Cork Oak Forests Soil Bacteria: Potential for Sustainable Agroforest Production
Abstract
:1. Introduction
2. Material and Methods
2.1. Sample Collection and Bacterial Isolation
2.2. Plant Growth Promoting Biochemical Assays
2.3. PGPR Effects on Arabidopsis Thaliana Root-Architecture
2.4. PGPR Antagonistic Activity against B. mediterranea and D. corticola
2.5. Data and Statistical Analyses
3. Results
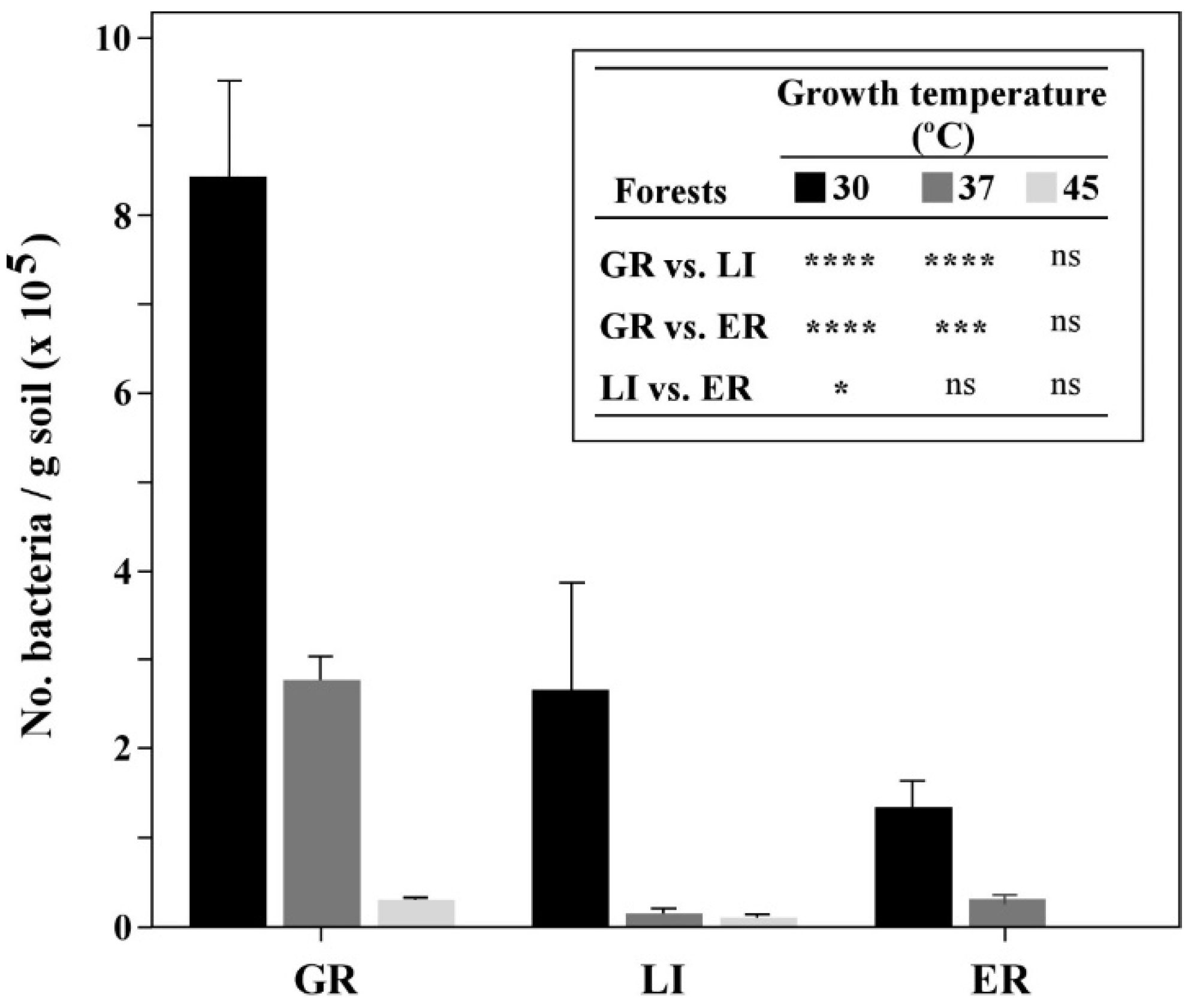
3.1. Identification of PGPR from Cork Oak Forests Soils
3.2. Arabidopsis Thaliana Root Modulation by Cork Oak Soil PGPR
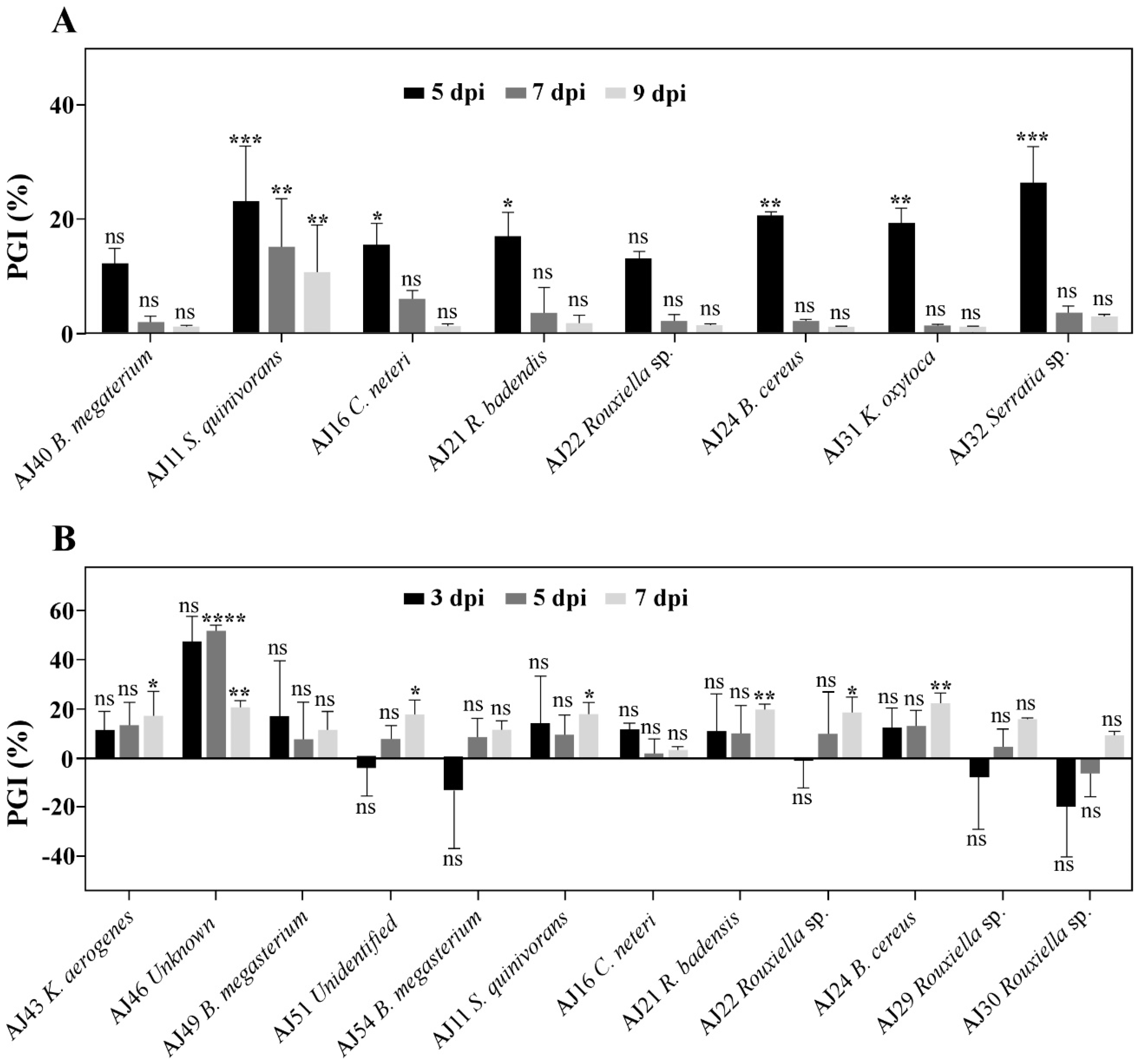
3.3. Serratia spp. and Bacillus spp. as Key Genera for Controlling Cork Oak Bark Pathogens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, H.N.; Jin, H.Y.; Kwak, M.J.; Khaine, I.; You, H.N.; Lee, T.Y.; Woo, S.Y. Why does Quercus suber species decline in Mediterranean areas? J. Asia Pac. Biodivers. 2017, 10, 337–341. [Google Scholar] [CrossRef]
- APCOR. Available online: https://www.apcor.pt/wp-content/uploads/2019/12/boletim_estatistico_apcor_2019.pdf (accessed on 1 November 2020).
- Bugalho, M.N.; Caldeira, M.C.; Pereira, J.S.; Aronson, J.; Pausas, J.G. Mediterranean cork oak savannas require human use to sustain biodiversity and ecosystem services. Front. Ecol. Environ. 2011, 9, 278–286. [Google Scholar] [CrossRef] [Green Version]
- Mendes, M.P.; Cherubini, P.; Plieninger, T.; Ribeiro, L.; Costa, A. Climate effects on stem radial growth of Quercus suber L.: Does tree size matter? Int. J. For. Res. 2018, 92, 73–84. [Google Scholar] [CrossRef]
- Desprez-Loustau, M.L.; Marçais, B.; Nageleisen, L.M.; Piou, D.; Vannini, A. Interactive effects of drought and pathogens in forest trees. Ann. For. Sci. 2006, 63, 597–612. [Google Scholar] [CrossRef] [Green Version]
- Moricca, S.; Linaldeddu, B.T.; Ginetti, B.; Scanu, B.; Franceschini, A.; Ragazzi, A. Endemic and emerging pathogens threatening cork oak trees: Management options for conserving a unique forest ecosystem. Plant Dis. 2016, 100, 2184–2193. [Google Scholar] [CrossRef] [Green Version]
