Naturally Produced Lovastatin Modifies the Histology and Proteome Profile of Goat Skeletal Muscle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Management
2.2. Slaughtering and Sample Collection
2.3. Histology
2.4. Liquid Chromatography Mass Spectrometry
3. Results
3.1. Histology
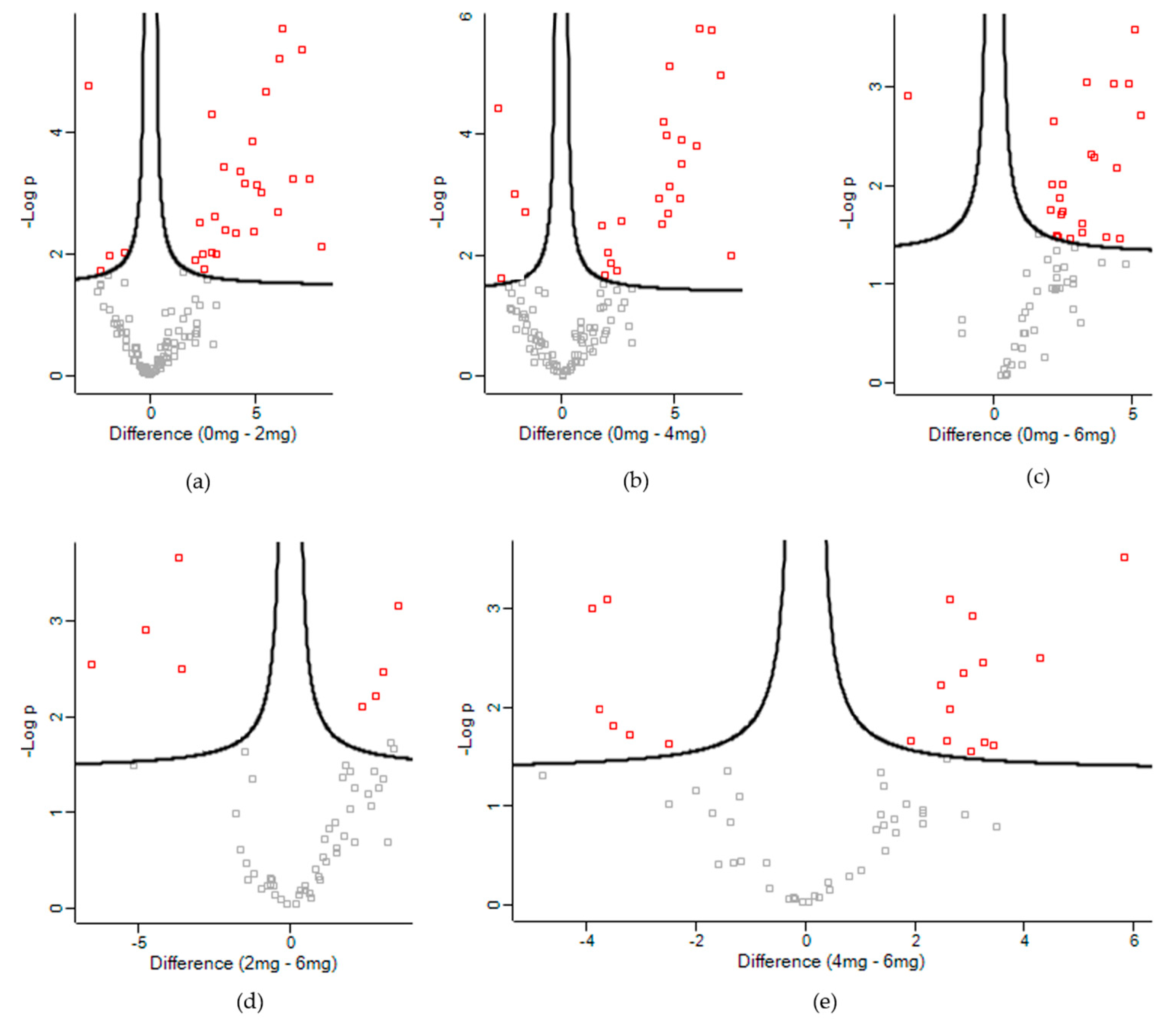
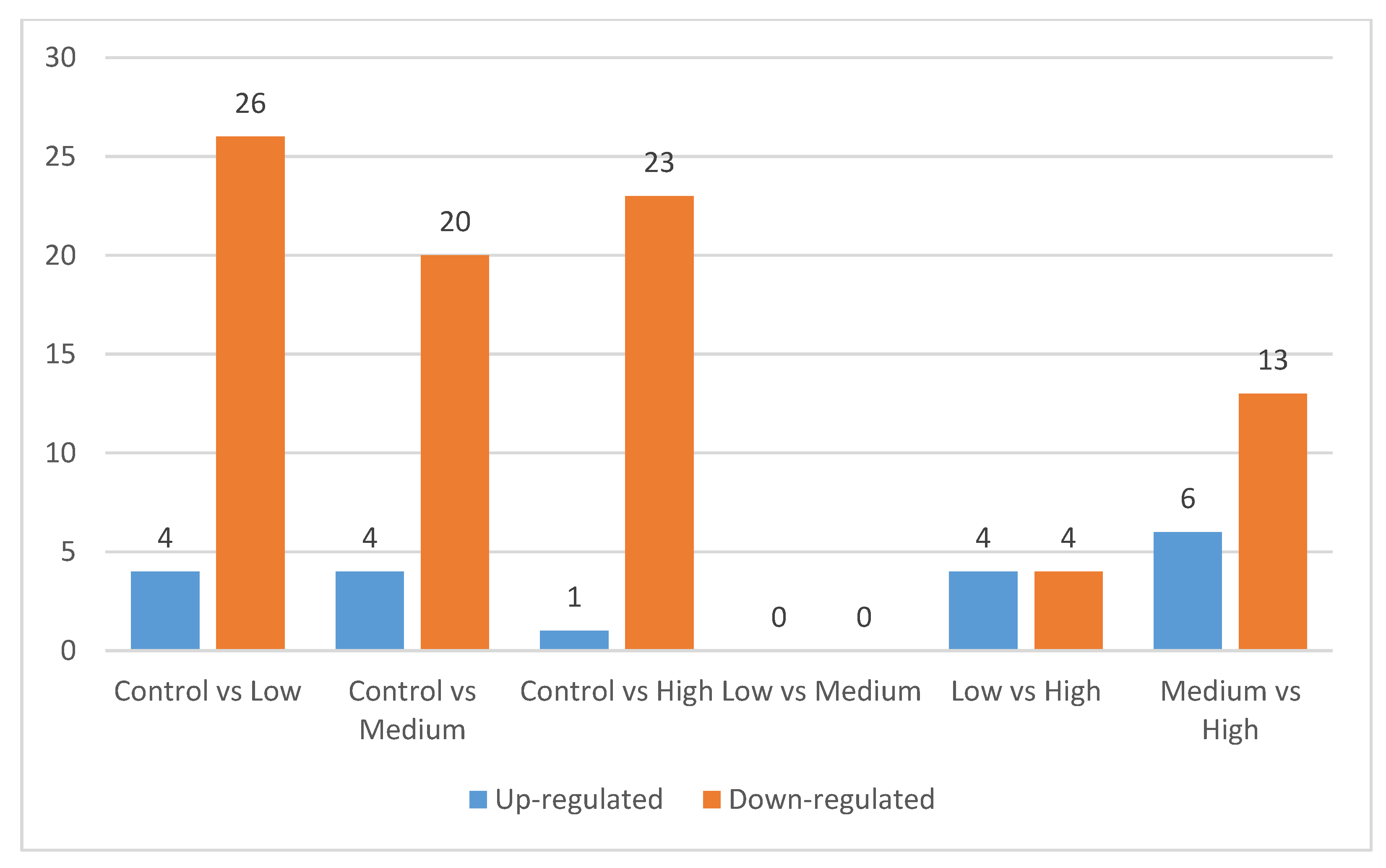
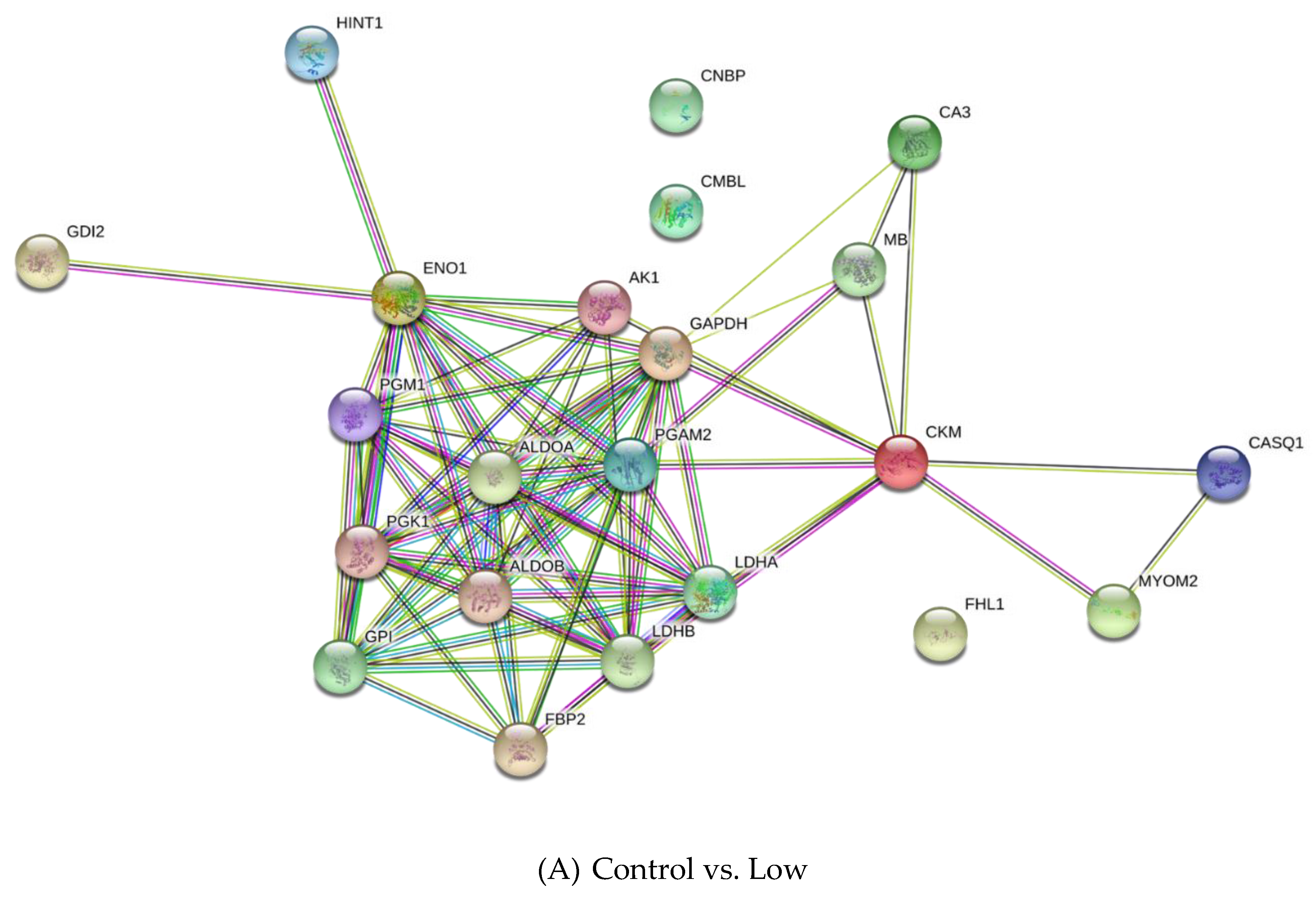
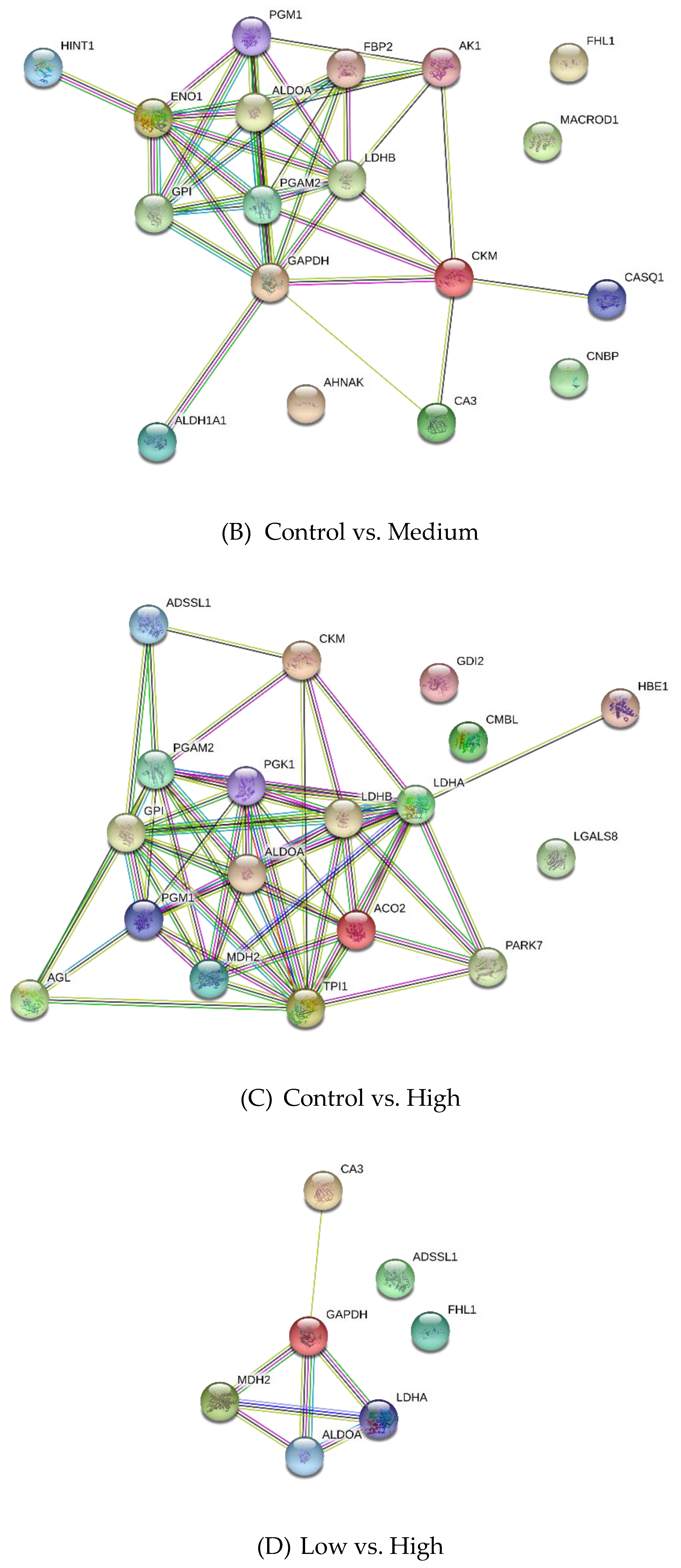
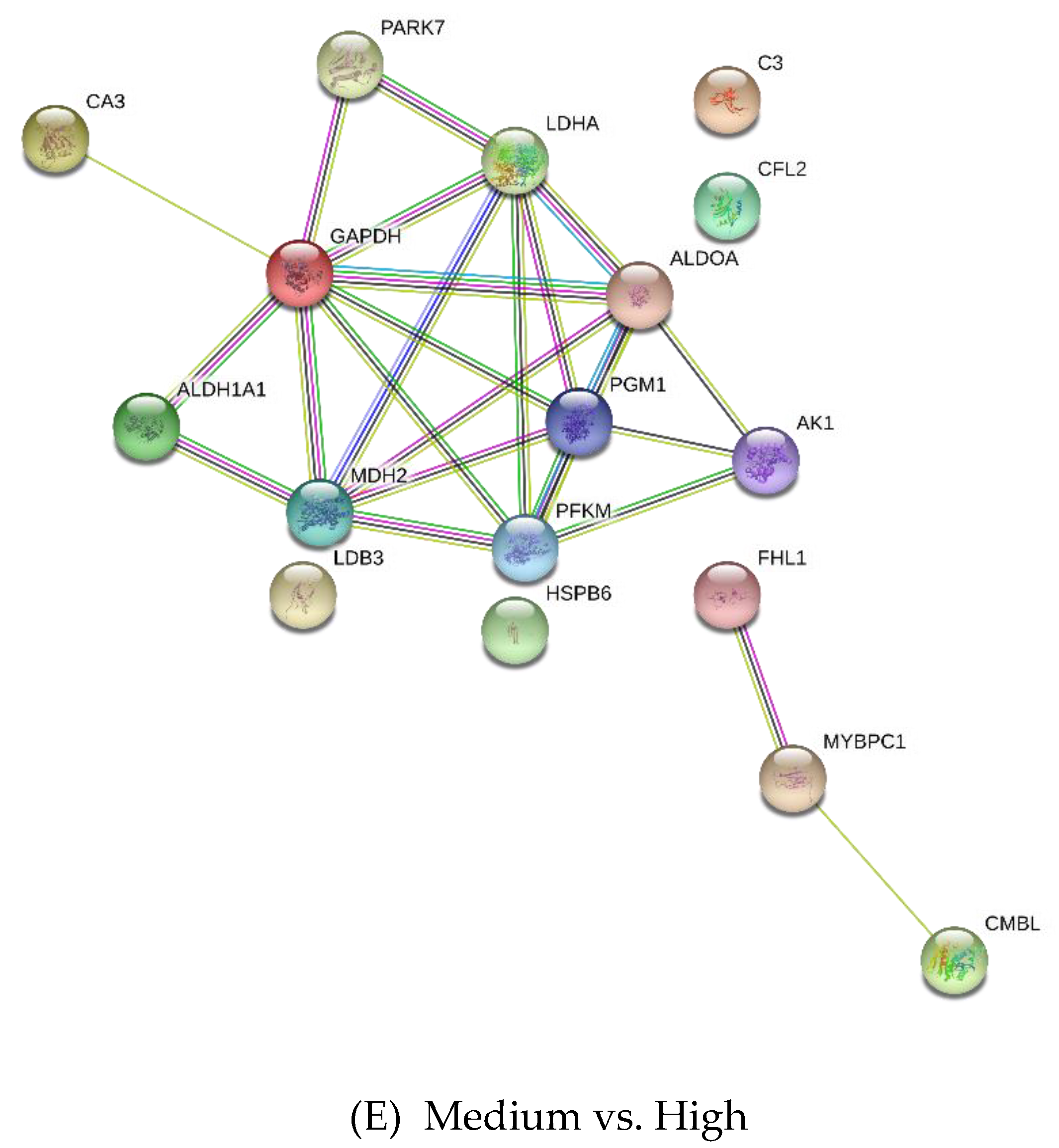
