Fungi in Permafrost-Affected Soils of the Canadian Arctic: Horizon- and Site-Specific Keystone Taxa Revealed by Co-Occurrence Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Site Description and Soil Sampling
2.2. Measurement of Environmental Factors
2.3. Extracellular Enzymes Activities
2.4. DNA Extraction and Quantitative Assessment of Fungal Community by qPCR
2.5. Barcoded Amplicon Sequencing
2.6. Network Construction
2.7. Statistical Analyses
3. Results
3.1. Environmental Variables
3.2. Fungal Gene Abundance, Community Composition, and Diversity Differed between Horizons and Sites
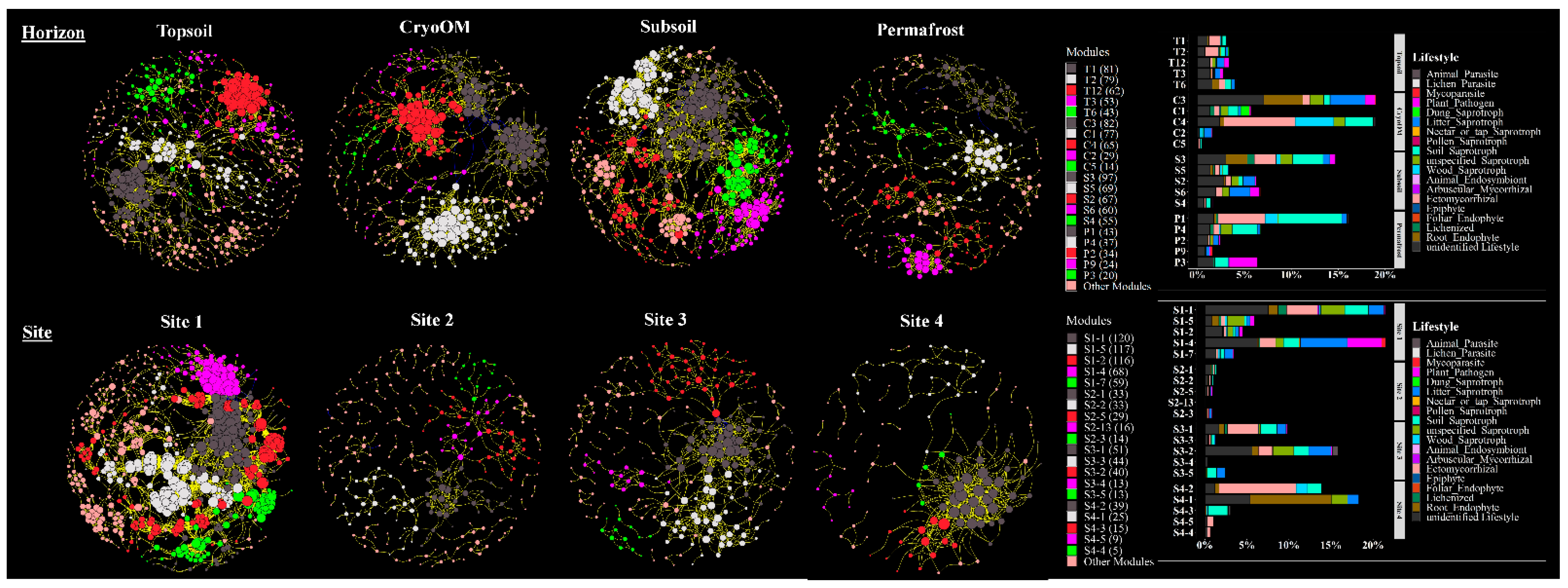
3.3. Key Topological Properties of Co-Occurrence Network
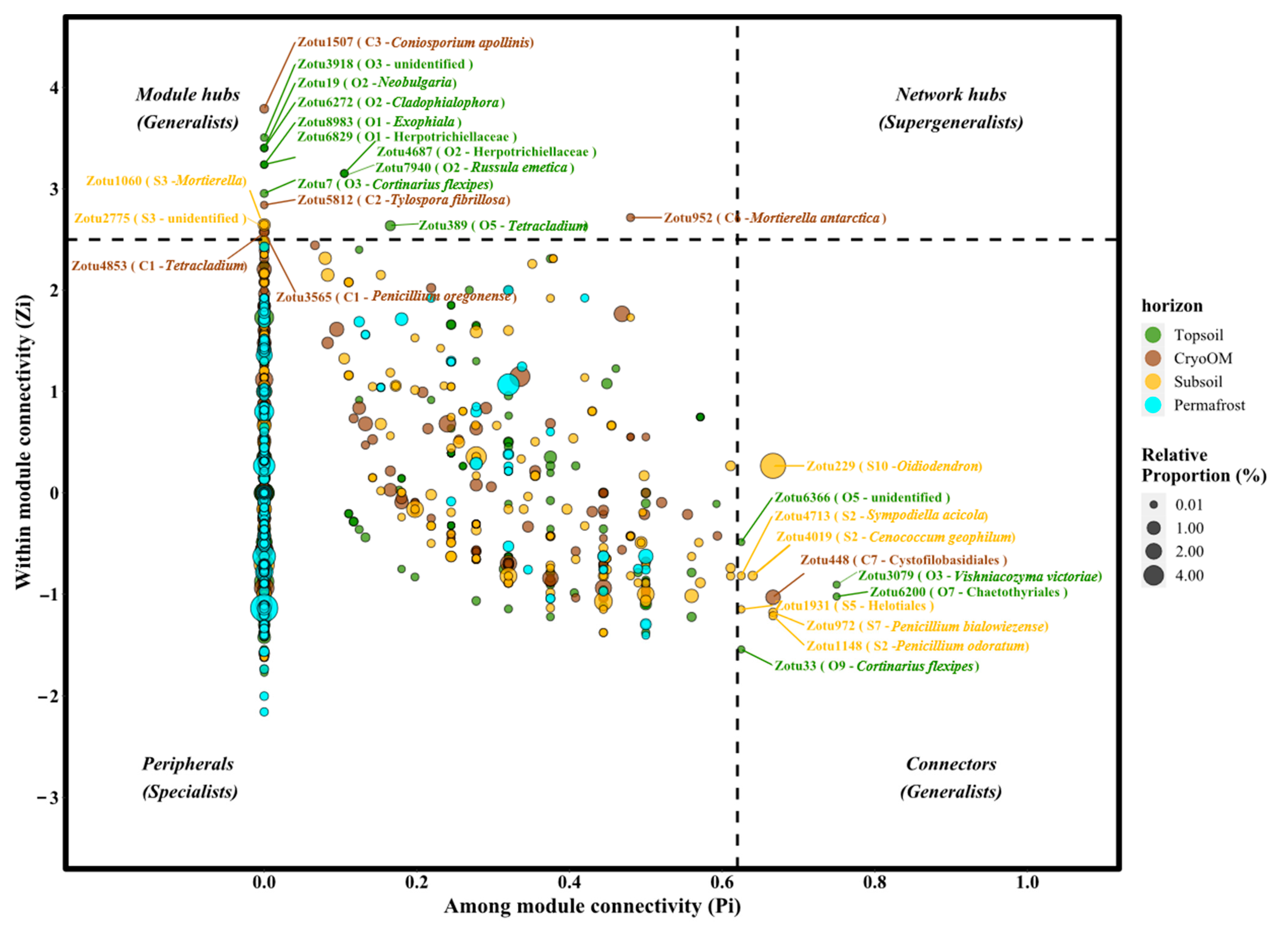
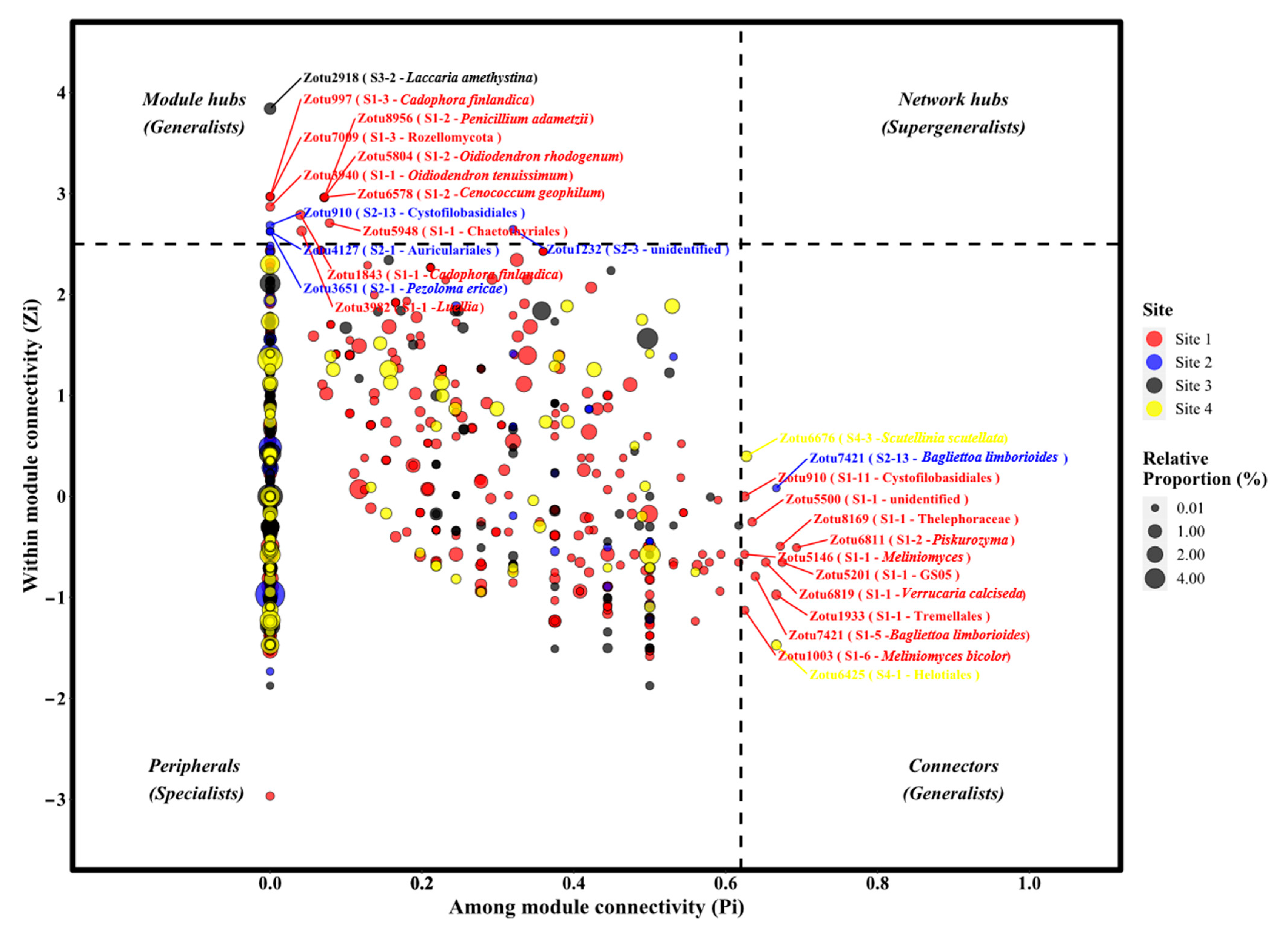
3.4. The Topological Roles of Nodes and Generalist-Specialist Shift
4. Discussion
4.1. Horizon and Tundra Specific Fungal Lifestyle
4.2. Co-Occurrence Networks Reveal More Complex Interactions in Deeper Soil Horizons
4.3. Greater Connectivity but Lower Specialization
4.4. Environmental Condition Associated with Topological Role Shift
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Payer, D.; Barry, T.; Berteaux, D.; Bültmann, H.; Cristiansen, J.S.; Cook, J.S.; Dahlberg, A.; Daniëls, F.J.A.; Ehrich, D.; Fjeldså, J.; et al. Arctic biodiversity assessment. Status and trends in Arctic biodiversity. In Fungi; Meltofte, H., Ed.; Narayana Press: Odder, Denmark, 2013; pp. 303–319. [Google Scholar]
- Barea, J.-M.; Pozo, M.J.; Azcón, R.; Azcón-Aguilar, C. Microbial co-operation in the rhizosphere. J. Exp. Bot. 2005, 56, 1761–1778. [Google Scholar] [CrossRef] [Green Version]
- Hestrin, R.; Hammer, E.C.; Mueller, C.W.; Lehmann, J. Synergies between mycorrhizal fungi and soil microbial communities increase plant nitrogen acquisition. Commun. Biol. 2019, 2, 233. [Google Scholar] [CrossRef] [PubMed]
- de Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geml, J.; Timling, I.; Robinson, C.H.; Lennon, N.; Nusbaum, H.C.; Brochmann, C.; Noordeloos, M.E.; Taylor, D.L. An arctic community of symbiotic fungi assembled by long-distance dispersers: Phylogenetic diversity of ectomycorrhizal basidiomycetes in Svalbard based on soil and sporocarp DNA. J. Biogeogr. 2012, 39, 74–88. [Google Scholar] [CrossRef]
- Blaud, A.; Phoenix, G.K.; Osborn, A.M. Variation in bacterial, archaeal and fungal community structure and abundance in High Arctic tundra soil. Polar Biol. 2015, 38, 1009–1024. [Google Scholar] [CrossRef]
- Wallenstein, M.D.; McMahon, S.; Schimel, J. Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol. Ecol. 2007, 59, 428–435. [Google Scholar] [CrossRef] [Green Version]
- Deslippe, J.R.; Hartmann, M.; Simard, S.W.; Mohn, W.W. Long-term warming alters the composition of Arctic soil microbial communities. FEMS Microbiol. Ecol. 2012, 82, 303–315. [Google Scholar] [CrossRef] [Green Version]
- Perini, L.; Gostinčar, C.; Anesio, A.M.; Williamson, C.; Tranter, M.; Gunde-Cimerman, N. Darkening of the Greenland Ice Sheet: Fungal Abundance and Diversity Are Associated With Algal Bloom. Front. Microbiol. 2019, 10, 557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyling, N.V.; Schmidt, N.M.; Eilenberg, J. Occurrence and diversity of fungal entomopathogens in soils of low and high Arctic Greenland. Polar Biol. 2012, 35, 1439–1445. [Google Scholar] [CrossRef]
