Technological Parameters, Anti-Listeria Activity, Biogenic Amines Formation and Degradation Ability of L. plantarum Strains Isolated from Sheep-Fermented Sausage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Genotyping and Identification
2.2. Technological Characterization of L. plantarum Strains
2.2.1. Acidifying and Growth Activity
2.2.2. Proteolytic and Lipolytic Activity
2.2.3. Anti-Listeria Activity of L. plantarum Strains
2.3. Proof of Strains Relevant Safety Properties
2.3.1. Determination of Amino Acids Decarboxylase Activity
2.3.2. Detection of the Genes Related to Biogenic Amines (BAs) and BAs Degradation Ability
2.4. Statistical Analysis
3. Results
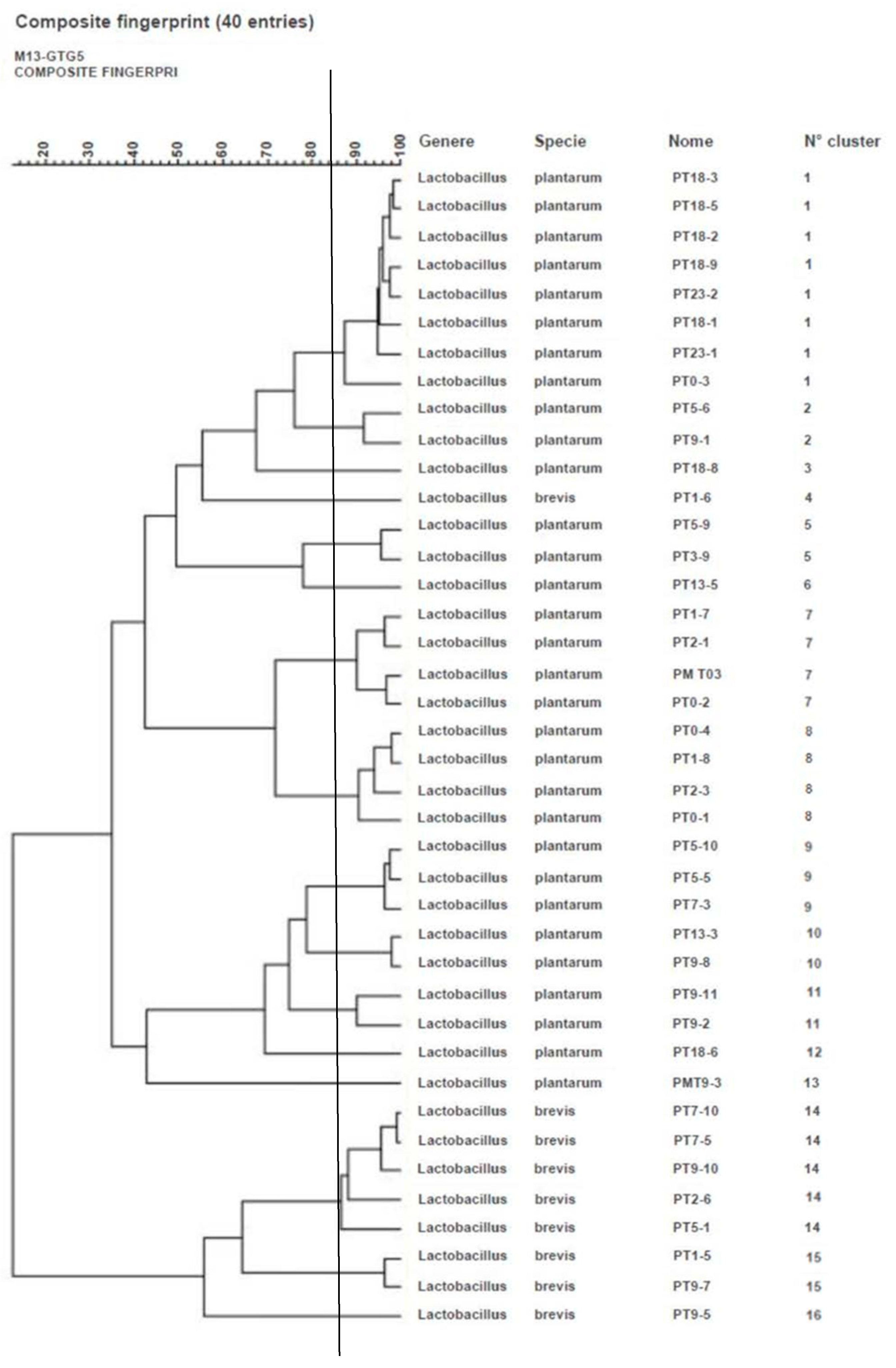
3.1. Bacteria Identification and Genotyping
3.2. Technological Characterization of L. plantarum Strains
3.2.1. Acidifying and Growth Ability of L. plantarum Strains
3.2.2. Proteolytic and Lipolytic Activity of L. plantarum Strains
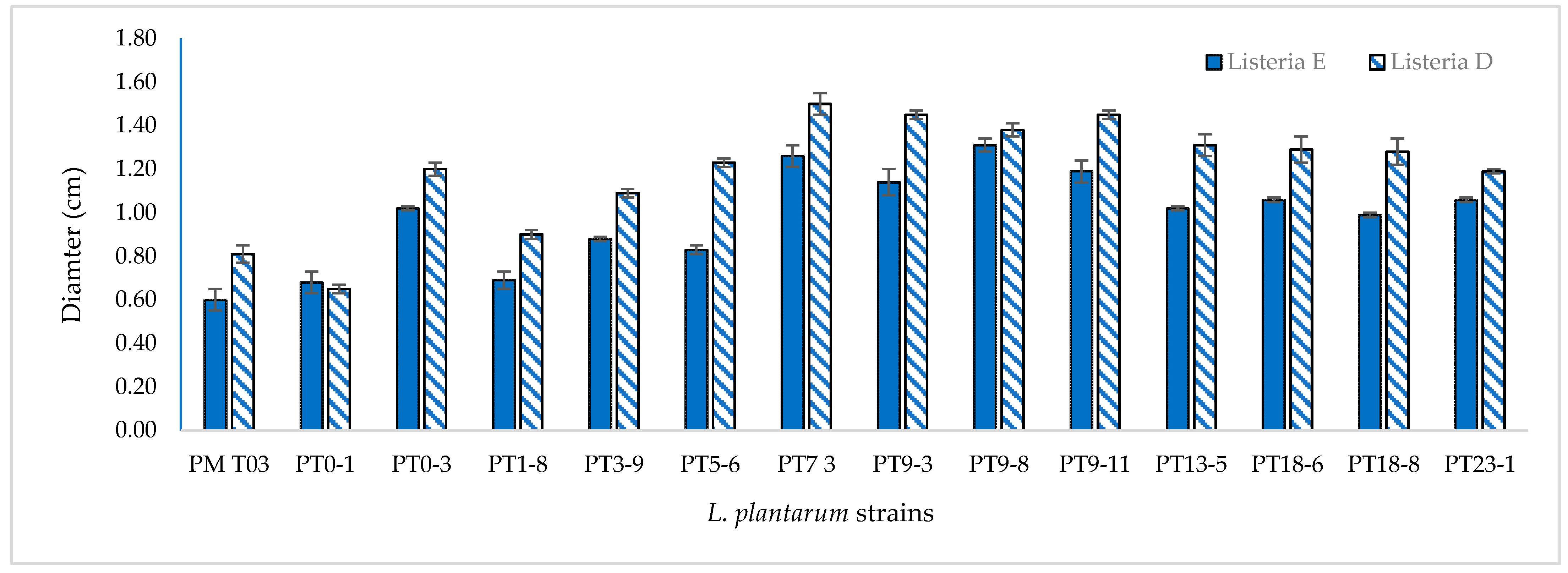
3.3. Anti-Listeria Activity
3.4. Safety-Related Properties
3.4.1. Amino Acid Decarboxylase Activity
3.4.2. Detection of BA-Producing Genes of the Strains
3.4.3. BAs degradation ability
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iaccarino, T.; Di Monaco, R.; Mincione, A.; Cavella, S.; Masi, P. Influence of information on origin and technology on the consumer response: The case of soppressata salami. Food Qual. Prefer. 2006, 17, 76–84. [Google Scholar] [CrossRef]
- Martuscelli, M.; Serio, A.; Capezio, O.; Mastrocola, D. Safety, Quality and Analytical Authentication of ḥalāl Meat Products, with Particular Emphasis on Salami: A Review. Foods 2020, 9, 1111. [Google Scholar] [CrossRef]
- Mangia, N.P.; Trani, A.; Di Luccia, A.; Faccia, M.; Gambacorta, G.; Fancello, F.; Deiana, P. Effect of the use of autochthonous Lactobaccillus curvatus, Lactobacillus plantarum and Staphylococcus xylosus strains on microbiological and biochemical properties of the Sardinian fermented sausage. Eur. Food Res. Tech. 2013, 236, 557–566. [Google Scholar] [CrossRef]
- Mangia, N.P.; Murgia, M.A.; Garau, G.; Merella, R.; Deiana, P. Sardinian fermented sheep sausage: Microbial biodiversity resource for quality improvement. Opt. Méditerranéennes A 2008, 78, 273–277. [Google Scholar]