3.2. Differentially Expressed Proteins
4. Discussion
4.1. Histology
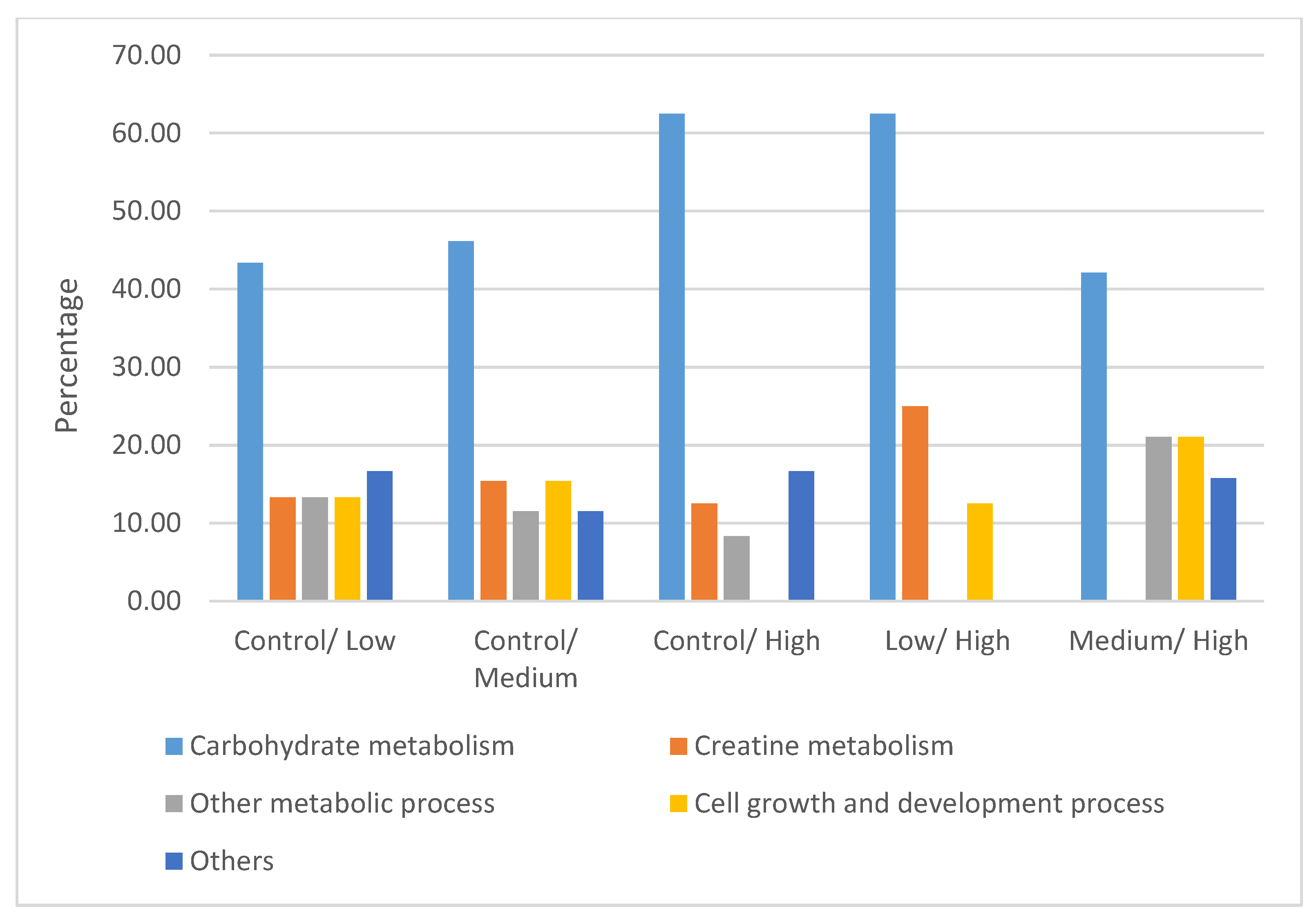
4.2. Differentially Expressed Proteins
4.2.1. Carbohydrate Metabolism
4.2.2. Creatine Metabolism
4.2.3. Other Metabolic Processes
4.2.4. Cell Growth and Development Process
4.2.5. Other Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). Livestock a Major Threat to the Environment: Remedies Urgently Needed. 2006. Available online: www.fao.org/newsroom/en/news/2006/1000448/index.html (accessed on 21 December 2017).
