC)17, (AC)18 | 154~226 | 6 |
| R: CGACTTGGGGTTTCCTAACA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Fu, Y.; Gu, P.; Zhang, Y.; Tu, W.; Chao, Z.; Wu, H.; Cao, J.; Zhou, X.; Liu, B.; et al. Genome-Wide Characterization and Comparative Analyses of Simple Sequence Repeats among Four Miniature Pig Breeds. Animals 2020, 10, 1792. https://doi.org/10.3390/ani10101792
Wang H, Fu Y, Gu P, Zhang Y, Tu W, Chao Z, Wu H, Cao J, Zhou X, Liu B, et al. Genome-Wide Characterization and Comparative Analyses of Simple Sequence Repeats among Four Miniature Pig Breeds. Animals. 2020; 10(10):1792. https://doi.org/10.3390/ani10101792
Chicago/Turabian StyleWang, Hongyang, Yang Fu, Peng Gu, Yingying Zhang, Weilong Tu, Zhe Chao, Huali Wu, Jianguo Cao, Xiang Zhou, Bang Liu, and et al. 2020. "Genome-Wide Characterization and Comparative Analyses of Simple Sequence Repeats among Four Miniature Pig Breeds" Animals 10, no. 10: 1792. https://doi.org/10.3390/ani10101792
APA StyleWang, H., Fu, Y., Gu, P., Zhang, Y., Tu, W., Chao, Z., Wu, H., Cao, J., Zhou, X., Liu, B., Michal, J. J., Fan, C., & Tan, Y. (2020). Genome-Wide Characterization and Comparative Analyses of Simple Sequence Repeats among Four Miniature Pig Breeds. Animals, 10(10), 1792. https://doi.org/10.3390/ani10101792