- Laranjo, M.; Talon, R.; Lauková, A.; Fraqueza, J.M.; Elias, M. Traditional Meat Products: Improvement of Quality and Safety. J. Food Qual. 2017, 2017, 2873793. [Google Scholar] [CrossRef] [Green Version]
- Teleky, B.-E.; Martău, A.G.; Ranga, F.; Chețan, F.; Vodnar, D.C. Exploitation of Lactic Acid Bacteria and Baker’s Yeast as Single or Multiple Starter Cultures of Wheat Flour Dough Enriched with Soy Flour. Biomolecules 2020, 10, 778. [Google Scholar] [CrossRef] [PubMed]
- Roseiro, C.; Santos, C.; Sol, M.; Silva, L.; Fernandes, I. Prevalence of biogenic amines during ripening of a traditional dry fermented pork sausage and its relation to the amount of sodium chloride added. Meat Sci. 2006, 74, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Latorre-Moratalla, M.L.; Bover-Cid, S.; Veciana-Nogués, M.T.; Vidal-Carou, M.C. Control of biogenic amines in fermented sausages: Role of starter cultures. Front. Microbiol. 2015, 3, 169. [Google Scholar] [CrossRef] [Green Version]
- Doeun, D.; Davaatseren, M.; Chung, M.S. Biogenic amines in foods. Food Sci. Biotech. 2017, 26, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Carou, M.C.; Latorre-Moratalla, M.L.; Veciana-Nogués, M.T.; Bover-Cid, S. Biogenic amines: Risks and control. In Handbook of Fermented Meat and Poultry; Toldrà, F., Ed.; Blackwell Publishing: Hoboken, NJ, USA, 2007; pp. 455–468. [Google Scholar]
- Gardini, F.; Ozogul, Y.; Suzzi, G.; Tabanelli, G.; Ozogul, F. Technological factors affecting biogenic amine content n foods: A review. Front. Microbiol. 2016, 7, 1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzzi, G.; Gardini, F. Biogenic amines in dry fermented sausages: A review. Int. J. Food Microbiol. 2003, 88, 41–54. [Google Scholar] [CrossRef]
- Galgano, M.C.; Condelli, N.; Favati, F. Focused Review: Agmatine in fermented foods. Front. Microbiol. 2012, 3, 199. [Google Scholar] [CrossRef] [Green Version]
- Callejón, S.; Sendra, R.; Ferrer, S.; Pardo, I. Identification of a novel enzymatic activity from lactic acid bacteria able to degrade biogenic amines in wine. Appl. Microbiol. Biotechnol. 2014, 98, 185–198. [Google Scholar] [CrossRef]