- Costa, D.; Tavares, R.; Baptista, P.; Lino-Neto, T. The influence of endophytes on cork oak forests under a changing climate. In Endophytes for a Growing World; Hodkinson, T., Doohan, F., Saunders, M., Murphy, B., Eds.; Cambridge University Press: Cambridge, UK, 2019; pp. 250–274. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maghnia, F.Z.; Abbas, Y.; Mahé, F.; Prin, Y.; El Ghachtouli, N.; Duponnois, R.; Sanguin, H. The rhizosphere microbiome: A key component of sustainable cork oak forests in trouble. For. Ecol. Manag. 2019, 434, 29–39. [Google Scholar] [CrossRef]
- Meena, R.S.; Kumar, S.; Datta, R.; Lal, R.; Vijayakumar, V.; Brtnicky, M.; Sharma, M.P.; Yadav, G.S.; Jhariya, M.K.; Jangir, C.K.; et al. Impact of Agrochemicals on Soil Microbiota and Management: A Review. Land 2020, 9, 34. [Google Scholar] [CrossRef] [Green Version]
- Bevivino, A.; Paganin, P.; Bacci, G.; Florio, A.; Pellicer, M.S.; Papaleo, M.C.; Mengoni, A.; Ledda, L.; Fani, R.; Benedetti, A.; et al. Soil bacterial community response to differences in agricultural management along with seasonal changes in a mediterranean region. PLoS ONE 2014, 9, e105515. [Google Scholar] [CrossRef] [Green Version]
- Reis, F.; Soares-Castro, P.; Costa, D.; Tavares, R.M.; Baptista, P.; Santos, P.M.; Lino-Neto, T. Climatic impacts on the bacterial community profiles of cork oak soils. Appl. Soil Ecol. 2019, 143, 89–97. [Google Scholar] [CrossRef] [Green Version]
- Lladó, S.; López-Mondéjar, R.; Baldrian, P. Forest soil bacteria: Diversity, involvement in ecosystem processes, and response to global change. Microbiol. Mol. Biol. Rev. 2017, 81, e00063-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tariq, M.; Noman, M.; Ahmed, T.; Hameed, A.; Manzoor, N.; Zafar, M. Antagonistic features displayed by plant growth-promoting rhizobacteria (PGPR): A Review. J. Plant Sci. Phytopathol. 2017, 1, 038–043. [Google Scholar] [CrossRef] [Green Version]
- Etesami, H.; Adl, S.M. Plant Growth-Promoting Rhizobacteria (PGPR) and Their Action Mechanisms in Availability of Nutrients to Plants; Springer: Singapore, 2020; pp. 147–203. [Google Scholar]