- Gittel, A.; Bárta, J.; Kohoutová, I.; Mikutta, R.; Owens, S.; Gilbert, J.; Schnecker, J.; Wild, B.; Hannisdal, B.; Maerz, J.; et al. Distinct microbial communities associated with buried soils in the Siberian tundra. ISME J. 2014, 8, 841–853. [Google Scholar] [CrossRef]
- Timling, I.; Walker, D.A.; Nusbaum, C.; Lennon, N.J.; Taylor, D.L. Rich and cold: Diversity, distribution and drivers of fungal communities in patterned-ground ecosystems of the North American Arctic. Mol. Ecol. 2014, 23, 3258–3272. [Google Scholar] [CrossRef] [Green Version]
- Louca, S.; Jacques, S.M.S.; Pires, A.P.F.; Leal, J.S.; Srivastava, D.S.; Parfrey, L.W.; Farjalla, V.F.; Doebeli, M. High taxonomic variability despite stable functional structure across microbial communities. Nat. Ecol. Evol. 2017, 1, 15. [Google Scholar] [CrossRef]
- Cernansky, R. Biodiversity moves beyond counting species. Nature 2017, 546, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.; Robson, G.D.; Trinci, A.P.J. 21st Century Guidebook to Fungi; Cambridge University Press: Cambridge, UK, 2011; ISBN 9780511977022. [Google Scholar]
- Alzarhani, A.K.; Clark, D.R.; Underwood, G.J.C.; Ford, H.; Cotton, T.E.A.; Dumbrell, A.J. Are drivers of root-associated fungal community structure context specific? ISME J. 2019, 13, 1330–1344. [Google Scholar] [CrossRef] [PubMed]
- Veach, A.M.; Stokes, C.E.; Knoepp, J.; Jumpponen, A.; Baird, R. Fungal Communities and Functional Guilds Shift Along an Elevational Gradient in the Southern Appalachian Mountains. Microb. Ecol. 2018, 76, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Fahey, C.; Koyama, A.; Antunes, P.M.; Dunfield, K.; Flory, S.L. Plant communities mediate the interactive effects of invasion and drought on soil microbial communities. ISME J. 2020, 14, 1396–1409. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Wild, B.; Schnecker, J.; Bárta, J.; Čapek, P.; Guggenberger, G.; Hofhansl, F.; Kaiser, C.; Lashchinsky, N.; Mikutta, R.; Mooshammer, M.; et al. Nitrogen dynamics in Turbic Cryosols from Siberia and Greenland. Soil Biol. Biochem. 2013, 67, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, K.W.; Buscot, F.; Canbäck, B.; Choi, C.; Cichocki, N.; Clum, A.; et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 2015, 47, 410–415. [Google Scholar] [CrossRef]
- Wutkowska, M.; Vader, A.; Mundra, S.; Cooper, E.J.; Eidesen, P.B. Dead or Alive; or Does It Really Matter? Level of Congruency Between Trophic Modes in Total and Active Fungal Communities in High Arctic Soil. Front. Microbiol. 2019, 9, 3243. [Google Scholar] [CrossRef]
- Mäkipää, R.; Rajala, T.; Schigel, D.; Rinne, K.T.; Pennanen, T.; Abrego, N.; Ovaskainen, O. Interactions between soil- and dead wood-inhabiting fungal communities during the decay of Norway spruce logs. ISME J. 2017, 11, 1964–1974. [Google Scholar] [CrossRef] [Green Version]
- Anthony, M.A.; Frey, S.D.; Stinson, K.A. Fungal community homogenization, shift in dominant trophic guild, and appearance of novel taxa with biotic invasion. Ecosphere 2017, 8, e01951. [Google Scholar] [CrossRef] [Green Version]
- Faust, K.; Sathirapongsasuti, J.F.; Izard, J.; Segata, N.; Gevers, D.; Raes, J.; Huttenhower, C. Microbial Co-occurrence Relationships in the Human Microbiome. PLoS Comput. Biol. 2012, 8, e1002606. [Google Scholar] [CrossRef] [PubMed]