- Guarcello, R.; de Angelis, M.; Settanni, L.; Formiglio, S.; Gaglio, R.; Minervini, F.; Moschetti, G.; Gobbetti, M. Selection of Amine-Oxidizing Dairy Lactic Acid Bacteria and Identification of the Enzyme and Gene Involved in the Decrease of Biogenic Amines. Appl. Environ. Microbiol. 2016, 82, 6870–6880. [Google Scholar] [CrossRef] [Green Version]
- Pištěková, H.; Jančová, P.; Berčíková, L.; Buňka, F.; Sokolová, I.; Šopík, T.; Maršálková, K.; de Amaral, O.M.R.P.; Buňková, L. Application of qPCR for multicopper oxidase gene (MCO) in biogenic amines degradation by Lactobacillus casei. Food Microbiol. 2020, 91, 103550. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, L.; Giraffa, G. Rapid identification of dairy lactic acid bacteria by M13 generated, RAPD-PCR fingerprint database. J. Microbiol. Methods 2005, 63, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Gallego, J.; Alessandria, V.; Fontana, M.; Bisotti, S.; Taricco, S.; Dolci, P.; Cocolin, L.; Rantsiou, K. Diversity and functional characterization of Lactobacillus spp. isolated throughout the ripening of a hard cheese. Int. J. Food Microbiol. 2014, 181, 60–66. [Google Scholar] [CrossRef]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus 25 plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis 22 1 and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essid, I.; Medini, M.; Hassouna, M. Technological and safety properties of Lactobacillus plantarum strains isolated from a Tunisian traditional salted meat. Meat Sci. 2009, 81, 203–208. [Google Scholar] [CrossRef]
- Mauriello, G.; Casaburi, A.; Blaiotta, G.; Villani, F. Isolation and technological properties of coagulase negative staphylococci from fermented sausages of Southern Italy. Meat Sci. 2004, 67, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Drosinos, E.H.; Paramithiotis, S.; Kolovos, G.; Tsikouras, I. Phenotypic and technological diversity of lactic acid bacteria and staphylococci isolated from traditionally fermented sausages in Southern Greece. Food Microbiol. 2007, 24, 260–270. [Google Scholar] [CrossRef]
- Mangia, N.P.; Saliba, L.; Deiana, P. Functional and safety characterization of autochthonous Lactobacillus paracasei FS103 isolated from sheep cheese and its survival in sheep and cow fermented milk during cold storage. Ann. Microbiol. 2019, 69, 161–170. [Google Scholar] [CrossRef]