- Reis, F.; Magalhães, A.P.; Tavares, R.M.; Baptista, P.; Lino-Neto, T. Bacteria could help ectomycorrhizae establishment under climate variations. Mycorrhiza 2021, 21, 395–401. [Google Scholar] [CrossRef]
- Reis, F.; Valdiviesso, T.; Varela, C.; Tavares, R.M.; Baptista, P.; Lino-Neto, T. Ectomycorrhizal fungal diversity and community structure associated with cork oak in different landscapes. Mycorrhiza 2018, 28, 357–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobti, S.; Belhadj, H.A.; Djaghoubi, A. Isolation and characterization of the native Rhizobia under hyper-salt edaphic conditions in Ouargla (southeast Algeria). Energy Procedia 2015, 74, 1434–1439. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- Pikovskaya, R.I. Mobilization of Phosphorus in Soil Connection with the Vital Activity of Some Microbial Species. Microbiology 1948, 17, 362–370. [Google Scholar]
- Mehta, S.; Nautiyal, C.S. An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr. Microbiol. 2001, 43, 51–56. [Google Scholar] [CrossRef]
- Joseph, B.; Patra, R.; Lawrence, R. Characterization of plant growth-promoting rhizobacteria associated with chickpea (Cicer arietinum L.). Int. J. Plant Prod. 2012, 1, 141–152. [Google Scholar] [CrossRef]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Luna, F.M.; López-Bucio, J.; Altamirano-Hernández, J.; Valencia-Cantero, E.; Reyes de la Cruz, H.; Macías-Rodríguez, L. Plant growth-promoting rhizobacteria modulate root-system architecture in Arabidopsis thaliana through volatile organic compound. Symbiosis 2010, 51, 75–83. [Google Scholar] [CrossRef]
- Costa, D.; Tavares, R.M.; Baptista, P.; Lino-Neto, T. Cork oak endophytic fungi as potential biocontrol agents against Biscogniauxia mediterranea and Diplodia corticola. J. Fungi 2020, 6, 287. [Google Scholar] [CrossRef] [PubMed]
- Idris, H.A.; Labuschagne, N.; Korsten, L. Screening rhizobacteria for biological control of Fusarium root and crown rot of sorghum in Ethiopia. Biol. Control 2007, 40, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighborneighbor-joining method: A new method for reconstructing phlylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Brock, T.D.; Madigan, M.T.; Martinko, J.M.; Parker, J. Biology of Microorganisms, 8th ed.; Prentice-Hall: Englewood Cliffs, NJ, USA; Hoboken, NJ, USA, 1994; p. 909. [Google Scholar]
