Effect of Soybean Oil and Fish Oil on Lipid-Related Transcripts in Subcutaneous Adipose Tissue of Dairy Cows
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Experimental Diets, and Tissue Sampling
2.2. Plasma Samples and Fatty Acid Analysis
2.3. Biopsies, RNA Extraction, and Reverse Transcription Quantitative Polymerase Chain Reaction (RTqPCR)
2.4. Statistical Analysis
3. Results
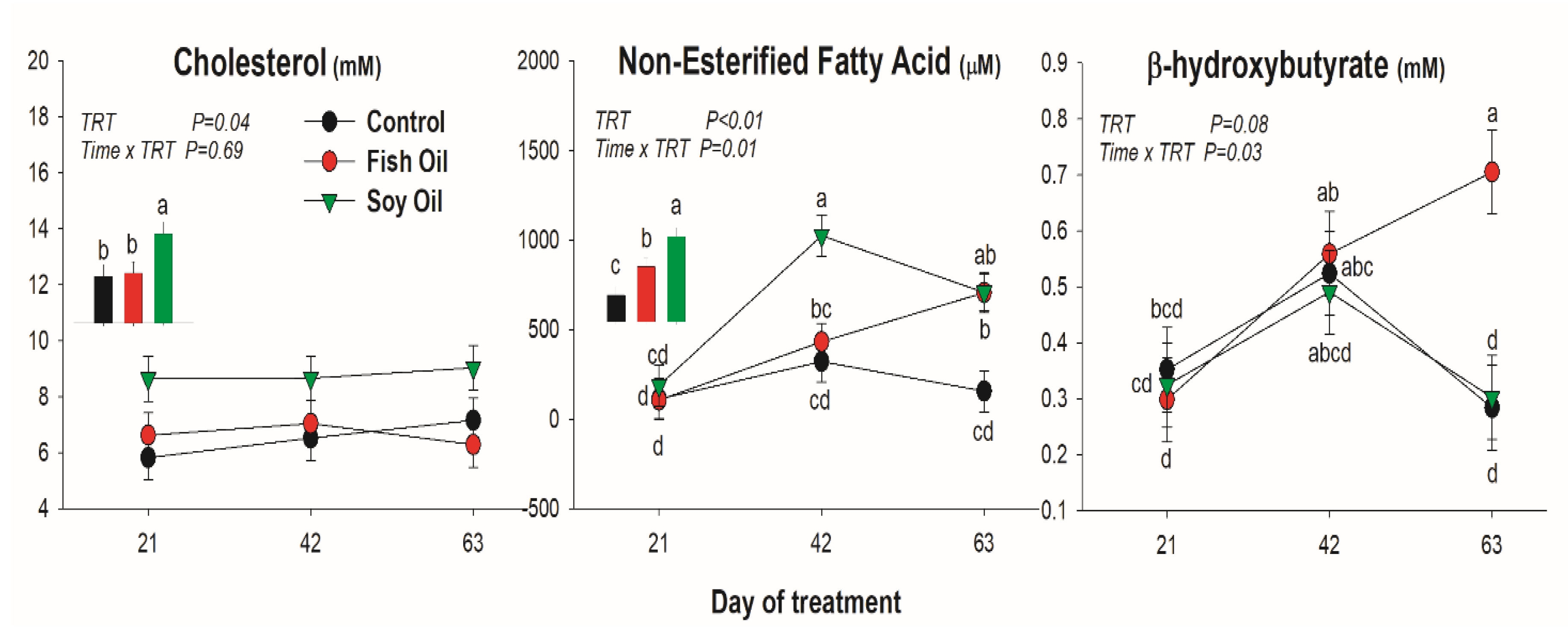
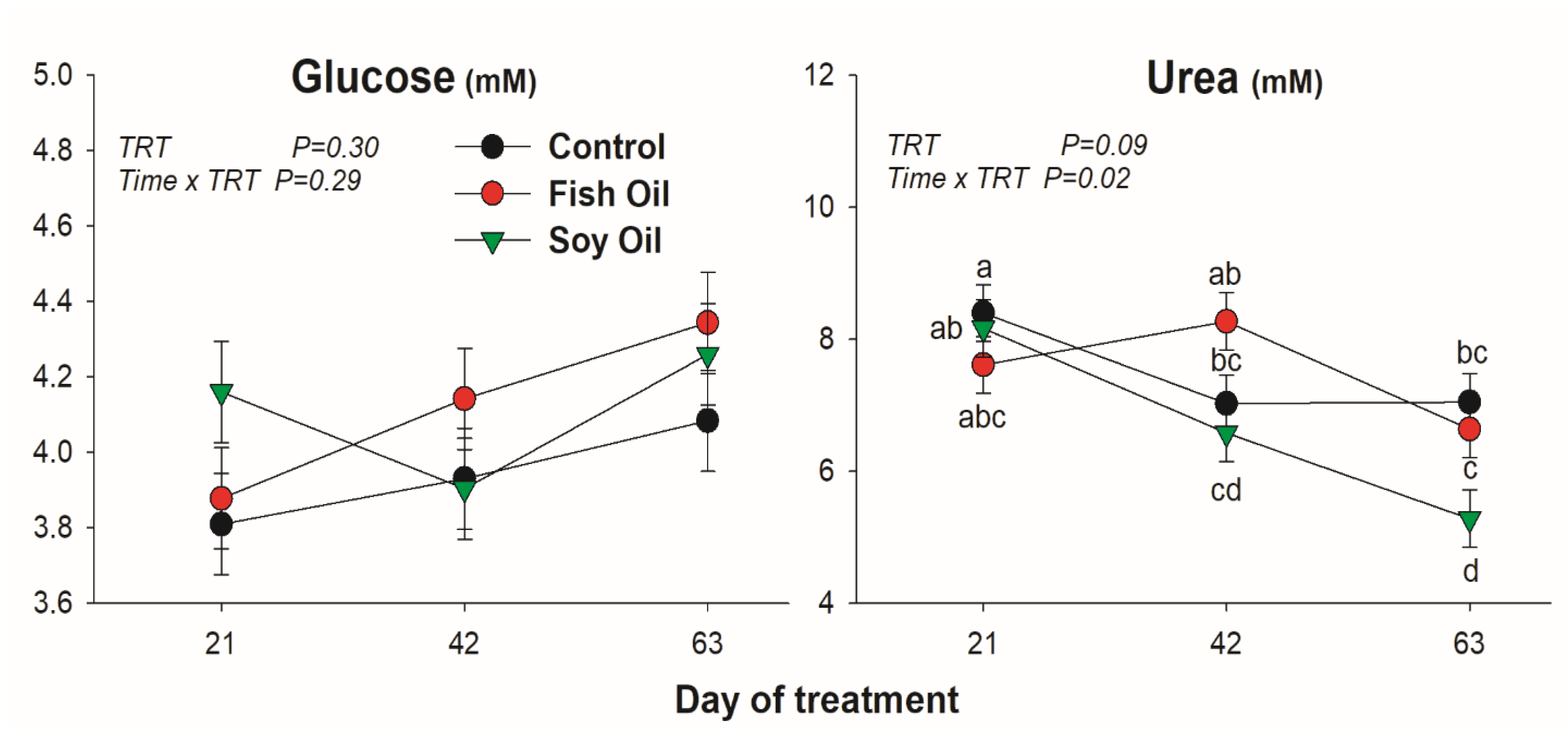
3.1. Blood Metabolites
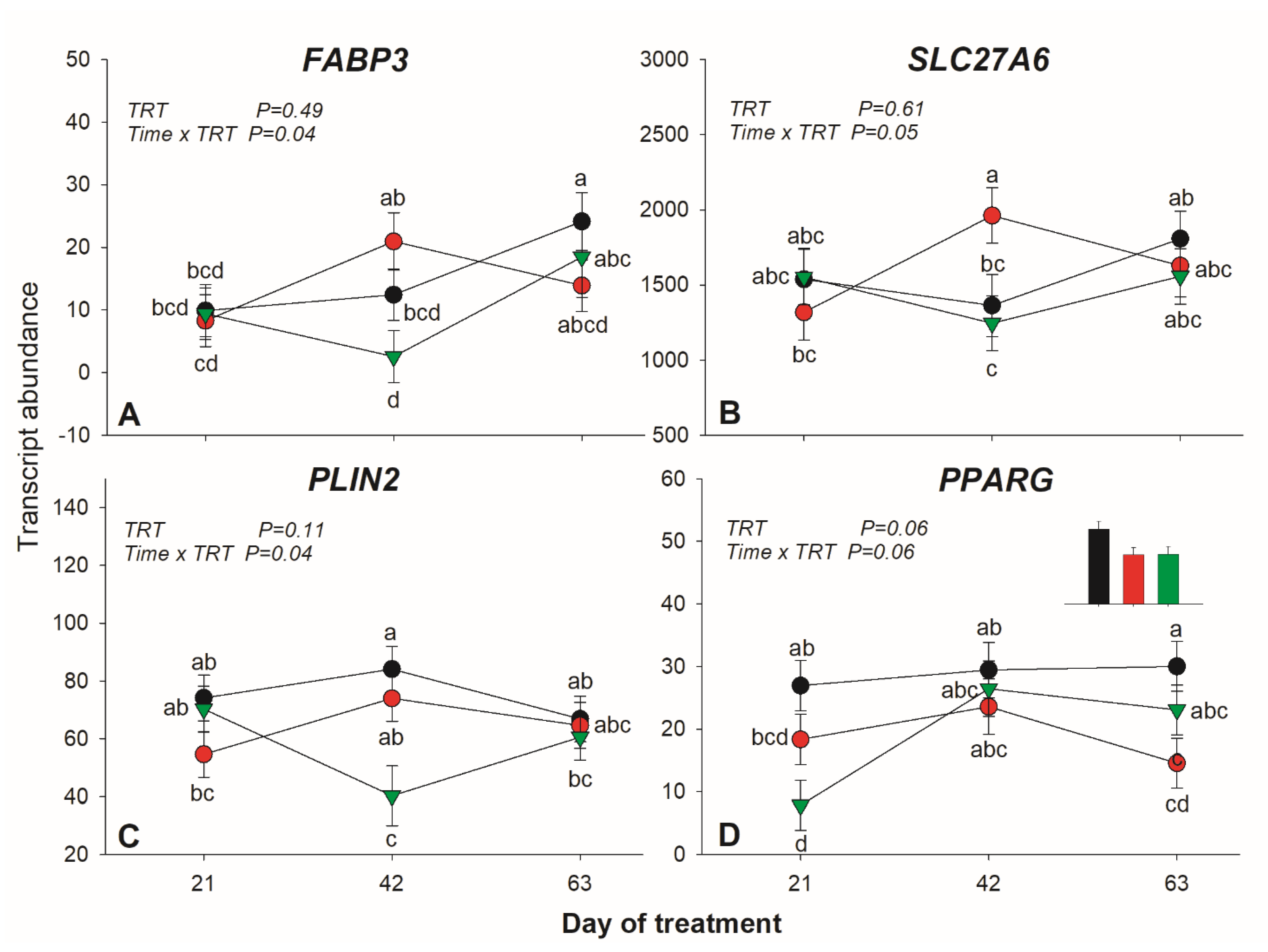
3.2. Transcription
3.2.1. Transcripts Related to Fatty Acid Transport and Activation
3.2.2. De Novo Synthesis and Fatty Acid Desaturation
3.2.3. Triacylglycerol Synthesis and Lipid Droplet Formation
3.2.4. Transcription Regulation
4. Discussion
4.1. Dairy Cow’s Performance and Milk Traits
4.2. Blood Metabolites
4.3. Effects of Dietary SO and FO on Lipid-Related Genes in Subcutaneous Adipose Tissue
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kliem, K.E.; Humphries, D.J.; Kirton, P.; Givens, D.I.; Reynolds, C.K. Differential effects of oilseed supplements on methane production and milk fatty acid concentrations in dairy cows. Animal 2019, 13, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bello-Perez, E.; Fehrmann-Cartes, K.; Iniguez-Gonzalez, G.; Toro-Mujica, P.; Garnsworthy, P.C. Short communication: Chemical composition, fatty acid composition, and sensory characteristics of Chanco cheese from dairy cows supplemented with soybean and hydrogenated vegetable oils. J. Dairy Sci. 2015, 98, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bello-Perez, E.; Iniguez-Gonzalez, G.; Fehrmann-Cartes, K.; Toro-Mujica, P.; Garnsworthy, P.C. Influence of fish oil alone or in combination with hydrogenated palm oil on sensory characteristics and fatty acid composition of bovine cheese. Anim. Feed Sci. Tech. 2015, 205, 60–68. [Google Scholar] [CrossRef]
