Epiphytic Bacteria from Sweet Pepper Antagonistic In Vitro to Ralstonia solanacearum BD 261, a Causative Agent of Bacterial Wilt
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites and Crop Management
2.2. Sample Collection, Processing, and Isolation of Potential Antagonists
2.3. Plant Bacterial Pathogen
2.4. Multiplication of Potential Antagonists and the Pathogen
2.5. In Vitro Screening of Isolates for Antagonism
2.6. DNA Extraction and PCR Amplification of Potential Antagonistic Strains
2.6.1. PCR Amplification of 16S rRNA Genes
2.6.2. Sequencing and Bioinformatics Analysis of the 16S rRNA Amplicons
2.6.3. Nucleotide Sequence Accession Numbers
2.7. Optimization for Improved Activity of Potential Antagonistic Strains
2.8. Determination of Potential Antimicrobial Traits
2.8.1. Cellulase Activity
2.8.2. Protease Activity
2.8.3. Detection of Phosphate Solubilization
2.8.4. Siderophore Production
2.9. Statistical Data Processing
3. Results
3.1. Isolation and Identification of Potent Bacterial Strains
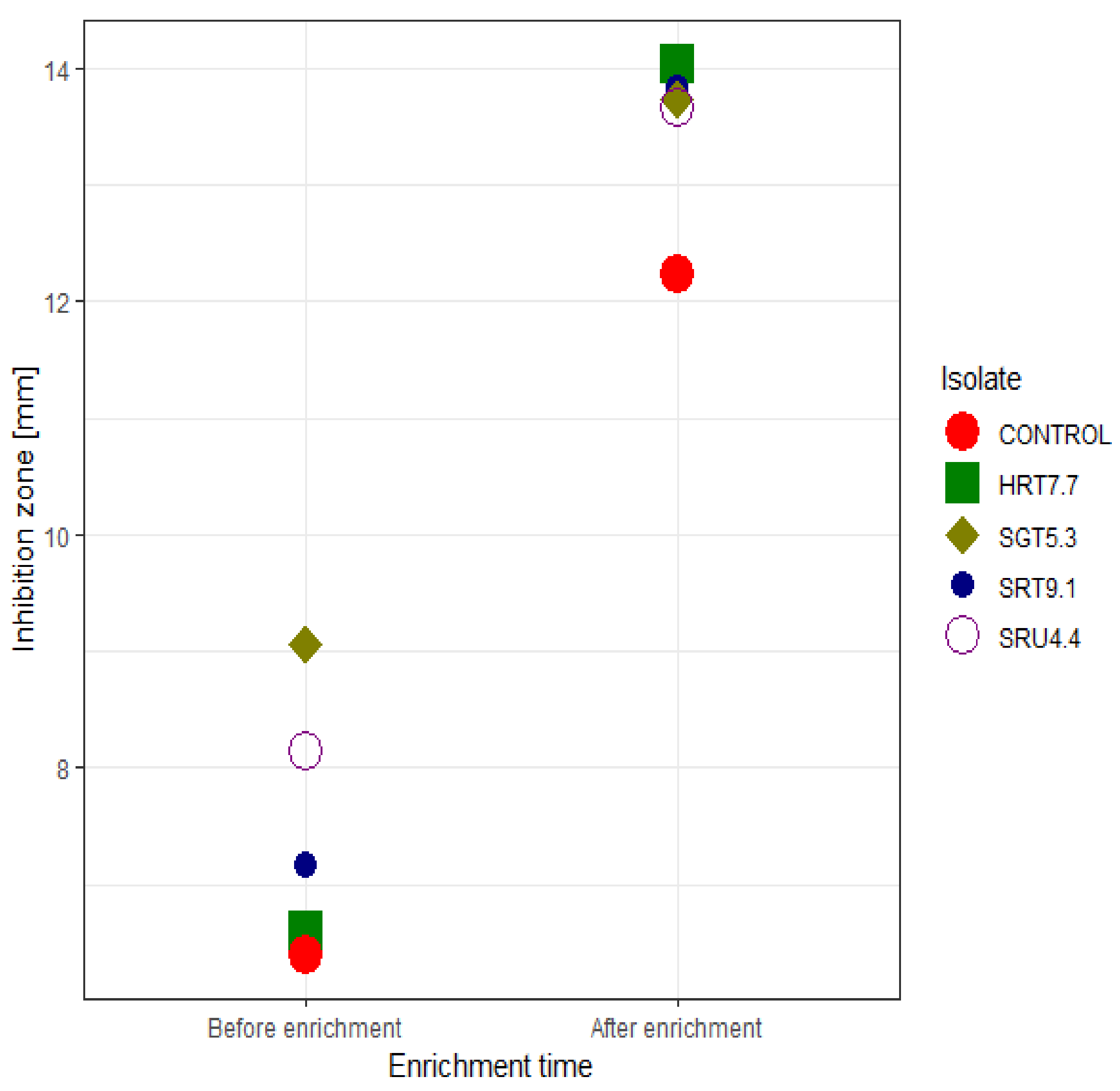
3.2. Optimization for Enhanced Antagonistic Activity
3.3. Determination of Antimicrobial Traits of the Antagonists
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Food and Agricultural Commodities Production. 2014. Available online: http://faostat.fao.org (accessed on 22 February 2017).