- Marulanda-Aguirre, A.; Azcón, R.; Ruiz-Lozano, J.M.; Aroca, R. Differential effects of a Bacillus megaterium strain on Lactuca sativa plant growth depending on the origin of the arbuscular mycorrhizal fungus coinoculated: Physiologic and biochemical traits. J. Plant Growth Regul. 2007, 27, 10–18. [Google Scholar] [CrossRef]
- Tan, I.S.; Ramamurthi, K.S. Spore formation in Bacillus subtilis. Environ. Microbiol. Rep. 2013, 6, 212–225. [Google Scholar] [CrossRef] [Green Version]
- Salazar-Badillo, F.; Salas-Muñoz, S.; Mauricio-Castillo, J.; Sáenz-Mata, J.; Mendoza, A.; Nieto Jacobo, M.; Steyaert, J. The rhizospheres of arid and semi-arid ecosystems are a source of microorganisms with growth-promoting potential. In Advances in PGPR Research; CABI: Wallingford, UK, 2017. [Google Scholar] [CrossRef]
- Hanna, A.L.; Youssef, H.H.; Amer, W.M.; Monib, M.; Fayez, M.; Hegazi, N.A. Diversity of bacteria nesting the plant cover of north Sinai deserts, Egypt. J. Adv. Res. 2013, 4, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Bano, A.; Ali, S.; Ali Babar, M.D. Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 2020, 90, 189–203. [Google Scholar] [CrossRef]
- Alarcón, M.V.; Salguero, J.; Lloret, P.G. Auxin modulated initiation of lateral roots is linked to pericycle cell length in maize. Front. Plant Sci. 2019, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Chen, F.; Du, H.; Zhang, X.; Wang, X.; Yao, G.; Xu, B. Graphene oxide and indole-3-acetic acid cotreatment regulates the root growth of Brassica napus L. via multiple phytohormone pathways. BMC Plant Biol. 2020, 20, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahn, L.; Hofmann, U.; Ludwig-Müller, J. Indole-3-Acetic acid is synthesized by the endophyte Cyanodermella asteris via a tryptophan-dependent and -independent way and mediates the interaction with a non-host plant. Int. J. Mol. Sci. 2021, 22, 2651. [Google Scholar] [CrossRef] [PubMed]
