Effects of Ensiling Oxytropis glabra with Whole-Plant Corn at Different Proportions on Fermentation Quality, Alkaloid Swainsonine Content, and Lactic Acid Bacteria Populations
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Silage Preparation
2.2. Microbial and Chemical Analysis
2.3. Species Identification by 16S rRNA Sequencing
2.4. Determination of the SW Removal Rate in OG Broth by LAB
2.5. Statistical Analysis
3. Results
3.1. Microbial Populations of OG and Whole-Plant Corn Mixtures Prior to and after 60 d of Ensiling
3.2. Chemical Compositions of OG and Whole-Plant Corn Mixtures Prior to and after 60 d of Ensiling
3.3. Fermentation Characteristics of OG and Whole-Plant Corn Mixture Silages
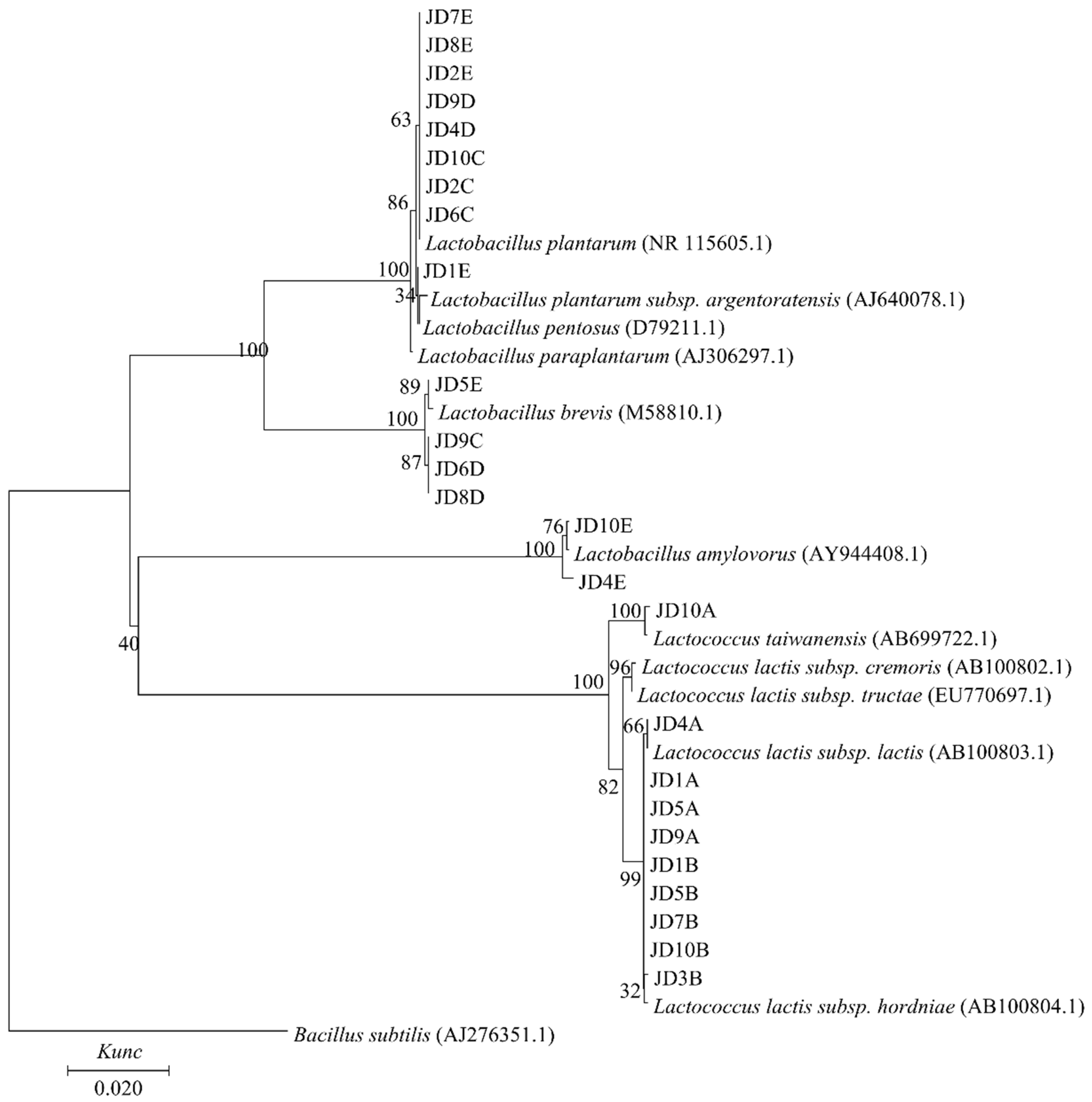
3.4. Isolation, 16S rRNA Gene Sequencing, and Phylogenetic Analysis of LAB
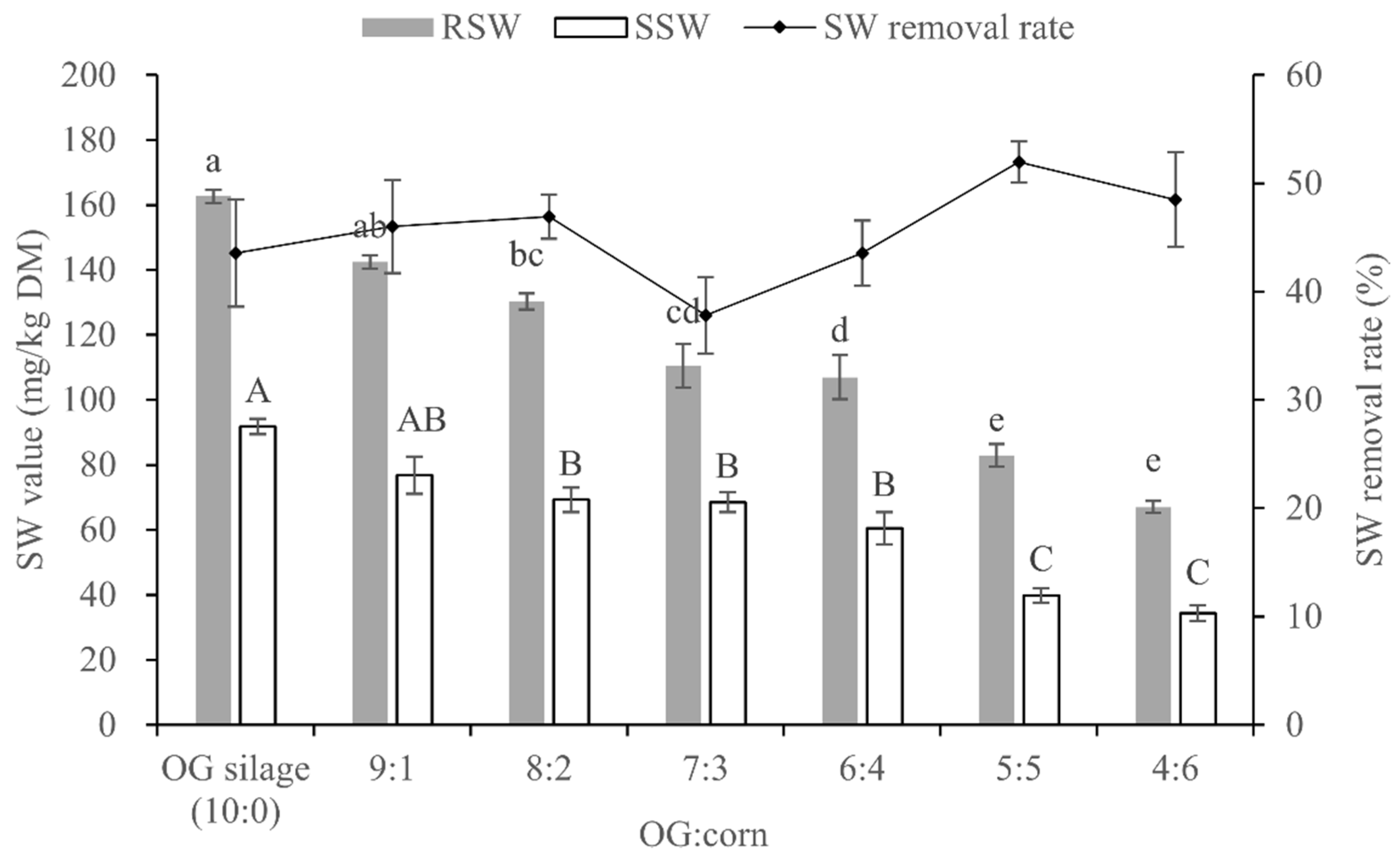
3.5. SW Content and Removal Rate of OG and Whole-Plant Corn Mixture Silages
3.6. The SW Removal Rate in OG Broth by LAB Strains
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wang, S.; Hu, J.; Ali, M.; Ma, C. Analysis on nutrients in Oxytropis glabra in southern Xinjiang. Prat. Sci. 2010, 27, 136–139. [Google Scholar] [CrossRef]
- Lu, P.; Zhao, M.; Han, G. Hazards and utilization of toxicity in Oxytropis glabra. Prat. Sci. 2009, 26, 97–101. [Google Scholar]
- Wang, S.; Tao, D.; Liu, Q.; Jiang, X. Progress on chemical constituents and pharmacological effects of Oxytropis glabra DC. Progr. Vet. Med. 2019, 40, 104–107. [Google Scholar] [CrossRef]
- Colegate, S.M.; Dorling, P.R.; Huxtable, C.R. A spectroscopic investigation of swainsonine: An α-mannosidase inhibitor isolated from swainsona canescens. Aust. J. Chem. 1979, 32. [Google Scholar] [CrossRef]
- Tulsiani, D.R.; Broquist, H.P.; James, L.F.; Touster, O. Production of hybrid glycoproteins and accumulation of oligosaccharides in the brain of sheep and pigs administered swainsonine or locoweed. Arch. Biochem. Biophys. 1988, 264, 607–617. [Google Scholar] [CrossRef]
- Huisden, C.; Szabo, N.; Ogunade, I.; Adesogan, A. Mucuna pruriens detoxification: Effects of ensiling duration and particle size. Anim. Feed Sci. Technol. 2014, 198, 20–27. [Google Scholar] [CrossRef]
- Makkar, H.P.S.; Becker, K. Plant toxins and detoxification methods to improve feed quality of tropical seeds-Review. Asian Austral. J. Anim. 1999, 12, 467–480. [Google Scholar] [CrossRef]