- Hristov, A.N.; Oh, J.; Firkins, J.L.; Dijkstra, J.; Kebreab, E.; Waghorn, G.; Makkar, H.P.S.; Adesogan, A.T.; Yang, W.; Lee, C.; et al. SPECIAL TOPICS—Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options1. J. Anim. Sci. 2013, 91, 5045–5069. [Google Scholar] [CrossRef]
- McAllister, T.A.; Newbold, C.J. Redirecting rumen fermentation to reduce methanogenesis. Aust. J. Exp. Agric. 2008, 48, 7–13. [Google Scholar] [CrossRef]
- Johnson, D.E.; Hill, T.M.; Ward, G.M.; Johnson, K.A.; Branine, M.R.; Carmean, B.R.; Lodman, D.W. Ruminants and other animals. In Atmospheric Methane: Sources, Sinks, and Role in Global Change. NATO ASI Series (Series I: Global Environmental Change); Khalil, M.A.K., Ed.; Springer: Heideberg, Germany, 1993; pp. 199–229. [Google Scholar]
- Elghandour, M.M.Y.; Vázquez, J.C.; Salem, A.Z.M.; Kholif, A.E.; Cipriano, M.M.; Camacho, L.M.; Marquez, O. In Vitro gas and methane production of two mixed rations influenced by three different cultures of Saccharomyces cerevisiae. J. Appl. Anim. Res. 2017, 45, 389–395. [Google Scholar] [CrossRef]
- Salem, A.Z.; Kholif, A.E.A.A.; Elghandour, M.M.; Buendía, G.; Mariezcurrena, M.D.; Hernandez, S.R.; Camacho, L.M. Influence of Oral Administration ofSalix BabylonicaExtract on Milk Production and Composition in Dairy Cows. Ital. J. Anim. Sci. 2014, 13, 2978. [Google Scholar] [CrossRef]
- Wildenauer, F.X.; Blotevogel, K.H.; Winter, J. Effect of monensin and 2-bromoethanesulfonic acid on fatty acid metabolism and methane production from cattle manure. Appl. Microbiol. Biotechnol. 1984, 19, 125–130. [Google Scholar] [CrossRef]
- Martin, S.A. Manipulation of ruminal fermentation with organic acids: A review. J. Anim. Sci. 1998, 76, 3123–3132. [Google Scholar] [CrossRef]
- Dohme, F.; Machmuller, A.; Wasserfallen, A.; Kreuzer, M. Ruminal methanogenesis as influenced by individual fatty acids supplemented to complete ruminant diets. Lett. Appl. Microbiol. 2001, 32, 47–51. [Google Scholar] [CrossRef]
- Lee, S.Y.; Yang, S.H.; Lee, W.S.; Kim, H.S.; Shin, D.E.; Ha, J.K. Effect of 2-Bromoethanesulfonic Acid on In vitro Fermentation Characteristics and Methanogen Population. Asian-Australas. J. Anim. Sci. 2009, 22, 42–48. [Google Scholar] [CrossRef]
- Williams, Y.J.; Popovski, S.; Rea, S.M.; Skillman, L.C.; Toovey, A.F.; Northwood, K.S.; Wright, A.-D.G. A Vaccine against Rumen Methanogens Can Alter the Composition of Archaeal Populations. Appl. Environ. Microbiol. 2009, 75, 1860–1866. [Google Scholar] [CrossRef]
- Mohammed, N.; Ajisaka, N.; Lila, Z.A.; Mikuni, K.; Kanda, S.; Itabashi, H.; Hara, K.; Hara, K. Effect of Japanese horseradish oil on methane production and ruminal fermentation in vitro and in steers1. J. Anim. Sci. 2004, 82, 1839–1846. [Google Scholar] [CrossRef]
- Jahromi, M.F.; Liang, J.B.; Ho, Y.W.; Mohamad, R.; Goh, Y.M.; Shokryazdan, P. Lovastatin Production by Aspergillus terreus Using Agro-Biomass as Substrate in Solid State Fermentation. J. Biomed. Biotechnol. 2012, 2012, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Alberts, A.W.; Chen, J.; Kuron, G.; Hunt, V.; Huff, J.; Hoffman, C.; Rothrock, J.; Lopez, M.; Joshua, H.; Harris, E.; et al. Mevinolin: A highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc. Natl. Acad. Sci. USA 1980, 77, 3957–3961. [Google Scholar] [CrossRef] [PubMed]
- Wolin, M.J.; Miller, T.L. Control of rumen methanogenesis by inhibiting the growth and activity of methanogens with hydroxymethylglutaryl-CoA inhibitors. Int. Congr. Ser. 2006, 1293, 131–137. [Google Scholar] [CrossRef]
- Azlan, P.M.; Jahromi, M.F.; Ariff, M.O.; Ebrahimi, M.; Candyrine, S.C.L.; Liang, J.B. Aspergillus terreus treated rice straw suppresses methane production and enhances feed digestibility in goats. Trop. Anim. Health Prod. 2018, 50, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Restrepo, C.A.; López-Villalobos, N.; Padmanabha, J.; McSweeney, C.; O’Neill, C.J.; O’Neill, C.J. Tropical cattle methane emissions: The role of natural statins supplementation. Anim. Prod. Sci. 2014, 54, 1294–1299. [Google Scholar] [CrossRef]
- Candyrine, S.C.L.; Mahadzir, M.F.; Garba, S.; Jahromi, M.F.; Ebrahimi, M.; Goh, Y.M.; Samsudin, A.A.; Sazili, A.Q.; Chen, W.L.; Ganesh, S.; et al. Effects of naturally-produced lovastatin on feed digestibility, rumen fermentation, microbiota and methane emissions in goats over a 12-week treatment period. PLoS ONE 2018, 13, e0199840. [Google Scholar] [CrossRef]
- Du Souich, P.; Roederer, G.; Dufour, R. Myotoxicity of statins: Mechanism of action. Pharmacol. Ther. 2017, 175, 1–16. [Google Scholar] [CrossRef]
- Camerino, G.M.; Pellegrino, M.A.; Brocca, L.; Digennaro, C.; Camerino, D.C.; Pierno, S.; Bottinelli, R. Statin or fibrate chronic treatment modifies the proteomic profile of rat skeletal muscle. Biochem. Pharmacol. 2011, 81, 1054–1064. [Google Scholar] [CrossRef]
- Department of Standards Malaysia. MS1500:2009, Halal Food-Production, Preparation, Handling and Storage-General Guideline, 1st Revision; Department of Standards Malaysia: Cyberjaya, Malaysia, 2009; pp. 1–13.