- Swain, M.R.; Naskar, S.K.; Ray, R.C. Indole-3-acetic acid production and effect on sprouting of yam (Dioscorea rotundata L.) minisetts by Bacillus subtilis isolated from culturable cowdung microflora. Pol. J. Microbiol. 2007, 56, 103. [Google Scholar] [PubMed]
- Hariprasad, P.; Niranjana, S.R. Isolation and characterization of phosphate solubilizing rhizobacteria to improve plant health of tomato. Plant Soil 2009, 316, 13–24. [Google Scholar] [CrossRef]
- Perrig, D.; Boiero, M.L.; Masciarelli, O.A.; Penna, C.; Ruiz, O.A.; Cassán, F.D.; Luna, M.V. Plant-growth-promoting compounds produced by two agronomically important strains of Azospirillum brasilense, and implications for inoculant formulation. Appl. Microbiol. Biotechnol. 2007, 75, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016, 2, 1. [Google Scholar] [CrossRef]
- Fernando, W.G.D.; Nakkeeran, S.; Zhang, Y. Biosynthesis of Antibiotics by PGPR and its Relation in Biocontrol of Plant Diseases. In PGPR: Biocontrol and Biofertilization; Siddiqui, Z.A., Ed.; Springer: Dordrecht, The Netherlands, 2005; pp. 67–109. [Google Scholar] [CrossRef]
- Tabli, N.; Rai, A.; Bensidhoum, L.; Palmieri, G.; Gogliettino, M.; Cocca, E.; Consiglio, C.; Cillo, F.; Bubici, G.; Nabti, E. Plant growth promoting and inducible antifungal activities of irrigation well water-bacteria. Biol. Control 2018, 117, 78–86. [Google Scholar] [CrossRef]
- Ali, S.; Hameed, S.; Shahid, M.; Iqbal, M.; Lazarovits, G.; Imran, A. Functional characterization of potential PGPR exhibiting broad-spectrum antifungal activity. Microbiol. Res. 2019, 232, 126389. [Google Scholar] [CrossRef] [PubMed]
- Deora, A.; Hashidoko, Y.; Islam, M.T.; Tahara, S. Antagonistic rhizoplane bacteria induce diverse morphological alterations in Peronosporomycete hyphae during in vitro interaction. Eur. J. Plant Pathol. 2005, 112, 311–322. [Google Scholar] [CrossRef]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant Growth Promoting Rhizobacteria (PGPR) as Green Bioinoculants: Recent Developments, Constraints, and Prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
| Forest | Temp | Code | Identification | At Root Development | Antifungal Activity | |||
|---|---|---|---|---|---|---|---|---|