- Ma, Z.X.; Amaro, F.X.; Romero, J.J.; Pereira, O.G.; Jeong, K.C.; Adesogan, A.T. The capacity of silage inoculant bacteria to bind aflatoxin B1 in vitro and in artificially contaminated corn silage. J. Dairy Sci. 2017, 100, 7198–7210. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, T.; Yang, Y.; Zhu, C.; Ding, C.; Dai, C. Binding and detoxification of chlorpyrifos by lactic acid bacteria on rice straw silage fermentation. J. Environ. Health Sci. B 2016, 51, 316–325. [Google Scholar] [CrossRef]
- Nowak, A.; Kuberski, S.; Libudzisz, Z. Probiotic lactic acid bacteria detoxify N-nitrosodimethylamine. Food Addit. Contam. A 2014, 31, 1678–1687. [Google Scholar] [CrossRef]
- Franco, T.; Garcia, S.; Hirooka, E.; Ono, Y.; Dos Santos, J. Lactic acid bacteria in the inhibition of Fusarium graminearum and deoxynivalenol detoxification. J. Appl. Microbiol. 2011, 111, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; He, X.; Wang, J.; Song, Y.; Geng, G.; Wang, J. Biodegradation of Swainsonine by Acinetobacter calcoaceticus strain YLZZ-1 and its isolation and identification. Biodegradation 2009, 20, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; He, X.; Zhang, T.; Wang, J. Isolation and characterization of a Swainsonine degrading bacteria strain Stenotrophomonas maltophilia. Afr. J. Microbiol. Res. 2012, 6, 112–119. [Google Scholar] [CrossRef]
- McDonald, P.; Henderson, N.; Heron, S. The Biochemistry of Silage, 2nd ed.; Chalcombe Publications: Berkshire, UK, 1991; pp. 12, 84–85, 108–111, 167. [Google Scholar]
- Chen, J.; Huang, W.; Zhao, B.; Mo, C. Effect of fermentation time on the quality and swainsonine content of Oxytropis ochrocephala silage. Progr. Vet. Med. 2019, 40, 77–81. [Google Scholar] [CrossRef]
- Mahanna, W.; Chase, L.E. Practical applications and solutions to silage problems. In Silage Science and Technology; Agronomy Monograph No. 42; ASA-CSSA-SSA: Madison, WI, USA, 2003; pp. 855–895. [Google Scholar]
- Serrapica, F.; Uzun, P.; Masucci, F.; Napolitano, F.; Braghieri, A.; Genovese, A.; Sacchi, R.; Romano, R.; Barone, C.M.A.; Di Francia, A. Hay or silage? How the forage preservation method changes the volatile compounds and sensory properties of Caciocavallo cheese. J. Dairy Sci. 2020, 10, 1391–1403. [Google Scholar] [CrossRef]
- Lin, C.; Bolsen, K.; Brent, B.; Fung, D. Epiphytic lactic acid bacteria succession during the pre-ensiling and ensiling periods of alfalfa and maize. J. Appl. Bacteriol. 1992, 73, 375–387. [Google Scholar] [CrossRef]