- Govender, L.; Pillay, K.; Siwela, M.; Modi, A.; Mabhaudhi, T. Food and nutrition insecurity in selected rural communities of KwaZulu-Natal, South Africa—Linking human nutrition and agriculture. Int. J. Environ. Res. Public Health 2016, 14, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kairiza, T.; Kembo, G.D. Coping with food and nutrition insecurity in Zimbabwe: Does household head gender matter? Agric. Food Econ. 2019, 7, 1–16. [Google Scholar] [CrossRef]
- Serpeloni, J.M.; Specian, A.F.; Ribeiro, D.L.; Tuttis, K.; Vilegas, W.; Martínez-López, W.; Dokkedal, A.L.; Saldanha, L.L.; Cólus, I.M.; Varanda, E.A. Antimutagenicity and induction of antioxidant defense by flavonoid rich extract of Myrcia bella Cambess. in normal and tumor gastric cells. J. Ethnopharmacol. 2015, 176, 345–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahidi, F.; Ambigaipalan, P. Phenolics and polyphenolics in foods, beverages and spices: Antioxidant activity and health effects—A review. J. Funct. Foods 2015, 18, 820–897. [Google Scholar] [CrossRef]
- Deepa, N.; Kaur, C.; George, B.; Sing, B.; Kapoor, H.C. Antioxidant constituents in some sweet pepper (Capsicum annuum L.) genotypes during maturity. LWT Food Sci. Technol. 2017, 40, 121–129. [Google Scholar] [CrossRef]
- Chen, L.; Kang, Y.-H. Anti-inflammatory and antioxidant activities of red pepper (Capsicum annuum L.) stalk extracts: Comparison of pericarp and placenta extracts. J. Funct. Foods 2013, 5, 1724–1731. [Google Scholar] [CrossRef]
- Sreeramulu, D.; Raghunath, M. Antioxidant activity and phenolic content of roots, tubers and vegetables commonly consumed in India. Food Res. Int. 2010, 43, 1017–1020. [Google Scholar] [CrossRef]
- Luning, P.A.; Vries, R.V.D.V.D.; Yuksel, D.; Ebbenhorst-Seller, T.; Wichers, H.; Roozen, J.P. Combined instrumental and sensory evaluation of flavor of fresh bell peppers (Capsicum annuum) harvested at three maturation stages. J. Agric. Food Chem. 1994, 42, 2855–2861. [Google Scholar] [CrossRef]
- Tundis, R.; Menichini, F.; Bonesi, M.; Conforti, F.; Statti, G.; Menichini, F.; Loizzo, M.R. Antioxidant and hypoglycaemic activities and their relationship to phytochemicals in Capsicum annuum cultivars during fruit development. LWT Food Sci. Technol. 2013, 53, 370–377. [Google Scholar] [CrossRef]
- López, A.; Fenoll, J.; Hellín, P.; Flores, P. Cultivation approach for comparing the nutritional quality of two pepper cultivars grown under different agricultural regimes. LWT Food Sci. Technol. 2014, 58, 299–305. [Google Scholar] [CrossRef]
- Pagán, I.; García-Arenal, F. Tolerance to plant pathogens: Theory and experimental evidence. Int. J. Mol. Sci. 2018, 19, 810. [Google Scholar] [CrossRef] [Green Version]