- Castro-Carrera, T.; Frutos, P.; Leroux, C.; Chilliard, Y.; Hervas, G.; Belenguer, A.; Bernard, L.; Toral, P.G. Dietary sunflower oil modulates milk fatty acid composition without major changes in adipose and mammary tissue fatty acid profile or related gene mRNA abundance in sheep. Animal 2015, 9, 582–591. [Google Scholar] [CrossRef] [Green Version]
- Ibeagha-Awemu, E.M.; Li, R.; Ammah, A.A.; Dudemaine, P.L.; Bissonnette, N.; Benchaar, C.; Zhao, X. Transcriptome adaptation of the bovine mammary gland to diets rich in unsaturated fatty acids shows greater impact of linseed oil over safflower oil on gene expression and metabolic pathways. BMC Genom. 2016, 17, 104. [Google Scholar] [CrossRef] [Green Version]
- Vargas-Bello-Pérez, E.; Zhao, W.; Bionaz, M.; Luo, J.; Loor, J.J. Nutrigenomic Effect of Saturated and Unsaturated Long Chain Fatty Acids on Lipid-Related Genes in Goat Mammary Epithelial Cells: What Is the Role of PPARγ? Vet. Sci. 2019, 6, 54. [Google Scholar] [CrossRef] [Green Version]
- Vargas-Bello-Pérez, E.; Bionaz, M.; Sciarresi-Arechabala, P.; Cancino-Padilla, N.; Morales, M.S.; Romero, J.; Leskinen, H.; Garnsworthy, P.C.; Loor, J.J. Long-Term Effects of Dietary Olive Oil and Hydrogenated Vegetable Oil on Expression of Lipogenic Genes in Subcutaneous Adipose Tissue of Dairy Cows. Vet. Sci. 2019, 6, 74. [Google Scholar] [CrossRef] [Green Version]
- Harvatine, K.J.; Perfield, J.W.; Bauman, D.E. Expression of Enzymes and Key Regulators of Lipid Synthesis Is Upregulated in Adipose Tissue during CLA-Induced Milk Fat Depression in Dairy Cows. J. Nutr. 2009, 139, 849–854. [Google Scholar] [CrossRef] [Green Version]