- Zhang, S.J.; Chaudhry, A.S.; Osman, A.; Shi, C.Q.; Edwards, G.R.; Dewhurst, R.J.; Cheng, L. Associative effects of ensiling mixtures of sweet sorghum and alfalfa on nutritive value, fermentation and methane characteristics. Anim. Feed Sci. Technol. 2015, 206, 29–38. [Google Scholar] [CrossRef]
- Ni, K.; Zhao, J.; Zhu, B.; Su, R.; Pan, Y.; Ma, J.; Zhou, G.; Tao, Y.; Liu, X.; Zhong, J. Assessing the fermentation quality and microbial community of the mixed silage of forage soybean with crop corn or sorghum. Bioresour. Technol. 2018, 265, 563–567. [Google Scholar] [CrossRef]
- Zhu, Y.; Bai, C.S.; Guo, X.S.; Xue, Y.L.; Ataku, K. Nutritive value of corn silage in mixture with vine peas. Anim. Prod. Sci. 2011, 51, 1117–1122. [Google Scholar] [CrossRef]
- Tao, Y.; Sun, Q.; Li, F.; Xu, C.; Cai, Y. Comparative analysis of ensiling characteristics and protein degradation of alfalfa silage prepared with corn or sweet sorghum in semiarid region of Inner Mongolia. Anim. Sci. J. 2020, 91. [Google Scholar] [CrossRef]
- Yang, J.; Tan, H.; Cai, Y. Characteristics of lactic acid bacteria isolates and their effect on silage fermentation of fruit residues. J. Dairy Sci. 2016, 99, 5325–5334. [Google Scholar] [CrossRef] [PubMed]
- Broderick, G.; Kang, J. Automated simultaneous determination of ammonia and total amino acid in ruminal fluid and in vitro media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis, 15th ed.; Association of Official Analytical Chemistry: Arlington, VA, USA, 1990. [Google Scholar]
- Van Soest, P.; Robertson, J.; Lewis, B. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Murphy, R. A method for the extraction of plant samples and the determination of total soluble carbohydrates. J. Sci. Food. Agric. 1958, 9, 714–717. [Google Scholar] [CrossRef]
- Hao, B.; Yang, X.; Wang, X.; Hu, Y.; Liang, J. Extraction and GC study on Swainsonine from locoweed Astragalus strictus in Tibet. Legume Res. 2013, 36, 49–55. [Google Scholar] [CrossRef]
- Cai, Y.; Benno, Y.; Ogaw, M.; Kumai, S. Effect of applying lactic acid bacteria isolated from forage crops on fermentation characteristics and aerobic deterioration of silage. J. Dairy Sci. 1999, 82, 520–526. [Google Scholar] [CrossRef]