- Lupatini, M.; Suleiman, A.K.A.; Jacques, R.J.S.; Antoniolli, Z.I.; de Siqueira Ferreira, A.; Kuramae, E.E.; Roesch, L.F.W. Network topology reveals high connectance levels and few key microbial genera within soils. Front. Environ. Sci. 2014, 2, 10. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G.A. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef]
- Banerjee, S.; Thrall, P.H.; Bissett, A.; Heijden, M.G.A.; Richardson, A.E. Linking microbial co-occurrences to soil ecological processes across a woodland-grassland ecotone. Ecol. Evol. 2018, 8, 8217–8230. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Disentangling Interactions in the Microbiome: A Network Perspective. Trends Microbiol. 2017, 25, 217–228. [Google Scholar] [CrossRef] [PubMed]
- de Vries, F.T.; Wallenstein, M.D. Below-ground connections underlying above-ground food production: A framework for optimising ecological connections in the rhizosphere. J. Ecol. 2017, 105, 913–920. [Google Scholar] [CrossRef] [Green Version]
- de Vries, F.T.; Liiri, M.E.; Bjørnlund, L.; Bowker, M.A.; Christensen, S.; Setälä, H.M.; Bardgett, R.D. Land use alters the resistance and resilience of soil food webs to drought. Nat. Clim. Chang. 2012, 2, 276–280. [Google Scholar] [CrossRef]
- Feng, J.; Wang, C.; Lei, J.; Yang, Y.; Yan, Q.; Zhou, X.; Tao, X.; Ning, D.; Yuan, M.M.; Qin, Y.; et al. Warming-induced permafrost thaw exacerbates tundra soil carbon decomposition mediated by microbial community. Microbiome 2020, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [Green Version]
- Benedek, Z.; Jordán, F.; Báldi, A. Topological keystone species complexes in ecological interaction networks. Community Ecol. 2007, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Wang, N.; Yu, L. Soil fungal community composition differs significantly among the Antarctic, Arctic, and Tibetan Plateau. Extremophiles 2020, 24, 821–829. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, N.-F.; Liu, H.-Y.; Zhang, Y.-Q.; Yu, L.-Y. Soil pH is a Key Determinant of Soil Fungal Community Composition in the Ny-Ålesund Region, Svalbard (High Arctic). Front. Microbiol. 2016, 7, 227. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Deng, Y.; Ding, J.-Z.; Hu, H.-W.; Xu, T.-L.; Li, F.; Yang, G.-B.; Yang, Y.-H. Distinct microbial communities in the active and permafrost layers on the Tibetan Plateau. Mol. Ecol. 2017, 26, 6608–6620. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Li, S.; Avera, B.N.; Strahm, B.D.; Badgley, B.D. Soil Bacterial and Fungal Communities Show Distinct Recovery Patterns during Forest Ecosystem Restoration. Appl. Environ. Microbiol. 2017, 83, e00966-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herren, C.M.; McMahon, K.D. Keystone taxa predict compositional change in microbial communities. Environ. Microbiol. 2018, 20, 2207–2217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín González, A.M.; Dalsgaard, B.; Olesen, J.M. Centrality measures and the importance of generalist species in pollination networks. Ecol. Complex. 2010, 7, 36–43. [Google Scholar] [CrossRef]
- Qi, G.; Ma, G.; Chen, S.; Lin, C.; Zhao, X. Microbial Network and Soil Properties Are Changed in Bacterial Wilt-Susceptible Soil. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [Green Version]