- Thering, B.J.; Graugnard, D.E.; Piantoni, P.; Loor, J.J. Adipose tissue lipogenic gene networks due to lipid feeding and milk fat depression in lactating cows. J. Dairy Sci. 2009, 92, 4290–4300. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, E.; Ballou, M.A.; Correa, M.N.; DePeters, E.J.; Drackley, J.K.; Loor, J.J. Dietary lipid during the transition period to manipulate subcutaneous adipose tissue peroxisome proliferator-activated receptor-gamma co-regulator and target gene expression. J. Dairy Sci. 2011, 94, 5913–5925. [Google Scholar] [CrossRef] [Green Version]
- Bionaz, M.; Osorio, J.; Loor, J.J. Triennial Lactation Symposium: Nutrigenomics in dairy cows: Nutrients, transcription factors, and techniques. J. Anim. Sci. 2015, 93, 5531–5553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas-Bello-Perez, E.; Cancino-Padilla, N.; Geldsetzer-Mendoza, C.; Vyhmeister, S.; Morales, M.S.; Leskinen, H.; Romero, J.; Garnsworthy, P.C.; Ibanez, R.A. Effect of Feeding Cows with Unsaturated Fatty Acid Sources on Milk Production, Milk Composition, Milk Fatty Acid Profile, and Physicochemical and Sensory Characteristics of Ice Cream. Animals 2019, 9, 568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [Green Version]
- Rosa, F.; Osorio, J.S.; Trevisi, E.; Yanqui-Rivera, F.; Estill, C.T.; Bionaz, M. 2,4-Thiazolidinedione Treatment Improves the Innate Immune Response in Dairy Goats with Induced Subclinical Mastitis. PPAR Res. 2017. [Google Scholar] [CrossRef] [Green Version]
- Leskinen, H.; Ventto, L.; Kairenius, P.; Shingfield, K.J.; Vilkki, J. Temporal changes in milk fatty acid composition during diet-induced milk fat depression in lactating cows. J. Dairy Sci. 2019, 102, 5148–5160. [Google Scholar] [CrossRef]
- Mathews, A.T.; Rico, J.E.; Sprenkle, N.T.; Lock, A.L.; McFadden, J.W. Increasing palmitic acid intake enhances milk production and prevents glucose-stimulated fatty acid disappearance without modifying systemic glucose tolerance in mid-lactation dairy cows. J. Dairy Sci. 2016, 99, 8802–8816. [Google Scholar] [CrossRef] [Green Version]