- OIE. Guidelines for the Slaughter of Animals; Appendix 3.7.5; Terrestrial Animal Health Code World Organization for Animal Health: Paris, France, 2007. [Google Scholar]
- Gall, A.; Treuting, P.; Elkon, K.B.; Loo, Y.-M.; Gale, M., Jr.; Barber, G.N.; Stetson, D.B. Autoimmunity initiates in non-hematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 2012, 36, 120–131. [Google Scholar] [CrossRef]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004, 117, 399–412. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt. Biochem. 1972, 72, 248–254. [Google Scholar] [CrossRef]
- Huang, D.W.; Lempicki, R.A.; Sherman, B.T. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Bruckert, E.; Hayem, G.; Dejager, S.; Yau, C.; Begaud, B. Mild to Moderate Muscular Symptoms with High-Dosage Statin Therapy in Hyperlipidemic Patients —The PRIMO Study. Cardiovasc. Drugs Ther. 2005, 19, 403–414. [Google Scholar] [CrossRef]
- Norata, G.D.; Tibolla, G.; Catapano, A.L. Statins and skeletal muscles toxicity: From clinical trials to everyday practice. Pharmacol. Res. 2014, 88, 107–113. [Google Scholar] [CrossRef]
- Cao, P.; Hanai, J.-I.; Tanksale, P.; Imamura, S.; Sukhatme, V.P.; Lecker, S.H. Statin-induced muscle damage and atrogin-1 induction is the result of a geranylgeranylation defect. FASEB J. 2009, 23, 2844–2854. [Google Scholar] [CrossRef]
- Westwood, F.R.; Scott, R.C.; Marsden, A.M.; Bigley, A.; Randall, K. Rosuvastatin: Characterization of Induced Myopathy in the Rat. Toxicol. Pathol. 2008, 36, 345–352. [Google Scholar] [CrossRef]
- Kim, Y.S.; Lee, Y.B.; Dalrymple, R.H. Effect of the Repartitioning Agent Cimaterol on Growth, Carcass and Skeletal Muscle Characteristics in Lambs. J. Anim. Sci. 1987, 65, 1392–1399. [Google Scholar] [CrossRef]
- Strydom, P.E.; Frylinck, L.; Montgomery, J.L.; Smith, M.F. The comparison of three β-agonists for growth performance, carcass characteristics and meat quality of feedlot cattle. Meat Sci. 2008, 81, 557–564. [Google Scholar] [CrossRef]
- Hanai, J.-I.; Cao, P.; Tanksale, P.; Imamura, S.; Koshimizu, E.; Zhao, J.; Kishi, S.; Yamashita, M.; Phillips, P.S.; Sukhatme, V.P.; et al. The muscle-specific ubiquitin ligase atrogin-1/MAFbx mediates statin-induced muscle toxicity. J. Clin. Investig. 2007, 117, 3940–3951. [Google Scholar] [CrossRef]
- Maron, D.J.; Fazio, S.; Linton, M.F. Current perspectives on statins. Circulation 2000, 101, 207–213. [Google Scholar] [CrossRef]
- Silva, M.; Matthews, M.L.; Jarvis, C.; Nolan, N.M.; Belliveau, P.; Malloy, M.; Gandhi, P. Meta-analysis of drug-induced adverse events associated with intensive-dose statin therapy. Clin. Ther. 2007, 29, 253–260. [Google Scholar] [CrossRef]
- Jacobson, T.A. Toward “Pain-Free” Statin Prescribing: Clinical Algorithm for Diagnosis and Management of Myalgia. Mayo Clin. Proc. 2008, 83, 687–700. [Google Scholar] [CrossRef]
- DiMauro, S.; Lamperti, C. Muscle glycogenoses. Muscle Nerve 2001, 24, 984–999. [Google Scholar] [CrossRef] [PubMed]
- Gamberi, T.; Fiaschi, T.; Valocchia, E.; Modesti, A.; Mantuano, P.; Rolland, J.-F.; Sanarica, F.; De Luca, A.; Magherini, F. Proteome analysis in dystrophic mdx mouse muscle reveals a drastic alteration of key metabolic and contractile proteins after chronic exercise and the potential modulation by anti-oxidant compounds. J. Proteom. 2018, 170, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Päivä, H.; Thelen, K.M.; Coster, R.; Smet, J.; Paepe, B.; Mattila, K.M.; Laakso, J.; Lehtimäki, T.; Von Bergmann, K.; Lütjohann, D. High-dose statins and skeletal muscle metabolism in humans: A randomized, controlled trial. Clin. Pharmacol. Ther. 2005, 78, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Wallimann, T.; Wyss, M.; Brdiczka, D.; Nicolay, K.; Eppenberger, H.M. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: The ‘phosphocreatine circuit’ for cellular energy homeostasis. Biochem. J. 1992, 281, 21–40. [Google Scholar] [CrossRef]
- Momken, I.; Lechêne, P.; Koulmann, N.; Fortin, D.; Mateo, P.; Doan, B.T.; Hoerter, J.; Bigard, X.; Veksler, V.; Ventura-Clapier, R. Impaired voluntary running capacity of creatine kinase-deficient mice. J. Physiol. 2005, 565, 951–964. [Google Scholar] [CrossRef]