- Kimura, M.; Ohta, T. On the stochastic model for estimation of mutational distance between homologous proteins. J. Mol. Evol. 1972, 2, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, J.; Pan, X.; Pei, J.; Zhang, B. Binding of benzo (a) pyrene by Lactobacilli strains. Acta Microbiol. Sin. 2011, 51, 956–964. [Google Scholar] [CrossRef]
- Yu, A.O.; Leveau, J.H.; Marco, M.L. Abundance, diversity and plant-specific adaptations of plant-associated lactic acid bacteria. Environ. Microbiol. Rep. 2020, 12, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Hellings, P.; Bertin, G.; Vanbelle, M. Effect of lactic acid bacteria on silage fermentation. In Proceedings of the 15th International Grassland Congress, Kyotosyppan, Kyoto, Japan, 24 August–7 September 1985; pp. 932–933. [Google Scholar]
- Smith, L.H. Theoretical carbohydrate requirement for alfalfa silage production. Agron. J. 1962, 54, 291–293. [Google Scholar] [CrossRef]
- Chen, L.; Guo, G.; Yuan, X.J.; Zhang, J.; Wen, A.Y.; Sun, X.H.; Shao, T. Effect of ensiling whole crop oat with lucerne in different ratios on fermentation quality, aerobic stability and in vitro digestibility on the Tibetan plateau. J. Anim. Physiol. Anim. Nutr. 2017, 101, e144–e153. [Google Scholar] [CrossRef] [PubMed]
- Pahlow, G.; Muck, R.E.; Driehuis, F.; Oude Elferink, S.J.W.H.; Spoelstra, S.F. Microbiology of ensiling. In Silage Science and Technology; Buxton, D.R., Muck, R.E., Harrison, J.H., Eds.; ASA, CSSA and SSSA: Madison, WI, USA, 2003; pp. 31–94. [Google Scholar]
- Cai, Y. Identification and characterization of Enterococcus species isolated from forage crops and their influence on silage fermentation. J. Dairy Sci. 1999, 82, 2466–2471. [Google Scholar] [CrossRef] [PubMed]
- Teuber, M.; Geis, A. The genus lactococcus. In The Prokaryotes; Bacteria: Firmicutes, Cyanobacteria, 2006; Volume 4, pp. 205–228. [Google Scholar] [CrossRef]
- Schleifer, K.; Kraus, J.; Dvorak, C.; Kilpper-Bälz, R.; Collins, M.; Fischer, W. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst. Appl. Microbiol. 1985, 6, 183–195. [Google Scholar] [CrossRef]
- Nakamura, L. Lactobacillus amylovorus, a new starch-hydrolyzing species from cattle waste-corn fermentations. Int. J. Syst. Evol. Micr. 1981, 31, 56–63. [Google Scholar] [CrossRef]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef]
- Wang, S.; Jia, Q.; Chen, G.; Zhang, L.; Hu, J.; Ma, C. Effect of Oxytropis glabra DC addition on the utilization of silage in karakul rams. Feed Ind. 2011, 32, 55–58. [Google Scholar] [CrossRef]