- Castro, T.; Martinez, D.; Isabel, B.; Cabezas, A.; Jimeno, V. Vegetable Oils Rich in Polyunsaturated Fatty Acids Supplementation of Dairy Cows’ Diets: Effects on Productive and Reproductive Performance. Animals 2019, 9, 205. [Google Scholar] [CrossRef] [Green Version]
- Shearer, G.C.; Savinova, O.V.; Harris, W.S. Fish oil—How does it reduce plasma triglycerides? Biochim. Biophys. Acta 2012, 1821, 843–851. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.R.; Oguejiofor, C.F.; Swangchan-Uthai, T.; Carr, S.; Wathes, D.C. Relationships between Circulating Urea Concentrations and Endometrial Function in Postpartum Dairy Cows. Animals 2015, 5, 748–773. [Google Scholar] [CrossRef] [PubMed]
- Laven, R.A.; Scaramuzzi, R.J.; Wathes, D.C.; Peters, A.R.; Parkinson, T.J. Recent research on the effects of excess dietary nitrogen on the fertility of dairy cows. Vet. Rec. 2007, 160, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.D.; Muino, R.; Pereira, V.; Campos, R.; Benedito, J.L. Relationship among blood indicators of lipomobilization and hepatic function during early lactation in high-yielding dairy cows. J. Vet. Sci. 2011, 12, 251–255. [Google Scholar] [CrossRef] [Green Version]
- McPherson, R.; Gauthier, A. Molecular regulation of SREBP function: The Insig-SCAP connection and isoform-specific modulation of lipid synthesis. Biochem. Cell Biol. 2004, 82, 201–211. [Google Scholar] [CrossRef]
- Strable, M.S.; Ntambi, J.M. Genetic control of de novo lipogenesis: Role in diet-induced obesity. Crit. Rev. Biochem. Mol. 2010, 45, 199–214. [Google Scholar] [CrossRef] [Green Version]
- Brownsey, R.W.; Boone, A.N.; Elliott, J.E.; Kulpa, J.E.; Lee, W.M. Regulation of acetyl-CoA carboxylase. Biochem. Soc. Trans. 2006, 34, 223–227. [Google Scholar] [CrossRef]
| Ingredient Composition (% DM) | Diet | ||
|---|---|---|---|
| Control | SO | FO | |
| Corn silage | 32.0 | 31.1 | 31.1 |