- Liu, M.; Walter, G.A.; Pathare, N.C.; Forster, R.E.; Vandenborne, K. A quantitative study of bioenergetics in skeletal muscle lacking carbonic anhydrase III using 31P magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 371–376. [Google Scholar] [CrossRef]
- Räisänen, S.R.; Lehenkari, P.; Tasanen, M.; Rahkila, P.; Härkönen, P.L.; Väänänen, H.K. Carbonic anhydrase III protects cells from hydrogen peroxide-induced apoptosis. FASEB J. 1999, 13, 513–522. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Prill, K.; Carlisle, C.; Stannard, M.; Reid, P.J.W.; Pilgrim, D.B. Myomesin is part of an integrity pathway that responds to sarcomere damage and disease. PLoS ONE 2019, 14, e0224206. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.L.; Li, A.; Mun, J.Y.; Previs, M.J.; Previs, S.B.; Campbell, S.G.; dos Remedios, C.G.; Tombe, P.P.; Craig, R.; Warshaw, D.M.; et al. Skeletal myosin binding protein-C isoforms regulate thin filament activity in a Ca2+-dependent manner. Sci. Rep. 2018, 8, 2604. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.; Gupton, S.L. Chapter three—Building blocks of functioning brain: Cytoskeletal dynamics in neuronal development. Int. Rev. Cel. Mol. Bio. 2016, 322, 183–245. [Google Scholar]
- Agrawal, P.B.; Greenleaf, R.S.; Tomczak, K.K.; Lehtokari, V.; Wallgren-Pettersson, C.; Wallefeld, W.; Laing, N.G.; Darras, B.T.; Maciver, S.K.; Cormitzer, P.R.; et al. Nemaline myopathy with minicores caused by mutation of the CFL2 gene encoding the skeletal muscle actin-binding protein, cofilin-2. Am. J. Hum. Genet. 2007, 80, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Doran, P.; Dowling, P.; Lohan, J.; Mcdonnell, K.; Poetsch, S.; Ohlendieck, K. Subproteomics analysis of Ca2+-binding proteins demonstrates decreased calsequestrin expression in dystrophic mouse skeletal muscle. Eur. J. Biochem. 2004, 271, 3943–3952. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Stock, L.; Schneider-Gold, C.; Sommer, C.; Timchenko, N.A.; Timchenko, L. Reduction of Cellular Nucleic Acid Binding Protein Encoded by a Myotonic Dystrophy Type 2 Gene Causes Muscle Atrophy. Mol. Cell. Boil. 2018, 38, e00649-17. [Google Scholar] [CrossRef] [PubMed]
- Bächner, D.; Korn, B.; Hameister, H.; Poustka, A.; Sedlacek, Z. Expression patterns of two human genes coding for different rab GDP-dissociation inhibitors (GDIs), extremely conserved proteins involved in cellular transport. Hum. Mol. Genet. 1995, 4, 701–708. [Google Scholar] [CrossRef]
- Kuwabara, I.; Sano, H.; Liu, F.-T. Functions of Galectins in Cell Adhesion and Chemotaxis. Methods Enzymol. 2003, 363, 532–552. [Google Scholar]
- Clements, C.M.; McNally, R.S.; Conti, B.J.; Mak, T.W.; Ting, J.P. DJ-1, a cancer and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. USA 2006, 103, 15091–15096. [Google Scholar] [CrossRef]
- Williams, R.S.; Neufer, P.D. Regulation of Gene Expression in Skeletal Muscle by Contractile Activity. In The Handbook of Physiology: Integration of Motor, Circulatory, Respiratory and Metabolic Control during Exercise; Rowell, L.B., Shepard, J.T., Eds.; American Physiological Society: Bethesda, MD, USA, 1996; pp. 1124–1150. [Google Scholar]
- Powers, S.K.; Smuder, A.; Judge, A. Oxidative stress and disuse muscle atrophy: Cause or consequence? Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 240–245. [Google Scholar] [CrossRef]

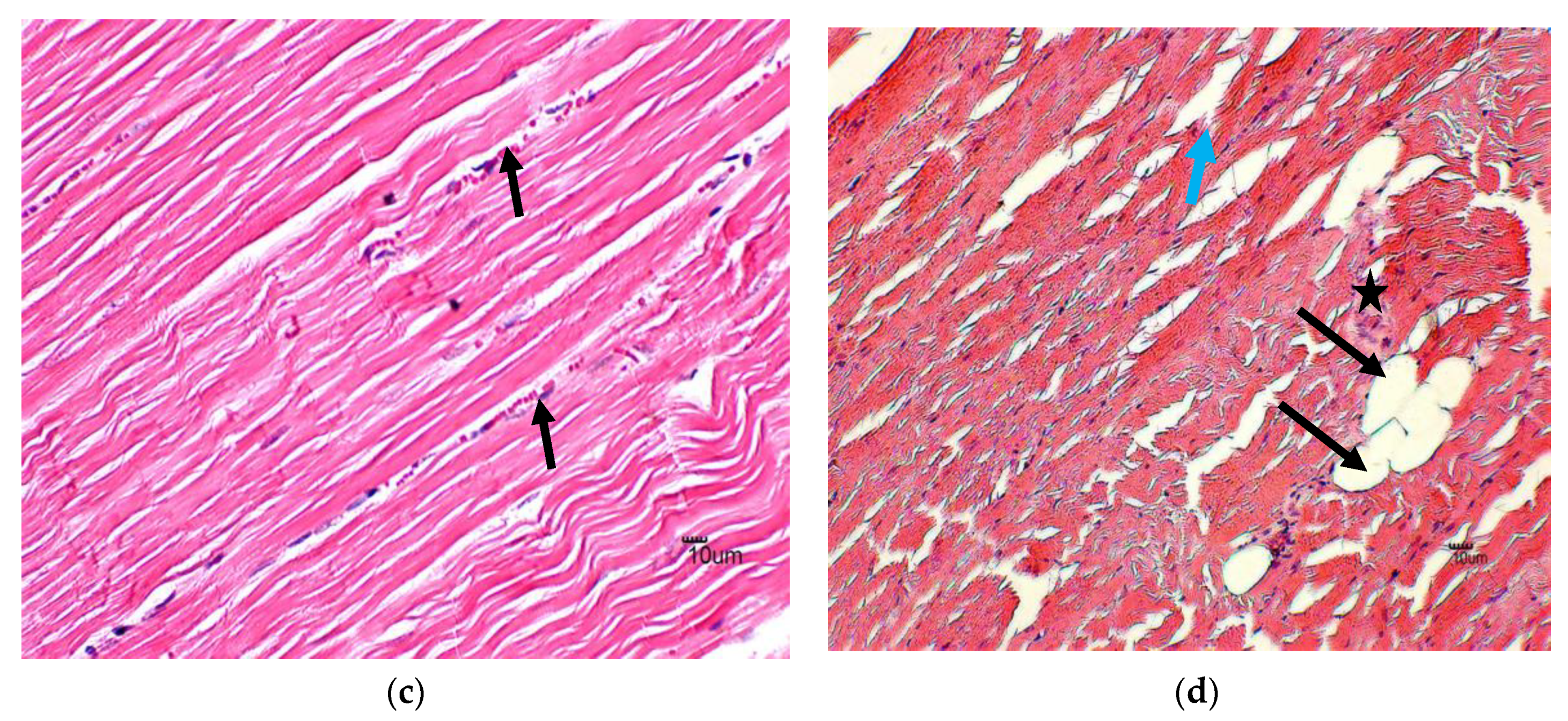

| Score | Histopathologic Injury | Percentage |
|---|---|---|
| 0 | Normal | 0% |
| 1 | Minimal | <15% |
| 2 | Mild | ≤25% |
| 3 | Moderate | ≤40% |
| 4 | Marked | ≥50 |
| Parameters | Control | Low | Medium | High | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | Min | Max | Median | Min | Max | Median | Min | Max | Median | Min | Max | |
| Necrosis | 0 a | 0 | 0 | 0 a | 0 | 0 | 1 b | 0 | 2 | 3 | 1 | 4 |
| Degeneration | 0 a | 0 | 0 | 0 b | 0 | 1 | 0 b | 0 | 2 | 2 | 0 | 3 |
| Interstitial space | 0 a | 0 | 0 | 0 a | 0 | 0 | 1 b | 0 | 3 | 1 b | 0 | 3 |
| Vacuolization | 0 a | 0 | 0 | 0 a | 0 | 0 | 1 b | 0 | 2 | 1 b | 0 | 3 |
| UniProt Accession | Description | -Log p-Value | Difference 1 |
|---|---|---|---|