- Kover, P.X.; Schaal, B.A. Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proc. Natl. Acad. Sci. USA 2002, 99, 11270–11274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cernava, T.; Müller, H.; Aschenbrenner, I.A.; Grube, M.; Berg, G. Analyzing the antagonistic potential of the lichen microbiome against pathogens by bridging metagenomic with culture studies. Front. Microbiol. 2015, 6, 620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, Z.; McLenachan, P.A.; Lockhart, P.J.; Waipara, N.; Er, O.; Reynolds, C.; Blanchon, D. Metagenome profiling identifies potential biocontrol agents for Selaginella kraussiana in New Zealand. Genes 2019, 10, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [Green Version]
- Knapp, S.; Bohs, L.; Nee, M.; Spooner, D.M. Solanaceae—A model for linking genomics with biodiversity. Comp. Funct. Genomics 2004, 5, 285–291. [Google Scholar] [CrossRef] [Green Version]
- Swanson, J.K.; Yao, J.; Tans-Kersten, J.; Allen, C. Behavior of Ralstonia solanacearum Race 3 Biovar 2 during latent and active infection of geranium. Phytopathology 2005, 95, 136–143. [Google Scholar] [CrossRef] [Green Version]
- Kelman, A. One hundred and one years of research on bacterial wilt. In Bacterial Wilt: Molecular and Ecological Aspects; Prior, P., Allen, C., Eds.; Springer: Berlin, Germany, 1998; pp. 1–5. [Google Scholar]
- Nion, Y.A.; Toyota, K. Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ. 2015, 30, 1–11. [Google Scholar]
- Bannihatti, R.K.; Suryawanshi, A.P. Integrated management of bacterial wilt of tomato caused by Ralstonia solanacearum. Int. J. Chem. Stud. 2019, 7, 1599–1603. [Google Scholar]
- Kamal, R.; Gusan, Y.S.; Kumar, V.; Sharma, A. Disease management through biological control agents: An eco-friendly and cost effective approach for sustainable agriculture—A Review. Agric. Rev. 2015, 36, 37–45. [Google Scholar] [CrossRef]
- Sharma, M.; Tarafdar, A.; Ghosh, R.; Gopalakrishanan, S. Biological control as a tool for eco-friendly management of plant pathogens. In Advances in Soil Microbiology: Recent Trends and Future Prospects; Adhya, T., Mishra, B., Annapurna, K., Verma, D., Kumar, U., Eds.; Springer: Singapore, 2017; pp. 153–188. [Google Scholar]
- Mamphogoro, T.P.; Babalola, O.O.; Aiyegoro, O.A. Sustainable management strategies for bacterial wilt of sweet peppers (Capsicum annuum) and other Solanaceous crops. J. Appl. Microbiol. 2020, 129, 496–508. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, L.; Wu, K.; Shi, J.; Wang, M.; Yang, X.; Shen, Q.; Shen, B. Evaluation of Bacillus-fortified organic fertilizer for controlling tobacco bacterial wilt in greenhouse and field experiments. Appl. Soil Ecol. 2013, 75, 86–94. [Google Scholar] [CrossRef]