| Fresh alfalfa | 24.0 | 23.3 | 23.3 |
| Malt distillers | 19.2 | 18.6 | 18.6 |
| Corn grain | 7.6 | 7.4 | 7.4 |
| Canola meal | 6.2 | 6.0 | 6.0 |
| Alfalfa hay | 5.0 | 4.9 | 4.9 |
| Soybean grain | 4.0 | 3.9 | 3.9 |
| Wheat bran | 1.6 | 1.6 | 1.6 |
| Vitamin and mineral premix 1 | 0.4 | 0.4 | 0.4 |
| Soybean oil | 0 | 2.9 | 0 |
| Fish oil | 0 | 0 | 2.9 |
| Symbol | Name | Function |
|---|---|---|
| ACACA | Acetyl-CoA carboxylase alfa | Catalyzes the rate-limiting reaction in the de novo synthesis of LCFA |
| ACSL1 | Acyl-CoA Synthetase Long Chain Family Member 1 | Convert LCFA into acyl-CoA esters, transport of exogenous FA |
| ACSS2 | Acyl-CoA Synthetase Short Chain Family Member 2 | The chemical reactions and pathways resulting in the formation of acetyl-CoA from acetate |
| PLIN2 | Adipose Differentiation-Related Protein | Involved in formation and maintenance of lipid droplets |
| DGAT1 and 2 | Diacylglycerol O-acyltransferase Homolog 1 and 2 | Acyltransferase that catalyzes the terminal and only committed step in triacylglycerol synthesis |
| FABP3 and 4 | Fatty Acid Binding Protein 3 and 4 | Intracellular transport of acyl-CoA; regulation of gene expression by providing LCFA to PPARγ |
| FADS2 | Fatty acid desaturase 2 | Desaturase introducing a cis double bond at carbon 6 of the fatty acyl chain |
| FASN | Fatty acid synthase | Fatty acid synthetase catalyzes the formation of long-chain fatty acids from acetyl-CoA, malonyl-CoA and NADPH |
| SLC27A6 | Soluble Carrier Protein 27A | LCFA translocation (high uptake); Convert LCFA into acyl-CoA esters |
| INSIG1 | Insulin Induced Gene 1 | Mediates feedback control of cholesterol synthesis by controlling SCAP and HMGCR |
| LPIN1 | Lipin 1 | Dephosphorylation of phosphatidate yielding diacylglycerol; Transcription (PPARα co- factor) |