- Mete, A.; Coşansu, S.; Demirkol, O.; Ayhan, K. Amino acid decarboxylase activities and biogenic amine formation abilities of lactic acid bacteria isolated from shalgam. Int. J. Food Prop. 2017, 20, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Le Jeune, C.; Lonvaud-Funel, A.; ten Brink, B.; Hofstra, H.; Van der Vossen, J.M.B.M. Development of a detection system for histamine decarboxylating lactic acid bacteria based on DNA probes, PCR and activity test. J. Appl. Bacteriol. 1995, 78, 316–326. [Google Scholar] [CrossRef]
- Coton, M.; Coton, E.; Lucas, P.; Lonvaud, A. Identification of the gene encoding a putative tyrosine decarboxylase of Carnobacterium divergens 508. Development of molecular tools for the detection of tyramine-producing bacteria. Food Microbiol. 2004, 21, 125–130. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Jan, S.; Yang, Q.; Wang, M. Expression and functional analysis of the lysine decarboxylase and copper amine oxidase genes from the endophytic fungus Colletotrichum gloeosporioides ES026. Sci. Rep. 2017, 7, 2766. [Google Scholar] [CrossRef] [Green Version]
- Nannelli, F.; Claisse, O.; Gindreau, E.; de Revel, G.; Lonvaud-Funel, A.; Lucas, P.M. Determination of lactic acid bacteria producing biogenic amines in wine by quantitative PCR methods. Lett. Appl. Microbiol. 2008, 47, 594–599. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Ortu, S.; Felis, G.E.; Marzotto, M.; Deriu, A.; Molicotti, P.; Sechi, L.A.; Dellaglio, F.; Zanetti, S. Identification and functional characterization of Lactobacillus strains isolated from milk and Gioddu, a traditional Sardinian fermented milk. Int. Dairy J. 2007, 17, 1312–1320. [Google Scholar] [CrossRef]
- Martino, M.E.; Bayjanov, J.R.; Caffrey, B.E.; Wels, M.; Joncour, P.; Hughes, S.; Gillet, B.; Kleerebezem, M.; van Hijum, S.A.; Leulier, F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 2016, 18, 4974–4989. [Google Scholar] [CrossRef] [PubMed]
- Kozačinski, L.; Drosinos, E.; Čaklovica, F.; Cocolin, L.; Gasparik-Reichardt, J.; Veskovi, S. Investigation of microbial association of traditionally fermented sausages. Food Technol. Biotechnol. 2008, 46, 93–106. [Google Scholar]