- Burn, C.R. Herschel Island Qikiqtaryuk: A Natural and Cultural History of Yukon’s Arctic Island; University of Calgary Press: Whitehorse, YT, Canada, 2012; pp. 48–53. [Google Scholar]
- Siewert, M.B.; Lantuit, H.; Richter, A.; Hugelius, G. Permafrost Causes Unique Fine-Scale Spatial Variability Across Tundra Soils. Glob. Biogeochem. Cycles 2021, 35. [Google Scholar] [CrossRef]
- Schoeneberger, P.J.; Wysocki, D.A.; Benham, E.C. (Eds.) Field Book for Describing and Sampling Soils; National Soil Survey Center, Natural Resources Conservation Service: Lincoln, NE, USA, 2012. [CrossRef]
- Ping, C.-L.; Clark, M.H.; Kimble, J.M.; Michaelson, G.J.; Shur, Y.; Stiles, C.A. Sampling Protocols for Permafrost-Affected Soils. Soil Horiz. 2013, 54, 13. [Google Scholar] [CrossRef]
- Siewert, M.B.; Hugelius, G.; Heim, B.; Faucherre, S. Landscape controls and vertical variability of soil organic carbon storage in permafrost-affected soils of the Lena River Delta. Catena 2016, 147, 725–741. [Google Scholar] [CrossRef]
- Varsadiya, M.; Urich, T.; Hugelius, G.; Bárta, J. Microbiome structure and functional potential in permafrost soils of the Western Canadian Arctic. FEMS Microbiol. Ecol. 2021, 97, fiab008. [Google Scholar] [CrossRef]
- Bárta, J.; Šlajsová, P.; Tahovská, K.; Picek, T.; Šantrůčková, H. Different temperature sensitivity and kinetics of soil enzymes indicate seasonal shifts in C, N and P nutrient stoichiometry in acid forest soil. Biogeochemistry 2014, 117, 525–537. [Google Scholar] [CrossRef]
- Marx, M.-C.; Wood, M.; Jarvis, S. A microplate fluorimetric assay for the study of enzyme diversity in soils. Soil Biol. Biochem. 2001, 33, 1633–1640. [Google Scholar] [CrossRef]
- Borneman, J.; Hartin, R.J. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol. 2000, 66, 4356–4360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. [Google Scholar]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 2016. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahti, L.; Shetty, S. Tools for Microbiome Analysis in R 2017. Available online: http://microbiome.github.com/microbiome (accessed on 19 July 2021).
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2011; ISBN 3900051070. [Google Scholar]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Harrell, F.E.J. R package, version 4.0-1; Hmisc: Harrell Miscellaneous: Nashville, TN, USA, 2020.
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Yang, Y. Phylogenetic Molecular Ecological Network of Soil Microbial Communities in Response to Elevated CO2. MBio 2011, 2, e00122-11. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Yin, S.; Liu, X.; Zhang, W.; Gu, T.; Shen, Q.; Qiu, H. Fungal networks in yield-invigorating and -debilitating soils induced by prolonged potato monoculture. Soil Biol. Biochem. 2013, 65, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 2007, 1, 54. [Google Scholar] [CrossRef] [Green Version]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The modularity of pollination networks. Proc. Natl. Acad. Sci. USA 2007, 104, 19891–19896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’hara, B.; Henry, M.; Maintainer, H.S. The Vegan Package Title Community Ecology Package. 2007. Available online: http://Cran.R-Project.Org/; http://R-Forge.R-Project.Org/Projects/Vegan/ (accessed on 19 July 2021).