| LPL | Lipoprotein Lipase | Catalyzes the hydrolysis of triglycerides from circulating chylomicrons and very low-density lipoproteins |
| PPARG | Peroxisome Proliferator Activated Receptor Gamma | Regulate transcription of lipogenic and adipogenic genes. |
| SCAP | SREBP Chaperone | Protein required for cholesterol as well as lipid homeostasis. Chaperone for activation of SREBP1 |
| SCD1 | Stearoyl-CoA desaturase 1 | Desaturase introducing introduce the first double bond into saturated fatty acyl-CoA substrates |
| SREBP1 | Sterol Regulatory Element Binding Transcription Factor | Transcriptional regulation of cholesterol synthesis and lipogenesis genes |
| THRSP | Thyroid Hormone Responsive | Nuclear protein which is important in the regulation of lipid metabolism |
| VLDLR | Very Low-Density Lipoprotein Receptor | Binds very low-density lipoproteins assisting LPL |
| Gene | Treatment | SEM | p-Value | ||||
|---|---|---|---|---|---|---|---|
| Control | FO | SO | Diet (D) | Time (T) | D × T | ||
| Fatty acid transport and activation | |||||||
| ACSL1 | 77.0 a | 68.9 ab | 52.5 b | 6.72 | 0.04 | 0.39 | 0.60 |
| FABP3 | 15.5 | 14.4 | 10.1 | 3.40 | 0.49 | 0.03 | 0.04 |
| FABP4 | 525.9 | 379.6 | 477.9 | 100.7 | 0.59 | 0.04 | 0.31 |
| LPL | 131.5 | 138.6 | 154.4 | 30.4 | 0.86 | 0.98 | 0.90 |
| SLC27A6 | 1569.4 | 1635.7 | 1451.4 | 134.7 | 0.61 | 0.37 | 0.05 |
| VLDLR | 6.20 | 6.74 | 5.16 | 0.43 | 0.06 | 0.13 | 0.24 |
| De novo synthesis and fatty acid desaturation | |||||||
| ACACA | 27.9 b | 39.1 a | 20.3 b | 3.95 | <0.01 | 0.58 | 0.82 |
| ACSS2 | 53.1 | 67.3 | 67.2 | 19.8 | 0.85 | 0.07 | 0.53 |
| FADS2 | 2.40 | 1.77 | 1.87 | 0.43 | 0.54 | <0.01 | 0.56 |
| FASN | 33.4 | 129.2 | 106.9 | 36.6 | 0.18 | 0.31 | 0.90 |
| SCD1 | 356.6 | 545.9 | 500.8 | 160.7 | 0.69 | 0.16 | 0.75 |
| Triacylglycerol synthesis and lipid droplet formation | |||||||
| DGAT1 | 33.3 | 32.3 | 25.0 | 2.98 | 0.11 | <0.01 | 0.09 |