- Kleiner, D.; Johnston, M. Purification and properties of a secondary alcohol dehydrogenase from the parasitic protozoan Tritrichomonas foetus. J. Biol. Chem. 1985, 260, 8038–8043. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Qiu, K.; Wang, J.; Li, Q. Degradation of swainsonine by the NADP-dependent alcohol dehydrogenase A1R6C3 in Arthrobacter sp. hw08. Toxins 2016, 8, 145. [Google Scholar] [CrossRef]
- Lees, G.; Jago, G. Acetaldehyde: An intermediate in the formation of ethanol from glucose by lactic acid bacteria. J. Dairy Res. 1976, 43, 63–73. [Google Scholar] [CrossRef] [PubMed]
| OG:Corn (Percentage of FM) | Prior to Ensiling | 60 d of Ensiling | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LAB | CB | AB | Yeast | Mold | LAB | CB | AB | Yeast | Mold | |
| OG silage (10:0) | 4.00 b | 5.16 | 6.24 a | 4.15 b | 2.40 | 6.97 | ND | 6.30 | 6.40 | ND |
| 9:1 | 4.64 ab | 5.27 | 6.47 a | 5.38 a | 2.70 | 6.58 | ND | 6.02 | 5.06 | ND |
| 8:2 | 5.45 a | 5.17 | 6.51 a | 5.68 a | 2.55 | 6.28 | ND | 5.80 | 5.56 | ND |
| 7:3 | 5.67 a | 5.25 | 6.65 a | 5.87 a | 2.40 | 6.88 | ND | 6.56 | 5.21 | ND |
| 6:4 | 5.71 a | 4.79 | 6.46 a | 5.78 a | 2.55 | 6.16 | ND | 6.04 | 4.80 | ND |
| 5:5 | 5.56 a | 4.93 | 6.24 a | 5.76 a | 2.40 | 6.31 | ND | 5.98 | 4.86 | ND |
| 4:6 | 5.44 a | 4.81 | 5.15 b | 5.69 a | 2.55 | 6.23 | ND | 5.43 | 5.55 | ND |
| Corn silage (0:10) | 5.64 a | 4.70 | 5.33 b | 6.01 a | 2.85 | 6.24 | ND | 5.41 | 5.80 | ND |
| SEM | 0.162 | 0.104 | 0.143 | 0.152 | 0.048 | 0.101 | - | 0.125 | 0.175 | - |
| Linear | 0.001 | 0.169 | <0.001 | 0.001 | 0.165 | 0.048 | - | 0.061 | 0.493 | - |
| Quadratic | 0.005 | 0.770 | 0.001 | 0.005 | 0.119 | 0.521 | - | 0.299 | 0.029 | - |
| Cubic | 0.145 | 0.708 | 0.325 | 0.010 | 0.031 | 0.749 | - | 0.604 | 0.945 | - |
| OG:Corn (Percentage of FM) | Prior to Ensiling | 60 d of Ensiling | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DM | CP | NDF | ADF | WSC | DM | CP | NDF | ADF | WSC | |
| OG silage (10:0) | 27.11 a | 19.10 a | 43.04 e | 37.36 a | 5.26 c | 26.31 a | 18.17 a | 42.64 e | 36.36 | 1.35 c |
| 9:1 | 27.15 a | 18.55 ab | 45.66 d | 35.91 ab | 5.16 c | 26.42 a | 18.15 a | 43.69 e | 35.18 | 2.21 b |
| 8:2 | 26.22 b | 18.19 b | 46.36 d | 35.14 b | 6.11 bc | 25.07 b | 16.30 b | 45.63 de | 35.78 | 2.23 b |