- Parks, D.H.; Beiko, R.G. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 2010, 26, 715–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guimerà, R.; Nunes Amaral, L.A. Functional cartography of complex metabolic networks. Nature 2005, 433, 895–900. [Google Scholar] [CrossRef] [Green Version]
- Hoppe, B.; Purahong, W.; Wubet, T.; Kahl, T.; Bauhus, J.; Arnstadt, T.; Hofrichter, M.; Buscot, F.; Krüger, D. Linking molecular deadwood-inhabiting fungal diversity and community dynamics to ecosystem functions and processes in Central European forests. Fungal Divers. 2016, 77, 367–379. [Google Scholar] [CrossRef]
- Bani, A.; Pioli, S.; Ventura, M.; Panzacchi, P.; Borruso, L.; Tognetti, R.; Tonon, G.; Brusetti, L. The role of microbial community in the decomposition of leaf litter and deadwood. Appl. Soil Ecol. 2018, 126, 75–84. [Google Scholar] [CrossRef]
- Dickie, I.A. Host preference, niches and fungal diversity. New Phytol. 2007, 174, 230–233. [Google Scholar] [CrossRef]
- Bascompte, J. Networks in ecology. Basic Appl. Ecol. 2007, 8, 485–490. [Google Scholar] [CrossRef]
- Robinson, C.H.; Saunders, P.W.; Madan, N.J.; Janie Pryce-Miller, E.; Pentecost, A. Does nitrogen deposition affect soil microfungal diversity and soil N and P dynamics in a high Arctic ecosystem? Glob. Chang. Biol. 2004, 10, 1065–1079. [Google Scholar] [CrossRef]
- Lennon, J.T.; Aanderud, Z.T.; Lehmkuhl, B.K.; Schoolmaster, D.R. Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 2012, 93, 1867–1879. [Google Scholar] [CrossRef] [Green Version]
- Mundra, S.; Halvorsen, R.; Kauserud, H.; Müller, E.; Vik, U.; Eidesen, P.B. Arctic fungal communities associated with roots of Bistorta vivipara do not respond to the same fine-scale edaphic gradients as the aboveground vegetation. New Phytol. 2015, 205, 1587–1597. [Google Scholar] [CrossRef] [Green Version]
- Clemmensen, K.E.; Michelsen, A.; Jonasson, S.; Shaver, G.R. Increased ectomycorrhizal fungal abundance after long-term fertilization and warming of two arctic tundra ecosystems. New Phytol. 2006, 171, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yao, Y.-F. Endophytic Fungal Communities Associated with Vascular Plants in the High Arctic Zone Are Highly Diverse and Host-Plant Specific. PLoS ONE 2015, 10, e0130051. [Google Scholar] [CrossRef] [Green Version]
- Newsham, K.K.; Upson, R.; Read, D.J. Mycorrhizas and dark septate root endophytes in polar regions. Fungal Ecol. 2009, 2, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Walker, X.J.; Basinger, J.F.; Kaminskyj, S.G.W. Endorhizal Fungi in Ranunculus from Western and Arctic Canada: Predominance of Fine Endophytes at High Latitudes. Open Mycol. J. 2010, 4, 1–9. [Google Scholar] [CrossRef]
- Müller, M.M.; Valjakka, R.; Suokko, A.; Hantula, J. Diversity of endophytic fungi of single Norway spruce needles and their role as pioneer decomposers. Mol. Ecol. 2001, 10, 1801–1810. [Google Scholar] [CrossRef] [PubMed]
- Korkama-Rajala, T.; Müller, M.M.; Pennanen, T. Decomposition and Fungi of Needle Litter from Slow- and Fast-growing Norway Spruce (Picea abies) Clones. Microb. Ecol. 2008, 56, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Promputtha, I.; Hyde, K.D.; McKenzie, E.H.C.; Peberdy, J.F.; Lumyong, S. Can leaf degrading enzymes provide evidence that endophytic fungi becoming saprobes? Fungal Divers. 2010, 41, 89–99. [Google Scholar] [CrossRef]
- Caldwell, B.A.; Jumpponen, A.; Trappe, J.M. Utilization of Major Detrital Substrates by Dark-Septate, Root Endophytes. Mycologia 2000, 92, 230. [Google Scholar] [CrossRef] [Green Version]
- Upson, R.; Read, D.J.; Newsham, K.K. Nitrogen form influences the response of Deschampsia antarctica to dark septate root endophytes. Mycorrhiza 2009, 20, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Surono; Narisawa, K. The dark septate endophytic fungus Phialocephala fortinii is a potential decomposer of soil organic compounds and a promoter of Asparagus officinalis growth. Fungal Ecol. 2017, 28, 1–10. [Google Scholar] [CrossRef]
- Verbruggen, E.; Pena, R.; Fernandez, C.W.; Soong, J.L. Mycorrhizal Interactions With Saprotrophs and Impact on Soil Carbon Storage. In Mycorrhizal Mediation of Soil; Elsevier: Amsterdam, The Netherlands, 2017; pp. 441–460. [Google Scholar]
- Bödeker, I.T.M.; Lindahl, B.D.; Olson, Å.; Clemmensen, K.E. Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct. Ecol. 2016, 30, 1967–1978. [Google Scholar] [CrossRef] [Green Version]
- Talbot, J.M.; Allison, S.D.; Treseder, K.K. Decomposers in disguise: Mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Funct. Ecol. 2008, 22, 955–963. [Google Scholar] [CrossRef] [Green Version]