- Wei, Z.; Huang, J.; Tan, S.; Mei, X.; Shen, Q.; Xu, Y. The congeneric strain Ralstonia pickettii QL-A6 of Ralstonia solanacearum as an effective biocontrol agent for bacterial wilt of tomato. Biol. Control 2013, 65, 278–285. [Google Scholar] [CrossRef]
- Durairaj, K.; Velmurugan, P.; Park, J.H.; Chang, W.S.; Park, Y.J.; Senthilkumar, P.; Choi, K.M.; Lee, J.H.; Oh, B.T. Potential for plant biocontrol activity of isolated Pseudomonas aeruginosa and Bacillus stratosphericus strains against bacterial pathogens acting through both induced plant resistance and direct antagonism. FEMS Microbiol. Lett. 2017, 364, fnx225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mamphogoro, T.P.; Maboko, M.M.; Babalola, O.O.; Aiyegoro, O.A. Bacterial communities associated with the surface of fresh sweet pepper (Capsicum annuum) and their potential as biocontrol. Sci. Rep. 2020, 10, 8560. [Google Scholar] [CrossRef]
- Paulino, L.C.; Tseng, C.H.; Strober, B.E.; Blaser, M.J. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J. Clin. Microbiol. 2006, 44, 2933–2941. [Google Scholar] [CrossRef] [Green Version]
- Shutt, V.; Shin, G.; Van Der Waals, J.; Goszczynska, T.; Coutinho, T. Characterization of Ralstonia strains infecting tomato plants in South Africa. Crop Prot. 2018, 112, 56–62. [Google Scholar] [CrossRef]
- Facelli, E.; Taylor, C.; Scott, E.; Fegan, M.; Huys, G.; Noble, R.D.; Swings, J.; Sedgley, M. Identification of the causal agent of pistachio dieback in Australia. Eur. J. Plant Pathol. 2005, 112, 155–165. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Ranamukhaarachchi, S.L. Soil-borne antagonists for biological control of bacterial wilt disease caused by Ralstonia solanacearum in tomato and pepper. J. Plant Pathol. 2010, 92, 395–406. [Google Scholar]
- Doan, T.T.; Nguyen, T.H.; Zeller, H.; Ullrich, C. Status of research on biological control of tomato and groundnut bacterial wilt in Vietnam. In Proceedings of the 1st International Symposium Biological Control of Bacterial Plant Diseases, Seeheim/Darmstadt, Germany, 23–26 October 2005; Zeller, W., Ullrich, C., Eds.; Federal Biological Research Center for Agriculture and Forestry: Berlin, Germany, 2006; pp. 105–111. [Google Scholar]
- Arturo, A.M.; Oldelson, D.A.; Hichey, R.F.; Tiedje, J.M. Bacterial community fingerprinting of amplified 16-23S ribosomal DNA gene and restriction endonuclease analysis. In Molecular Microbial Ecology Manual; Akkermans, A.D.L., van Elsas, J.D., Bruijn, F.J., Eds.; Springer: Dordrecht, the Netherlands, 1995; pp. 1–8. [Google Scholar]