- Benito, M.J.; Martin, A.; Aranda, A.; Perez-Nevado, F.; Ruiz-Moyano, S.; Cordoba, M.G. Characterization and selection of autochthonous lactic acid bacteria isolated from traditional Iberian dry-fermented salchichón and chorizo sausages. J. Food Sci. 2007, 72, 193–201. [Google Scholar] [CrossRef] [PubMed]
- García-Fontán, M.C.; Lorenzo, J.M.; Parada, A.; Franco, I.; Carballo, J. Microbiological characteristics of “androlla”, a Spanish traditional pork sausage. Food Microbiol. 2007, 24, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Manzano, M.; Cantoni, C.; Comi, G. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 2001, 67, 5113–5121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comi, G.; Urso, R.; Iacumin, L.; Rantsiou, K.; Cattaneo, P.; Cantoni, C.; Cocolin, L. Characterisation of naturally fermented sausages produced in the North East of Italy. Meat Sci. 2005, 69, 381–392. [Google Scholar] [CrossRef]
- Parente, E.S.; Grieco, S.; Crudele, M.A. Phenotypic diversity of lactic acid bacteria isolated from fermented sausages produced in Basilicata (Southern Italy). J. Appl. Microbiol. 2001, 90, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Coppola, S.; Mauriello, G.; Aponte, M.; Moschetti, G.; Villani, F. Microbial succession during ripening of Naples-type salami, a southern Italian fermented sausage. Meat Sci. 2000, 56, 321–329. [Google Scholar] [CrossRef]
- Aquilanti, L.; Garofalo, C.; Osimani, A.; Clementi, F. Ecology of lactic acid bacteria and coagulase negative cocci in fermented dry sausages manufactured in Italy and other Mediterranean countries: An overview. Int. Food Res. J. 2016, 23, 429–445. [Google Scholar]
- Lee, J.Y.; Kim, C.J.; Kunz, B. Identification of lactic acid bacteria isolated from kimchi and studies on their suitability for application as starter culture in the production of fermented sausages. Meat Sci. 2006, 72, 437–445. [Google Scholar] [CrossRef]
- Bovolenta, S.; Boscolo, D.; Dovier, S.; Morgante, M.; Pallotti, A.; Piasentier, E. Effect of pork lard content on the chemical, microbiological and sensory properties of a typical fermented meat product (Pitina) obtained from Alpagota sheep. Meat Sci. 2008, 80, 771–779. [Google Scholar] [CrossRef]