| DGAT2 | 122.5 | 167.0 | 102.7 | 46.6 | 0.58 | 0.06 | 0.54 |
| LPIN1 | 8.81 | 6.89 | 8.25 | 1.90 | 0.76 | 0.22 | 0.26 |
| PLIN2 | 75.0 | 64.4 | 57.0 | 5.81 | 0.11 | 0.91 | 0.04 |
| Transcription regulation | |||||||
| INSIG1 | 2.07 ab | 1.71 b | 2.79 a | 0.28 | 0.04 | <0.01 | 0.96 |
| PPARG | 28.8 | 18.8 | 19.1 | 2.97 | 0.06 | 0.02 | 0.06 |
| SCAP | 6.90 | 5.57 | 8.17 | 0.81 | 0.12 | <0.01 | 0.63 |
| SREBP1 | 26.0 | 30.1 | 28.5 | 2.29 | 0.44 | 0.09 | 0.36 |
| THRSP | 37.8 | 85.6 | 78.9 | 25.1 | 0.45 | 0.37 | 0.66 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas-Bello-Pérez, E.; Bionaz, M.; Garrido-Sartore, M.; Cancino-Padilla, N.; Morales, M.S.; Romero, J.; Leskinen, H.; Garnsworthy, P.C.; Loor, J.J. Effect of Soybean Oil and Fish Oil on Lipid-Related Transcripts in Subcutaneous Adipose Tissue of Dairy Cows. Animals 2020, 10, 54. https://doi.org/10.3390/ani10010054
Vargas-Bello-Pérez E, Bionaz M, Garrido-Sartore M, Cancino-Padilla N, Morales MS, Romero J, Leskinen H, Garnsworthy PC, Loor JJ. Effect of Soybean Oil and Fish Oil on Lipid-Related Transcripts in Subcutaneous Adipose Tissue of Dairy Cows. Animals. 2020; 10(1):54. https://doi.org/10.3390/ani10010054
Chicago/Turabian StyleVargas-Bello-Pérez, Einar, Massimo Bionaz, Macarena Garrido-Sartore, Nathaly Cancino-Padilla, María Sol Morales, Jaime Romero, Heidi Leskinen, Philip C. Garnsworthy, and Juan J. Loor. 2020. "Effect of Soybean Oil and Fish Oil on Lipid-Related Transcripts in Subcutaneous Adipose Tissue of Dairy Cows" Animals 10, no. 1: 54. https://doi.org/10.3390/ani10010054
APA StyleVargas-Bello-Pérez, E., Bionaz, M., Garrido-Sartore, M., Cancino-Padilla, N., Morales, M. S., Romero, J., Leskinen, H., Garnsworthy, P. C., & Loor, J. J. (2020). Effect of Soybean Oil and Fish Oil on Lipid-Related Transcripts in Subcutaneous Adipose Tissue of Dairy Cows. Animals, 10(1), 54. https://doi.org/10.3390/ani10010054