- Turner, S.; Pryer, K.M.; Miao, V.P.; Palmer, J.D. Investigating deep phylogenetic relationships among cyanobacteria and plas-tids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighborjoining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Costa, E.; Teixidó, N.; Usall, J.; Atarés, E.; Vinas, I. The effect of nitrogen and carbon sources on growth of the biocontrol agent Pantoea agglomerans strain CPA-2. Lett. Appl. Microbiol. 2002, 35, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Teather, R.M.; Wood, P.J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 1982, 43, 777–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, P.A.; Ohman, D.E.; Iglewski, B.H. A more sensitive plate assay for detection of protease production by Pseudomanas aeruginosa. J. Clin. Microbiol. 1979, 9, 538–540. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate-solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal CAS assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Mendiburu, F.; Simon, R. Agricolae - Ten years of an open source statistical tool for experiments in breeding, agriculture and biology. Peer J. PrePrints 2015, 3, e1404v1. [Google Scholar]
- Wickham, H.; Dianne, C.; Heike, H. Visualizing statistical models: Removing the blindfold. Stat. Anal. Data Min. 2015, 8, 203–225. [Google Scholar] [CrossRef]
- Long, H.H.; Furuya, N.; Kurose, D.; Takeshita, M.; Takanami, Y. Isolation of endophyiic bacteria from Solawam sp. and their antibacterial activity against plant pathogenic bacteria. J. Fac. Agric. Kyushu Univ. 2003, 48, 21–28. [Google Scholar] [CrossRef]
- Lee, H.B.; Magan, N. Environmental factors and nutritional utilization patterns affect niche overlap indices between Aspergillus ochraceus and other spoilage fungi. Lett. Appl. Microbiol. 1999, 28, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Kok, C.J.; Papert, A. Effect of temperature on in vitro interactions between Verticillium chlamydosporium and other Meloidogyne-associated microorganisms. BioControl 2002, 47, 603–606. [Google Scholar] [CrossRef]
- Passari, A.K.; Mishra, V.K.; Leo, V.V.; Gupta, V.K.; Singh, B.P. Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol. Res. 2016, 193, 57–73. [Google Scholar] [CrossRef]
- Kim, Y.S.; Balaraju, K.; Jeon, Y.H. Biological characteristics of Bacillus amyloliquefaciens AK-0 and suppression of ginseng root rot caused by Cylindrocarpon destructans. J. Appl. Microbiol. 2017, 122, 166–179. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, W.S.; Akhkha, A.; El-Naggar, M.Y.; Elbadry, M. In vitro antagonistic activity, plant growth promoting traits and phylogenetic affiliation of rhizobacteria associated with wild plants grown in arid soil. Front. Microbiol. 2014, 5, 651. [Google Scholar] [CrossRef] [Green Version]