- Meloni, D.; Consolati, S.G.; Mazza, R.; Mureddu, A.; Fois, F.; Piras, F.; Mazzette, R. Presence and molecular characterization of the major serovars of Listeria monocytogenes in ten Sardinian fermented sausage processing plants. Meat Sci. 2014, 97, 443–450. [Google Scholar] [CrossRef]
- Papamanoli, E.; Tzanetakis, N.; Litopoulou-Tzanetakis, E.; Kotzekidou, P. Characterization of lactic acid bacteria isolated from a Greek dry-fermented sausage in respect of their technological and probiotic properties. Meat Sci. 2003, 65, 859–867. [Google Scholar] [CrossRef]
- Kamiloğlu, A.; Kaban, G.; Kaya, M. Effects of autochthonous Lactobacillus plantarum strains on Listeria monocytogenes in sucuk during ripening. J. Food Safety 2019, 39, 12618. [Google Scholar] [CrossRef]
- Belicová, A.; Mikulášová, M.; Dušinský, R. Probiotic potential and safety properties of Lactobacillus plantarum from Slovak Bryndza cheese. BioMed Res. Int. 2013, 760298, 8. [Google Scholar]
- Campaniello, D.; Speranza, B.; Bevilacqua, A.; Altieri, C.; Rosaria Corbo, M.; Sinigaglia, M. Industrial Validation of a Promising Functional Strain of Lactobacillus plantarum to Improve the Quality of Italian Sausages. Microorganisms 2020, 8, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G.; Fiocco, D. Use of Lactobacillus plantarum Strains as a Bio-Control Strategy against Food-Borne Pathogenic Microorganisms. Front. Microbiol. 2016, 7, 464. [Google Scholar] [CrossRef] [Green Version]
- Lazzeri, A.M.; Mangia, N.P.; Mura, M.E.; Floris, I.; Satta, A.; Ruiu, L. Potential of novel food-borne Lactobacillus isolates against the honeybee pathogen Paenibacillus larvae. Biocontrol Sci. Technol. 2000, 30, 897–908. [Google Scholar] [CrossRef]
- Fadda, S.; Sanz, Y.; Vignolo, G.M.; Aristoy, C.; Oliver, G.; Toldra, F. Characterization of muscle sarcoplasmic and myofibrillar protein hydrolysis caused by Lactobacillus plantarum. Appl. Environ. Microbiol. 1999, 65, 3540–3546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Xia, W.; Gao, P.; Xu, Y. Sarcoplasmic Protein Hydrolysis Activity of Lactobacillus plantarum 120 Isolated from Suanyu: A Traditional Chinese Low Salt Fermented Fish. J. Food Proc. Preserv. 2017, 41, e12821. [Google Scholar] [CrossRef]