| PRGI (%) | LRI (%) | IRH (%) | Bmed | Dcort | ||||
| GR | 30 °C | AJ40 | Bacillus megaterium | 95.8 | 602.1 | 59.4 | + | − |
| AJ41 | Bacillus megaterium | 81.3 | 442.6 | 50.0 | − | − | ||
| 37 °C | AJ42 | Bacillus megaterium | 64.1 | 527.7 | 75.0 | − | − | |
| AJ43 | Klebsiella aerogenes | 67.7 | 445.7 | 71.9 | − | + | ||
| AJ44 | Klebsiella aerogenes | 90.1 | 455.3 | 93.8 | − | − | ||
| AJ45 | Klebsiella aerogenes | 88.5 | 731.9 | 43.8 | − | − | ||
| AJ46 | Unknown | 106.3 | 829.8 | 66.7 | − | + | ||
| AJ47 | Bacillus megaterium | 66.2 | 538.3 | 37.5 | − | − | ||
| AJ48 | Bacillus megaterium | 63.0 | 394.7 | 41.7 | − | − | ||
| AJ49 | Bacillus megaterium | 71.4 | 425.5 | 41.7 | − | + | ||
| AJ50 | Bacillus megaterium | 64.0 | 531.9 | 96.9 | − | − | ||
| AJ51 | Unidentified | 64.6 | 658.5 | 40.6 | − | + | ||
| AJ52 | Bacillus sp. | 67.7 | 541.5 | 68.8 | − | − | ||
| AJ53 | Bacillus megaterium | 58.9 | 386.2 | 96.9 | − | − | ||
| 45 °C | AJ54 | Bacillus megaterium | 57.8 | 523.4 | 54.2 | − | + | |
| AJ55 | Bacillus megaterium | 67.2 | 535.1 | 78.1 | − | − | ||
| AJ56 | Unidentified | 64.6 | 404.3 | 58.3 | − | − | ||
| AJ57 | Bacillus megaterium | 55.2 | 456.4 | 87.5 | − | − | ||
| AJ58 | Bacillus megaterium | 57.3 | 552.1 | 90.6 | − | − | ||
| AJ59 | Unidentified | 60.9 | 525.5 | 59.4 | − | − | ||
| AJ60 | Unidentified | 56.8 | 519.1 | 50.0 | − | − | ||
| AJ61 | Bacillus megaterium | 88.5 | 671.3 | 50.0 | − | − | ||
| AJ62 | Unidentified | 72.9 | 625.5 | 58.3 | − | − | ||
| AJ63 | Unidentified | 56.8 | 528.7 | 56.3 | − | − | ||
| AJ64 | Bacillus megaterium | 57.3 | 498.9 | 68.8 | − | − | ||
| LI | 30 °C | AJ10 | Unidentified | 80.2 | 439.4 | 21.9 | − | − |
| AJ11 | Serratia quinivorans | 64.1 | 594.7 | 34.4 | + | + | ||
| AJ14 | Cedecea neteri | 59.9 | 435.1 | 53.1 | − | − | ||
| 37 °C | AJ8 | Cedecea sp. | 60.4 | 425.5 | 43.8 | − | − | |
| AJ9 | Bacillus megaterium | 71.9 | 445.7 | 37.5 | − | − | ||
| AJ12 | Cedecea neteri | 56.8 | 495.7 | 34.4 | − | − | ||
| AJ13 | Unidentified | 58.9 | 495.7 | 37.5 | − | − | ||
| AJ15 | Cedecea neteri | 63.5 | 469.1 | 53.1 | − | − | ||