- Rineau, F.; Shah, F.; Smits, M.M.; Persson, P.; Johansson, T.; Carleer, R.; Troein, C.; Tunlid, A. Carbon availability triggers the decomposition of plant litter and assimilation of nitrogen by an ectomycorrhizal fungus. ISME J. 2013, 7, 2010–2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colpaert, J.V.; Tichelen, K.K. Decomposition, nitrogen and phosphorus mineralization from beech leaf litter colonized by ectomycorrhizal or litter-decomposing basidiomycetes. New Phytol. 1996, 134, 123–132. [Google Scholar] [CrossRef]
- Gadgil, R.L.; Gadgil, P.D. Mycorrhiza and Litter Decomposition. Nature 1971, 233, 133. [Google Scholar] [CrossRef]
- Fontaine, S.; Barot, S.; Barré, P.; Bdioui, N.; Mary, B.; Rumpel, C. Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 2007, 450, 277–280. [Google Scholar] [CrossRef]
- Wild, B.; Schnecker, J.; Alves, R.J.E.; Barsukov, P.; Bárta, J.; Čapek, P.; Gentsch, N.; Gittel, A.; Guggenberger, G.; Lashchinskiy, N.; et al. Input of easily available organic C and N stimulates microbial decomposition of soil organic matter in arctic permafrost soil. Soil Biol. Biochem. 2014, 75, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Lindahl, B.D.; Ihrmark, K.; Boberg, J.; Trumbore, S.E.; Högberg, P.; Stenlid, J.; Finlay, R.D. Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phytol. 2007, 173, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Orwin, K.H.; Kirschbaum, M.U.F.; St John, M.G.; Dickie, I.A. Organic nutrient uptake by mycorrhizal fungi enhances ecosystem carbon storage: A model-based assessment. Ecol. Lett. 2011, 14, 493–502. [Google Scholar] [CrossRef]
- Averill, C.; Hawkes, C.V. Ectomycorrhizal fungi slow soil carbon cycling. Ecol. Lett. 2016, 19, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, C.; Meyer, H.; Biasi, C.; Rusalimova, O.; Barsukov, P.; Richter, A. Conservation of soil organic matter through cryoturbation in arctic soils in Siberia. J. Geophys. Res. Biogeosci. 2007, 112, 1–8. [Google Scholar] [CrossRef]
- Wild, B.; Schnecker, J.; Knoltsch, A.; Takriti, M.; Mooshammer, M.; Gentsch, N.; Mikutta, R.; Alves, R.J.E.; Gittel, A.; Lashchinskiy, N.; et al. Microbial nitrogen dynamics in organic and mineral soil horizons along a latitudinal transect in western Siberia. Glob. Biogeochem. Cycles 2015, 29, 567–582. [Google Scholar] [CrossRef] [Green Version]
- Talbot, J.M.; Martin, F.; Kohler, A.; Henrissat, B.; Peay, K.G. Functional guild classification predicts the enzymatic role of fungi in litter and soil biogeochemistry. Soil Biol. Biochem. 2015, 88, 441–456. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol. Lett. 2016, 19, 926–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upton, R.N.; Checinska Sielaff, A.; Hofmockel, K.S.; Xu, X.; Polley, H.W.; Wilsey, B.J. Soil depth and grassland origin cooperatively shape microbial community co-occurrence and function. Ecosphere 2020, 11, e02973. [Google Scholar] [CrossRef] [Green Version]
- Toju, H.; Kishida, O.; Katayama, N.; Takagi, K. Networks Depicting the Fine-Scale Co-Occurrences of Fungi in Soil Horizons. PLoS ONE 2016, 11, e0165987. [Google Scholar] [CrossRef] [Green Version]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef]
- Albert, R.; Barabási, A.-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002, 74, 47–97. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Tu, Q.; Zhi, X. Functional Molecular Ecological Networks. MBio 2010, 1, e00169-10. [Google Scholar] [CrossRef] [Green Version]
- Dupont, Y.L.; Olesen, J.M. Ecological modules and roles of species in heathland plant-insect flower visitor networks. J. Anim. Ecol. 2009, 78, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Wu, L.; Yang, Y.; Chen, S.; Zhao, M.; Zhu, Z.; Yang, S.; Qu, Y.; Ma, Q.; He, Z.; Zhou, J.; et al. Long-term successional dynamics of microbial association networks in anaerobic digestion processes. Water Res. 2016, 104, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Zhang, J.; Liu, Y.; Shi, P.; Wei, G. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol. Biochem. 2018, 118, 178–186. [Google Scholar] [CrossRef]
- Tao, J.; Meng, D.; Qin, C.; Liu, X.; Liang, Y.; Xiao, Y.; Liu, Z.; Gu, Y.; Li, J.; Yin, H. Integrated network analysis reveals the importance of microbial interactions for maize growth. Appl. Microbiol. Biotechnol. 2018, 102, 3805–3818. [Google Scholar] [CrossRef]
- Schnecker, J.; Wild, B.; Hofhansl, F.; Eloy Alves, R.J.; Bárta, J.; Čapek, P.; Fuchslueger, L.; Gentsch, N.; Gittel, A.; Guggenberger, G.; et al. Effects of Soil Organic Matter Properties and Microbial Community Composition on Enzyme Activities in Cryoturbated Arctic Soils. PLoS ONE 2014, 9, e94076. [Google Scholar] [CrossRef] [Green Version]
- Jurgens, J.A.; Blanchette, R.A.; Filley, T.R. Fungal diversity and deterioration in mummified woods from the ad Astra Ice Cap region in the Canadian High Arctic. Polar Biol. 2009, 32, 751–758. [Google Scholar] [CrossRef]