| 7:3 | 24. 70 c | 17.18 c | 47.13 cd | 36.21 ab | 7.27 b | 24.12 bc | 15.59 bc | 46.85 cd | 35.90 | 2.20 b |
| 6:4 | 23. 97 d | 15.27 d | 48.99 bc | 35.68 ab | 7.24 b | 23.24 cd | 14.45 cd | 49.04 bc | 35.12 | 2.60 b |
| 5:5 | 23.67 e | 13.79 e | 49.15 bc | 36.28 ab | 6.99 b | 22.40 de | 13.51 de | 50.22 b | 36.51 | 2.50 b |
| 4:6 | 23.33 f | 13.51 e | 50.31 b | 35.48 ab | 7.01 b | 22.30 de | 13.01 e | 50.75 b | 34.81 | 2.61 b |
| Corn silage (0:10) | 22.60 g | 8.23 f | 57.73 a | 36.27 ab | 10.31 a | 21.31 e | 8.65 f | 58.33 a | 35.43 | 3.59 a |
| SEM | 0.427 | 0.877 | 1.060 | 0.186 | 0.394 | 0.379 | 0.615 | 0.995 | 0.175 | 0.143 |
| Linear | <0.001 | < 0.001 | <0.001 | 0.167 | <0.001 | <0.001 | <0.001 | <0.001 | 0.224 | <0.001 |
| Quadratic | <0.001 | < 0.001 | <0.001 | 0.020 | 0.008 | 0.361 | <0.001 | 0.001 | 0.924 | 0.469 |
| Cubic | <0.001 | < 0.001 | <0.001 | 0.071 | <0.001 | 0.089 | 0.002 | 0.003 | 0.303 | <0.001 |
| OG: Corn (Percentage of FM) | pH | Lactic Acid | Acetic Acid | Propionic Acid | Butyric Acid | Ammonia-N (% TN) |
|---|---|---|---|---|---|---|
| (% DM) | ||||||
| OG silage (10:0) | 4.46 a | 2.20 | 1.71 | 0.15 a | ND | 4.86 a |
| 9:1 | 4.22 ab | 3.15 | 1.39 | 0.05 b | ND | 4.31 b |
| 8:2 | 4.24 ab | 3.59 | 1.41 | 0.02 b | ND | 4.20 b |
| 7:3 | 4.24 ab | 4.01 | 1.26 | 0.06 b | ND | 4.27 b |
| 6:4 | 4.22 ab | 4.46 | 1.19 | 0.00 b | ND | 4.21 b |
| 5:5 | 4.21 b | 3.79 | 1.47 | 0.00 b | ND | 4.16 b |
| 4:6 | 4.21 b | 4.56 | 1.19 | 0.00 b | ND | 4.06 bc |
| Corn silage (0:10) | 4.24 ab | 3.88 | 1.15 | 0.00 b | ND | 3.71 c |
| SEM | 0.023 | 0.229 | 0.086 | 0.011 | - | 0.072 |
| Linear | 0.022 | 0.013 | 0.199 | <0.001 | - | <0.001 |
| Quadratic | 0.016 | 0.057 | 0.632 | 0.001 | - | 0.373 |
| Cubic | 0.145 | 0.821 | 0.503 | 0.045 | - | <0.001 |
| Source | OG: Corn (Percentage of FM) | Strains | Group |
|---|---|---|---|
| Fresh material | OG (10:0) | JD10B | Lac. lactis subsp. hordniae |
| 9:1 | JD7B | Lac. lactis subsp. hordniae | |
| 8:2 | JD5B | Lac. lactis subsp. hordniae | |
| 7:3 | JD3B | Lac. lactis subsp. hordniae | |
| 6:4 | JD1B | Lac. lactis subsp. hordniae | |
| 5:5 | JD9A | Lac. lactis subsp. hordniae | |
| JD10A | Lac. taiwanensis | ||
| 4:6 | JD5A | Lac. lactis subsp. hordniae | |
| Corn (0:10) | JD1A | Lac. lactis subsp. hordniae | |
| JD4A | Lac. lactis subsp. lactis | ||
| 60 d silage | OG silage (10:0) | JD8E | L. plantarum |
| JD10E | L. amylovorus | ||
| 9:1 | JD4E | L. amylovorus | |