| AJ16 | Cedecea neteri | 59.9 | 412.8 | 31.3 | + | + | ||
| AJ17 | Bacillus megaterium | 83.9 | 564.9 | 94.0 | − | − | ||
| AJ18 | Bacillus simplex | 66.7 | 390.4 | 33.3 | − | − | ||
| 45 °C | AJ19 | Bacillus megaterium | 76.6 | 485.1 | 43.8 | − | − | |
| ER | 30 °C | AJ21 | Rouxiella badensis | 76.6 | 442.6 | 31.3 | + | + |
| AJ22 | Rouxiella sp. | 64.6 | 581.9 | 46.9 | + | + | ||
| AJ23 | Bacillus mycoides | 67.2 | 519.1 | 53.1 | − | − | ||
| AJ24 | Bacillus cereus | 87.5 | 588.3 | 65.6 | + | + | ||
| AJ25 | Pseudomonas mohnii | 117.2 | 788.3 | 71.9 | − | − | ||
| AJ26 | Bacillus cereus | 74.5 | 392.6 | 25.0 | − | − | ||
| AJ27 | Ewingella americana | 62.5 | 363.8 | 41.7 | − | − | ||
| AJ28 | Ewingella americana | 73.4 | 525.5 | 56.3 | − | − | ||
| AJ29 | Rouxiella sp. | 66.2 | 558.5 | 29.2 | − | + | ||
| 37 °C | AJ30 | Rouxiella sp. | 70.8 | 452.1 | 45.8 | − | + | |
| AJ31 | Klebsiella oxytoca | 50.0 | 219.1 | 0.0 | + | − | ||
| AJ32 | Serratia sp. | 52.6 | 292.6 | 20.8 | + | − | ||
| 45 °C | AJ33 | Bacillus megaterium | 49.0 | 359.6 | 21.9 | − | − | |
| AJ34 | Bacillus nakamurai | 53.7 | 475.5 | 28.1 | − | − | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reis, F.; Pereira, A.J.; Tavares, R.M.; Baptista, P.; Lino-Neto, T. Cork Oak Forests Soil Bacteria: Potential for Sustainable Agroforest Production. Microorganisms 2021, 9, 1973. https://doi.org/10.3390/microorganisms9091973
Reis F, Pereira AJ, Tavares RM, Baptista P, Lino-Neto T. Cork Oak Forests Soil Bacteria: Potential for Sustainable Agroforest Production. Microorganisms. 2021; 9(9):1973. https://doi.org/10.3390/microorganisms9091973
Chicago/Turabian StyleReis, Francisca, Ana João Pereira, Rui M. Tavares, Paula Baptista, and Teresa Lino-Neto. 2021. "Cork Oak Forests Soil Bacteria: Potential for Sustainable Agroforest Production" Microorganisms 9, no. 9: 1973. https://doi.org/10.3390/microorganisms9091973
APA StyleReis, F., Pereira, A. J., Tavares, R. M., Baptista, P., & Lino-Neto, T. (2021). Cork Oak Forests Soil Bacteria: Potential for Sustainable Agroforest Production. Microorganisms, 9(9), 1973. https://doi.org/10.3390/microorganisms9091973