- Obase, K.; Douhan, G.W.; Matsuda, Y.; Smith, M.E. Culturable fungal assemblages growing within Cenococcum sclerotia in forest soils. FEMS Microbiol. Ecol. 2014, 90, 708–717. [Google Scholar] [CrossRef] [Green Version]
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E.; Miadlikowska, J.; et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 2006, 443, 818–822. [Google Scholar] [CrossRef]



| Site | Horizon | N | Moisture (%) | pH | DOC (ug/g dw) | DN (ug/g dw) | Ctot (%) | Ntot (%) | C/N ratio | BG (nmol MUF g−1 dw h−1) | CBH (nmol MUF g−1 dw h−1) | LAP (nmol MUF g−1 dw h−1) | NAG (nmol MUF g−1 dw h−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site 1 | Topsoil | 9 | 76.6 ± 3.45 (a) | 6 ± 0.22 (a) | 751.08 ± 308.98 (a) | 2.56 ± 0.9 (b) | 40.06 ± 3.43 (a) | 1.15 ± 0.22 (a) | 44.76 ± 14.8 (a) | 1624.9 ± 294.78 (a) | 239.86 ± 76.91 (a) | 186.85 ± 79.3 (a) | 400.23 ± 83.42 (a) |
| CryoOM | 7 | 50.89 ± 6.91 (b) | 6.35 ± 0.19 (a) | 382.47 ± 84.75 (a) | 8.3 ± 3.38 (ab) | 11.87 ± 3.95 (b) | 0.77 ± 0.24 (ab) | 15.11 ± 0.57 (b) | 441.33 ± 375 (b) | 101.23 ± 107.23 (a) | 74.09 ± 66.4 (a) | 144.61 ± 40.54 (b) | |
| Subsoil | 5 | 30.44 ± 3.31 (c) | 5.81 ± 0.05 (a) | 205.97 ± 63.69 (a) | 9.92 ± 1.22 (a) | 4.63 ± 1.11 (b) | 0.33 ± 0.07 (b) | 13.48 ± 0.64 (b) | 75.32 ± 26.19 (b) | 13.47 ± 3.88 (a) | 15.39 ± 3.48 (a) | 76.08 ± 22.59 (b) | |
| Permafrost | 2 | 55.2 ± 5.89 (ab) | 5.93 ± 0.18 (a) | 695.72 ± 187.2 (a) | 7.68 ± 1.56 (ab) | 8.2 ± 0.33 (b) | 0.56 ± 0.03 (ab) | 14.78 ± 0.25 (b) | 115.51 ± 7.6 (b) | 13.16 ± 3.89 (a) | 10.67 ± 4.41 (a) | 73.9 ± 10.39 (b) | |
| Site 2 | Topsoil | 8 | 54.51 ± 4.08 (a) | 7.49 ± 0.29 (b) | 466.72 ± 142.64 (a) | 3.19 ± 1.46 (b) | 29.57 ± 4.66 (a) | 1.27 ± 0.13 (a) | 22.93 ± 2.25 (b) | 1595.35 ± 381.81 (a) | 268.63 ± 72.32 (a) | 704.2 ± 140.03 (a) | 389.05 ± 174.6 (a) |
| CryoOM | 2 | 54.26 ± 2.53 (a) | 8.05 ± 0.11 (ab) | 250.78 ± 11.86 (a) | 2.64 ± 0.48 (b) | 16.13 ± 1.29 (b) | 1.03 ± 0.03 (a) | 15.6 ± 0.75 (b) | 263.6 ± 96.89 (b) | 36.63 ± 18.79 (b) | 128.23 ± 24.4 (b) | 109.24 ± 2.79 (ab) | |
| Subsoil | 7 | 23.86 ± 3.87 (b) | 8.12 ± 0.23 (ab) | 326.18 ± 65.47 (a) | 19.58 ± 6.76 (ab) | 4.15 ± 0.71 (c) | 0.22 ± 0.06 (b) | 21.22 ± 2.2 (b) | 21.58 ± 17.59 (b) | 1.56 ± 1.33 (b) | 77.66 ± 45.09 (b) | 6.66 ± 5.14 (b) | |
| Permafrost | 9 | 29.04 ± 4.5 (b) | 8.24 ± 0.3 (a) | 374.77 ± 134.22 (a) | 31.19 ± 11.31 (a) | 3 ± 0.13 (c) | 0.1 ± 0.01 (b) | 29.52 ± 1.76 (a) | 1.02 ± 0.47 (b) | 0.15 ± 0.12 (b) | 25.35 ± 4.21 (b) | 0.58 ± 0.37 (b) | |
| Site 3 | Topsoil | 21 | 78.32 ± 4.17 (a) | 5.94 ± 0.38 (a) | 5104.09 ± 2839.27 (a) | 14.5 ± 7.45 (a) | 30.2 ± 4.74 (a) | 1.74 ± 0.25 (a) | 17.93 ± 3.19 (a) | 690.13 ± 399.39 (a) | 90.1 ± 75.82 (a) | 94.67 ± 73.1 (a) | 241.01 ± 116.29 (a) |
| CryoOM | 5 | 54.11 ± 9.86 (b) | 6.11 ± 0.39 (a) | 1066.41 ± 719.77 (a) | 22.09 ± 9.95 (a) | 14.57 ± 3.26 (b) | 1.06 ± 0.2 (b) | 13.72 ± 1.6 (a) | 175.59 ± 108.25 (a) | 20.53 ± 13.51 (a) | 16.18 ± 4.58 (a) | 119.29 ± 31.8 (a) | |
| Permafrost | 1 | 60.4 | 5.61 | 451.92 | 6.87 | 14.64 | 1.22 | 11.97 | 206.35 | 25 | 12.02 | 46 | |
| Site 4 | Topsoil | 12 | 75.57 ± 6.73 (a) | 5.35 ± 0.42 (b) | 2023.25 ± 628.9 (a) | 6.86 ± 3.88 (c) | 37.97 ± 6.07 (a) | 1 ± 0.26 (a) | 44.73 ± 12.06 (a) | 1336.83 ± 371.99 (a) | 204.45 ± 77.38 (a) | 59.27 ± 40.06 (a) | 271.86 ± 110.15 (a) |
| CryoOM | 16 | 59.85 ± 9.11 (a) | 5.93 ± 0.23 (b) | 1588.26 ± 1028.78 (a) | 13.9 ± 4.18 (bc) | 17.44 ± 3.81 (b) | 0.87 ± 0.12 (a) | 19.68 ± 2.59 (b) | 473.34 ± 422.09 (b) | 110.9 ± 134.8 (a) | 53.8 ± 25.98 (a) | 316.64 ± 104.08 (a) | |
| Subsoil | 10 | 29.9 ± 6.03 (b) | 5.91 ± 0.37 (b) | 442.22 ± 138.73 (a) | 38.54 ± 10.32 (b) | 4.23 ± 0.98 (c) | 0.22 ± 0.04 (b) | 18.65 ± 2.12 (b) | 99.28 ± 55.15 (b) | 11.69 ± 4.79 (a) | 35.73 ± 9.39 (a) | 37.7 ± 23.6 (b) | |
| Permafrost | 8 | 38.1 ± 11.77 (b) | 7.32 ± 0.56 (a) | 671.93 ± 477.72 (a) | 100.06 ± 24.95 (a) | 3.17 ± 0.64 (c) | 0.21 ± 0.04 (b) | 15.22 ± 0.88 (b) | 60.28 ± 52.38 (b) | 7.04 ± 7.78 (a) | 27.92 ± 9.69 (a) | 36.17 ± 33.42 (b) |
| Horizon | Topsoil | CryoOM | Subsoil | Permafrost | Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|---|---|---|---|---|
| Empirical network | Total zOTUs a | 558 | 413 | 479 | 479 | 688 | 458 | 324 | 212 |
| Abundance (%) b | 61.13 | 68.22 | 63.22 | 71.06 | 75.97 | 67.33 | 65 | 71.36 | |
| Total significant correlations | 3054 | 2454 | 3426 | 1110 | 6886 | 666 | 1152 | 814 | |
| Total node | 476 | 340 | 437 | 281 | 643 | 267 | 246 | 130 | |
| Total edge | 1527 | 1227 | 1713 | 555 | 3443 | 333 | 576 | 407 | |
| Positive edge | 1527 | 1216 | 1699 | 553 | 3277 | 331 | 568 | 407 | |
| Negative edge | 0 | 11 | 14 | 2 | 166 | 2 | 8 | 0 | |
| Average path length (APL) | 7.12 | 5.3 | 5.42 | 7.61 | 5.72 | 7.2 | 5.4 | 5.23 | |