- Avcı, A.; Çağrı-Mehmetoğlu, A.; Arslan, D. Production of antimicrobial substances by a novel Bacillus strain inhibiting Salmonella typhimurium. LWT-Food Sci. Technol. 2017, 80, 265–270. [Google Scholar] [CrossRef]
- Nutaratat, P.; Monprasit, A.; Srisuk, N. High-yield production of indole-3-acetic acid by Enterobacter sp. DMKU-RP206, a rice phyllosphere bacterium that possesses plant growth-promoting traits. 3 Biotech 2017, 7, 305. [Google Scholar] [CrossRef]
- Raza, W.; Yang, W.; Shen, Q. Paenibacillus polymyxa: Antibiotics, hydrolytic enzymes and hazard assessment. J. Plant. Pathol. 2008, 90, 419–430. [Google Scholar]
- Lin, C.; Jia, X.; Fang, Y.; Chen, L.; Zhang, H.; Lin, R.; Chen, J. Enhanced production of prodigiosin by Serratia marcescens FZSF02 in the form of pigment pellets. Electron. J. Biotechnol. 2019, 40, 58–64. [Google Scholar] [CrossRef]
- Dhar Purkayastha, G.; Mangar, P.; Saha, A.; Saha, D. Evaluation of the biocontrol efficacy of a Serratia marcescens strain indigenous to tea rhizosphere for the management of root rot disease in tea. PLoS ONE 2018, 13, e0191761. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.J.; Hong, S.J.; Choi, W.; Kim, B.S. Antifungal activity of Paenibacillus kribbensis strain T-9 isolated from soils against several plant pathogenic fungi. Plant Pathol. J. 2014, 30, 102–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.-H.; Shim, H.; Shin, J.H.; Kim, K.S. Antagonistic activities of bacillus spp. strains isolated from tidal flat sediment towards anthracnose pathogens Colletotrichum acutatum and C. gloeosporioides in South Korea. Plant Pathol. J. 2015, 31, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [Green Version]
- Nelson, L.M. Plant growth promoting rhizobacteria (PGPR): Prospects for new inoculants. Crop Manag. 2004, 3, 1–7. [Google Scholar] [CrossRef]
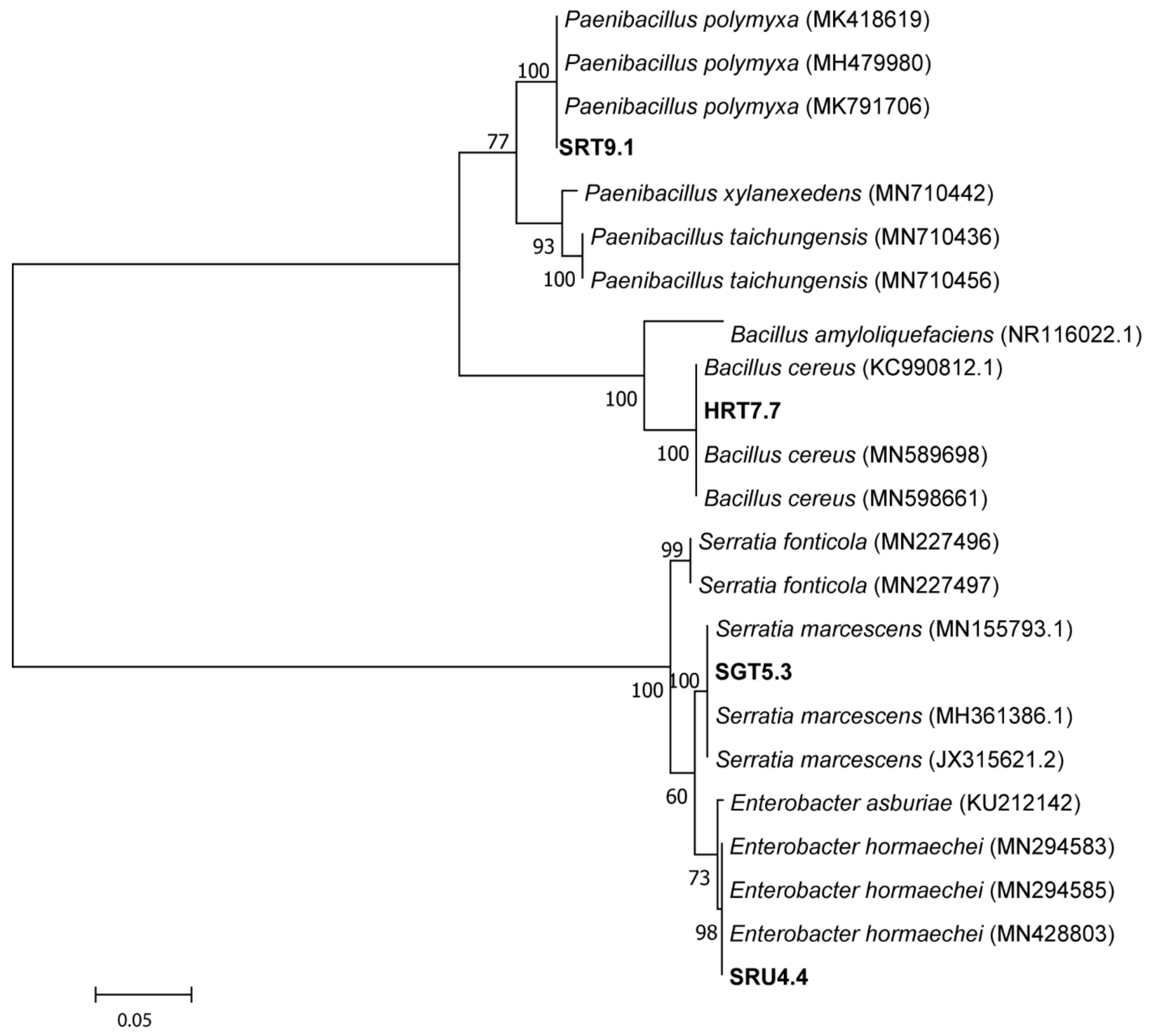


| Sample No | Strain Code a | Base Pair Length b | Species Name | Accession No c | Similarity (%) |
|---|---|---|---|---|---|
| 1 | HRT7.7 | 1264 bp | Bacillus cereus strain HRT7.7 | MN911398.1 | 99 |
| 2 | SGT5.3 | 1254 bp | Serratia marcescens strain SGT5.3 | MN911401.1 | 99 |
| 3 | SRT9.1 | 1265 bp | Paenibacillus polymyxa strain SRT9.1 | MN911399.1 | 99 |
| 4 | SRU4.4 | 1255 bp | Enterobacter hormaechei strain SRU4.4 | MN911400.1 | 98 |
| pH | Carbon Sources | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source of variation | Degrees of freedom | 5 | 6 | 7 | 8 | 9 | ─ | Glucose | Starch | Lactose | Maltose | Fructose | ─ | |
| Replication | 2 | 0.002 | 0.42467 | 0.32067 | 0.9613 | 0.4687 | ─ | 0.26467 | 0.162 | 0.05067 | 0.2167 | 0.42467 | ─ | |