| JD5E | L. brevis | ||
| JD7E | L. plantarum | ||
| 8:2 | JD1E | L. plantarum, L. pentosus, L. plantarum subsp. argentoratensis | |
| JD2E | L. plantarum | ||
| 7:3 | JD8D | L. brevis | |
| JD9D | L. plantarum | ||
| 6:4 | JD4D | L. plantarum | |
| JD6D | L. brevis | ||
| 5:5 | JD9C | L. brevis | |
| JD10C | L. plantarum | ||
| 4:6 | JD6C | L. plantarum | |
| Corn silage (0:10) | JD2C | L. plantarum |
| Source | OG: Corn (Percentage of FM) | Strains | Species | SW Removal Rate |
|---|---|---|---|---|
| Fresh material | 5:5 | JD9A | Lac. lactis subsp. hordniae | 93.35 c |
| 4:6 | JD5A | Lac. lactis subsp. hordniae | 88.10 d | |
| Corn (0:10) | JD4A | Lac. lactis subsp. lactis | 85.31 e | |
| 60 d silage | OG silage (10:0) | JD10E | L. amylovorus | 100.00 a |
| 6:4 | JD6D | L. brevis | 89.12 d | |
| 5:5 | JD9C | L. brevis | 94.96 bc | |
| 8:2 | JD1E | L. plantarum group | 96.72 b | |
| 8:2 | JD2E | L. plantarum | 100.00 a | |
| 6:4 | JD4D | L. plantarum | 100.00 a | |
| 5:5 | JD10C | L. plantarum | 100.00 a | |
| 4:6 | JD6C | L. plantarum | 100.00 a | |
| SEM | - | - | 0.591 | |
| p-value | - | - | <0.001 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, Y.; Niu, D.; Li, F.; Zuo, S.; Sun, Q.; Xu, C. Effects of Ensiling Oxytropis glabra with Whole-Plant Corn at Different Proportions on Fermentation Quality, Alkaloid Swainsonine Content, and Lactic Acid Bacteria Populations. Animals 2020, 10, 1733. https://doi.org/10.3390/ani10101733
Tao Y, Niu D, Li F, Zuo S, Sun Q, Xu C. Effects of Ensiling Oxytropis glabra with Whole-Plant Corn at Different Proportions on Fermentation Quality, Alkaloid Swainsonine Content, and Lactic Acid Bacteria Populations. Animals. 2020; 10(10):1733. https://doi.org/10.3390/ani10101733
Chicago/Turabian StyleTao, Ya, Dongze Niu, Feng Li, Sasa Zuo, Qizhong Sun, and Chuncheng Xu. 2020. "Effects of Ensiling Oxytropis glabra with Whole-Plant Corn at Different Proportions on Fermentation Quality, Alkaloid Swainsonine Content, and Lactic Acid Bacteria Populations" Animals 10, no. 10: 1733. https://doi.org/10.3390/ani10101733
APA StyleTao, Y., Niu, D., Li, F., Zuo, S., Sun, Q., & Xu, C. (2020). Effects of Ensiling Oxytropis glabra with Whole-Plant Corn at Different Proportions on Fermentation Quality, Alkaloid Swainsonine Content, and Lactic Acid Bacteria Populations. Animals, 10(10), 1733. https://doi.org/10.3390/ani10101733