| Control vs. Low | |||
| Q9XSC6 | Creatine kinase M-type | 5.68 | −6.27 |
| P06733 | Alpha-enolase | 5.20 | −6.08 |
| P54296 | Myomesin-2 | 4.75 | 2.93 |
| P00559 | Phosphoglycerate kinase 1 | 4.66 | −5.45 |
| P62633 | Cellular nucleic acid-binding protein | 4.28 | −2.88 |
| P02191 | Myoglobin | 3.83 | −4.87 |
| Q5S1S4 | Carbonic anhydrase 3 | 3.42 | −3.45 |
| P16290 | Phosphoglycerate mutase 2 | 3.34 | −4.27 |
| Q5E9B1 | L-lactate dehydrogenase B chain | 3.22 | −7.52 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 3.22 | −6.75 |
| Control vs. Medium | |||
| P15259 | Phosphoglycerate mutase 2 | 5.72 | −6.11 |
| P06733 | Alpha-enolase | 5.72 | −6.66 |
| P05065 | Fructose-bisphosphate aldolase A | 5.10 | −4.79 |
| P36871 | Phosphoglucomutase-1 | 4.95 | −7.02 |
| P62633 | Cellular nucleic acid-binding protein | 4.43 | 2.80 |
| P06732 | Creatine kinase M-type | 4.20 | −4.55 |
| Q13642 | Four and a half LIM domains protein 1 | 3.97 | −4.63 |
| P06744 | Glucose-6-phosphate isomerase | 3.90 | −5.34 |
| P00571 | Adenylate kinase isoenzyme 1 | 3.49 | −5.31 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 3.12 | −4.77 |
| Control vs. High | |||
| P19858 | L-lactate dehydrogenase A chain | 3.57 | −5.10 |
| P13707 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | 3.05 | −3.36 |
| Q1KZF3 | Beta A globin chain | 3.04 | −4.30 |
| Q96DG6 | Carboxymethylenebutenolidase homolog | 3.04 | −4.85 |
| P50397 | Rab GDP dissociation inhibitor beta | 2.91 | 3.13 |
| P36871 | Phosphoglucomutase-1 | 2.71 | −5.31 |
| I1 × 3V1 | Galectin | 2.65 | −2.16 |
| P00559 | Phosphoglycerate kinase 1 | 2.32 | −3.53 |
| Q5E9B1 | L-lactate dehydrogenase B chain | 2.18 | −4.43 |
| P06732 | Creatine kinase M-type | 2.02 | −2.50 |
| Low vs. High | |||
| Q5S1S4 | Carbonic anhydrase 3 | 3.65 | 3.63 |
| P19858 | L-lactate dehydrogenase A chain | 3.15 | −3.54 |
| P00883 | Fructose-bisphosphate aldolase A | 2.91 | 4.71 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 2.54 | 6.5 |
| Q13642 | Four and a half LIM domains protein 1 | 2.5 | 3.55 |
| K0J107 | Malate dehydrogenase, mitochondrial | 2.47 | −3.06 |
| A4Z6H0 | Adenylosuccinate synthetase isozyme 1 | 2.21 | −2.8 |
| P13707 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | 2.11 | −2.36 |
| Medium vs. High | |||
| Q96DG6 | Carboxymethylenebutenolidase homolog | 3.53 | −5.82 |
| Q5S1S4 | Carbonic anhydrase 3 | 3.09 | 3.63 |
| Q00872 | Myosin-binding protein C, slow-type | 3.09 | −2.64 |
| Q13642 | Four and a half LIM domains protein 1 | 3.01 | 3.9 |
| K0J107 | Malate dehydrogenase, mitochondrial | 2.93 | −3.06 |
| P48644 | Retinal dehydrogenase 1 | 2.5 | −4.3 |
| P19858 | L-lactate dehydrogenase A chain | 2.46 | −3.26 |
| Q148F1 | Cofilin-2 | 2.35 | −2.89 |
| P13707 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | 2.23 | −2.49 |
| Q99497 | Protein DJ-1 | 1.99 | −2.63 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leo, T.K.; Garba, S.; Abubakar, D.; Sazili, A.Q.; Candyrine, S.C.L.; Jahromi, M.F.; Goh, Y.M.; Ronimus, R.; Muetzel, S.; Liang, J.B. Naturally Produced Lovastatin Modifies the Histology and Proteome Profile of Goat Skeletal Muscle. Animals 2020, 10, 72. https://doi.org/10.3390/ani10010072
Leo TK, Garba S, Abubakar D, Sazili AQ, Candyrine SCL, Jahromi MF, Goh YM, Ronimus R, Muetzel S, Liang JB. Naturally Produced Lovastatin Modifies the Histology and Proteome Profile of Goat Skeletal Muscle. Animals. 2020; 10(1):72. https://doi.org/10.3390/ani10010072
Chicago/Turabian StyleLeo, Teik Kee, Sani Garba, Danmaigoro Abubakar, Awis Qurni Sazili, Su Chui Len Candyrine, Mohammad Faseleh Jahromi, Yong Meng Goh, Ron Ronimus, Stefan Muetzel, and Juan Boo Liang. 2020. "Naturally Produced Lovastatin Modifies the Histology and Proteome Profile of Goat Skeletal Muscle" Animals 10, no. 1: 72. https://doi.org/10.3390/ani10010072
APA StyleLeo, T. K., Garba, S., Abubakar, D., Sazili, A. Q., Candyrine, S. C. L., Jahromi, M. F., Goh, Y. M., Ronimus, R., Muetzel, S., & Liang, J. B. (2020). Naturally Produced Lovastatin Modifies the Histology and Proteome Profile of Goat Skeletal Muscle. Animals, 10(1), 72. https://doi.org/10.3390/ani10010072