| Treatment | 4 | 4.071 *** | 2.65 | 2.053 * | 27.907 *** | 7.036 *** | ─ | 1.278 ** | 1.562 ** | 2.489 *** | 5.304 *** | 1.674 ** | ─ | |
| Residual error | 8 | 0.032 | 0.733 | 0.35317 | 1.4853 | 0.2845 | ─ | 0.14967 | 0.14367 | 0.114 | 0.1525 | 0.2005 | ─ | |
| Nitrogen source | Temperature (°C) | |||||||||||||
| Degrees of freedom | Glycine | Yeast extract | Tryptone | (NH4)2SO4 | NH4CL | ─ | 25 | 28 | 30 | 35 | 37 | ─ | ||
| Replication | 2 | 0.1147 | 0.14467 | 0.042 | 0.15267 | 0.66467 | ─ | 0.1647 | 0.3247 | 0.1167 | 0.126 | 0.2847 | ─ | |
| Treatment | 4 | 6.631 *** | 0.34933 | 0.413 * | 0.676 * | 1.893 * | ─ | 20.464 *** | 14.142 *** | 8.259 *** | 14.451 *** | 3.424 ** | ─ | |
| Residual error | 8 | 0.1672 | 0.39883 | 0.08867 | 0.161 | 0.38217 | ─ | 0.2738 | 0.3163 | 0.0775 | 0.0677 | 0.3388 | ─ | |
| Starch concentration (%) | Tryptone concentration (%) | |||||||||||||
| Degrees of freedom | 0.5 | 1 | 1.5 | 2 | 2.5 | 3 | 0.5 | 1 | 1.5 | 2 | 2.5 | 3 | ||
| Replication | 2 | 0.0347 | 0.206 | 0.006 | 0.08867 | 0.0107 | 0.1607 | 0.0127 | 0.026 | 0.0507 | 0.1047 | 0.3227 | 0.1847 | |
| Treatment | 4 | 7.367 *** | 5.353 *** | 2.297 *** | 1.922 *** | 3.383 *** | 7.034 ** | 9.837 *** | 12.944 *** | 12.612 *** | 28.236 *** | 10.561 *** | 6.884 *** | |
| Residual error | 8 | 0.0563 | 0.0693 | 0.05933 | 0.04783 | 0.0557 | 0.544 | 0.1552 | 0.0443 | 0.1498 | 0.1755 | 0.2652 | 0.1563 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamphogoro, T.P.; Kamutando, C.N.; Maboko, M.M.; Aiyegoro, O.A.; Babalola, O.O. Epiphytic Bacteria from Sweet Pepper Antagonistic In Vitro to Ralstonia solanacearum BD 261, a Causative Agent of Bacterial Wilt. Microorganisms 2021, 9, 1947. https://doi.org/10.3390/microorganisms9091947
Mamphogoro TP, Kamutando CN, Maboko MM, Aiyegoro OA, Babalola OO. Epiphytic Bacteria from Sweet Pepper Antagonistic In Vitro to Ralstonia solanacearum BD 261, a Causative Agent of Bacterial Wilt. Microorganisms. 2021; 9(9):1947. https://doi.org/10.3390/microorganisms9091947
Chicago/Turabian StyleMamphogoro, Tshifhiwa Paris, Casper Nyaradzai Kamutando, Martin Makgose Maboko, Olayinka Ayobami Aiyegoro, and Olubukola Oluranti Babalola. 2021. "Epiphytic Bacteria from Sweet Pepper Antagonistic In Vitro to Ralstonia solanacearum BD 261, a Causative Agent of Bacterial Wilt" Microorganisms 9, no. 9: 1947. https://doi.org/10.3390/microorganisms9091947
APA StyleMamphogoro, T. P., Kamutando, C. N., Maboko, M. M., Aiyegoro, O. A., & Babalola, O. O. (2021). Epiphytic Bacteria from Sweet Pepper Antagonistic In Vitro to Ralstonia solanacearum BD 261, a Causative Agent of Bacterial Wilt. Microorganisms, 9(9), 1947. https://doi.org/10.3390/microorganisms9091947