| Degree distribution (DD) | 6.42 ± 7.88 | 7.22 ± 7.05 | 7.84 ± 6.05 | 3.95 ± 3.3 | 10.71 ± 9.83 | 2.49 ± 1.89 | 4.68 ± 4.66 | 6.26 ± 7.64 | |
| Average closeness (AC) | −4.42 ± 0.28 | −4.16 ± 0.28 | −3.82 ± 0.21 | −4.5 ± 0.24 | −4.33 ± 0.25 | −4.67 ± 0.16 | −4.23 ± 0.27 | −3.82 ± 0.23 | |
| Average betweenness (AB) | 1225.6 ± 2195.08 | 606.04 ± 1405.7 | 923.8 ± 1432.28 | 494.75 ± 1148.79 | 1404.42 ± 1920.9 | 246.18 ± 597.43 | 353.79 ± 728.76 | 153.46 ± 332.88 | |
| Edge density (ED) | 0.0135 | 0.0213 | 0.018 | 0.0141 | 0.0167 | 0.0094 | 0.0191 | 0.0485 | |
| Diameter (D) | 20 | 16 | 15 | 20 | 17 | 19 | 17 | 17 | |
| Clustering coefficient (CC) | 0.24 | 0.34 | 0.23 | 0.26 | 0.18 | 0.13 | 0.23 | 0.3 | |
| Number of modules | 31 | 21 | 12 | 34 | 21 | 40 | 27 | 18 | |
| Modularity (M) | 0.72 | 0.68 | 0.73 | 0.82 | 0.69 | 0.85 | 0.65 | 0.38 | |
| Random network c | Average path length (APL) | 3.52 ± 0.008 | 3.16 ± 0.007 | 3.18 ± 0.005 | 4.2 ± 0.042 | 2.44 ± 0.001 | 3.64 ± 0.022 | 2.7 ± 0.005 | 2.17 ± 0.005 |
| Clustering coefficient (CC) | 0.01 ± 0.002 | 0.02 ± 0.003 | 0.02 ± 0.002 | 0.01 ± 0.004 | 0.03 ± 0.001 | 0.02 ± 0.004 | 0.04 ± 0.003 | 0.1 ± 0.005 | |
| Modularity (M) | 0.32 ± 0.01 | 0.3 ± 0.011 | 0.28 ± 0.01 | 0.44 ± 0.016 | 0.15 ± 0.004 | 0.38 ± 0.014 | 0.26 ± 0.011 | 0.21 ± 0.012 |
| zOTUs ID | Genera | Species | Lifestyle | Topsoil | CryoOM | Subsoil | Permafrost |
|---|---|---|---|---|---|---|---|
| Zotu6272 | Cladophialophora | unidentified | Soil_Saprotroph | Module hubs | Peripheral | Peripheral | - |
| Zotu7 | Cortinarius | Cortinarius flexipes | Ectomycorrhizal | Module hubs | - | - | Peripheral |
| Zotu952 | Mortierella | Mortierella antarctica | Soil_Saprotroph | Peripheral | Module hubs | Peripheral | Peripheral |
| Zotu1060 | Mortierella | unidentified | Soil_Saprotroph | Peripheral | Peripheral | Module hubs | - |
| Zotu19 | Neobulgaria | unidentified | Wood_Saprotroph | Module hubs | - | Peripheral | Peripheral |
| Zotu229 | Oidiodendron | unidentified | Soil_Saprotroph | - | Peripheral | Connectors | Peripheral |
| Zotu972 | Penicillium | Penicillium bialowiezense | unspecified_Saprotroph | Peripheral | - | Connectors | Peripheral |
| Zotu1148 | Penicillium | Penicillium odoratum | unspecified_Saprotroph | Peripheral | Peripheral | Connectors | Peripheral |
| Zotu3565 | Penicillium | Penicillium oregonense | unspecified_Saprotroph | Peripheral | Module hubs | Peripheral | Peripheral |
| Zotu7940 | Russula | Russula emetica | Ectomycorrhizal | Module hubs | - | Peripheral | - |
| Zotu4713 | Sympodiella | Sympodiella acicola | Litter_Saprotroph | - | - | Connectors | Peripheral |
| Zotu389 | Tetracladium | unidentified | Litter_Saprotroph | Module hubs | - | - | Peripheral |
| Zotu4853 | Tetracladium | unidentified | Litter_Saprotroph | Peripheral | Module hubs | Peripheral | Peripheral |
| Zotu5812 | Tylospora | Tylospora fibrillosa | Ectomycorrhizal | - | Module hubs | Peripheral | - |
| Zotu3079 | Vishniacozyma | Vishniacozyma_victoriae | Soil_Saprotroph | Connectors | - | - | Peripheral |
| zOTUs ID | Genera | Species | Lifestyle | Site 1 | Site 2 | Site 3 | Site 4 |
|---|---|---|---|---|---|---|---|
| Zotu1843 | Cadophora | Cadophora finlandica | Litter_Saprotroph | Module hubs | Peripheral | Peripheral | Peripheral |
| Zotu6578 | Cenococcum | Cenococcum geophilum | Ectomycorrhizal | Module hubs | Peripheral | - | - |
| Zotu3982 | Luellia | unidentified | Wood_Saprotroph | Module hubs | Peripheral | Peripheral | - |
| Zotu5146 | Meliniomyces | unidentified | Root_Endophyte | Connectors | Peripheral | Peripheral | - |
| Zotu6819 | Verrucaria | Verrucaria calciseda | Lichenized | Connectors | Peripheral | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varsadiya, M.; Urich, T.; Hugelius, G.; Bárta, J. Fungi in Permafrost-Affected Soils of the Canadian Arctic: Horizon- and Site-Specific Keystone Taxa Revealed by Co-Occurrence Network. Microorganisms 2021, 9, 1943. https://doi.org/10.3390/microorganisms9091943
Varsadiya M, Urich T, Hugelius G, Bárta J. Fungi in Permafrost-Affected Soils of the Canadian Arctic: Horizon- and Site-Specific Keystone Taxa Revealed by Co-Occurrence Network. Microorganisms. 2021; 9(9):1943. https://doi.org/10.3390/microorganisms9091943
Chicago/Turabian StyleVarsadiya, Milan, Tim Urich, Gustaf Hugelius, and Jiří Bárta. 2021. "Fungi in Permafrost-Affected Soils of the Canadian Arctic: Horizon- and Site-Specific Keystone Taxa Revealed by Co-Occurrence Network" Microorganisms 9, no. 9: 1943. https://doi.org/10.3390/microorganisms9091943
APA StyleVarsadiya, M., Urich, T., Hugelius, G., & Bárta, J. (2021). Fungi in Permafrost-Affected Soils of the Canadian Arctic: Horizon- and Site-Specific Keystone Taxa Revealed by Co-Occurrence Network. Microorganisms, 9(9), 1943. https://doi.org/10.3390/microorganisms9091943