- Mejri, L.; Hassoun, M. Characterization and selection of Lactobacillus plantarum species isolated from dry fermented sausage reformulated with camel meat and hump fat. Appl. Biol. Chem. 2016, 59, 533–542. [Google Scholar] [CrossRef]
- European Commission (EC) Regulation N° 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2005:338:0001:0026:IT:PDF (accessed on 21 May 2021).
- Talon, R.; Leroy, S. Diversity and safety hazards of bacteria involved in meat fermentations. Meat Sci. 2011, 89, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Landeta, G.; Curiel, J.A.; Carrascosa, A.V.; Muñoz, R.; De Las Rivas, B. Technological and safety properties of lactic acid bacteria isolated from Spanish dry-cured sausages. Meat Sci. 2013, 95, 272–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladero, V.; Martín, M.C.; Redruello, B.; Mayo, B.; Flórez, A.B.; Fernández, M.; Alvarez, M.A. Genetic and functional analysis of biogenic amine production capacity among starter and non-starter lactic acid bacteria isolated from artisanal cheeses. Eur. Food Res. Technol. 2015, 24, 377–383. [Google Scholar] [CrossRef] [Green Version]
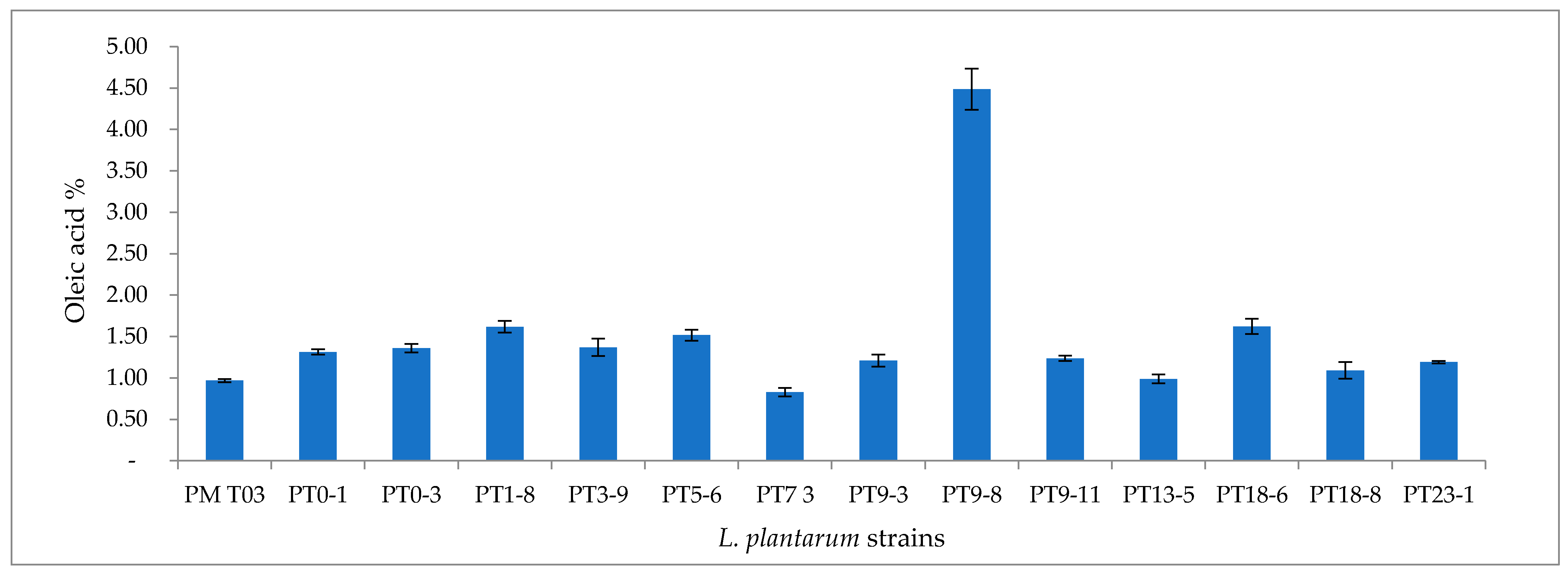


| GENE | PRIMER FORWARD | PRIMER REVERSE | T° | bp | Reference |
|---|---|---|---|---|---|
| hdc | 5′AGATGGTATTGTTTCTTATG3′ | 5′AGACCATACACCATAACCTT3′ | 46 | 367 | [26] |
| tyrdc | 5′CAAATGGAAGAAGAAGTTGG3′ | 5′ACATAGTCAACCATATTGAA3′ | 50 | 1100 | [27] |
| ldc | 5′TAGGTTCAGATTGGCCCTTAG3′ | 5′ACTTCAACACCTGCTGCTTTC3′ | 52 | 769 | [28] |
| agdi | 5′ATGCCCGGTGAATTTGAA3′ | 5′ TTGCGCTGGTTTAGCAC3′ | 48 | 90 | [29] |
| SufI | 5′TCGTTGATTTTGGTCAGTATCA3′ | 5′ATATGGCAGTGATACATGTAAAC3′ | 50 | 454 | * NC_004567.2 |
| L. plantarum | 0 h | 3 h | 6 h | 9 h | 24 h | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| pH | Log CFU mL−1 | pH | Log CFU mL−1 | pH | Log CFU mL−1 | pH | Log CFU mL−1 | pH | Log CFU mL−1 | |
| PM T03 | 5.88 ± 0.01 a | 4.75 ± 0.01 a | 5.62 ± 0.02 a | 5.53 ± 0.01 a | 5.54 ± 0.01 b | 5.63 ± 0.02 b | 4.87 ± 0.02 a | 6.27 ± 0.01 b | 4.27 ± 0.03 a | 7.99 ± 0.02 b |
| PT0-1 | 5.87 ± 0.01 a | 4.75 ± 0.01 a | 5.63 ± 0.04 b | 5.38 ± 0.03 a | 5.57 ± 0.01 b | 5.34 ± 0.02 a | 5.47 ± 0.03 b | 6.21 ± 0.00 b | 4.28 ± 0.01 b | 7.85 ± 0.05 a |
| PT0-3 | 5.87 ± 0.03 a | 4.52 ± 0.06 a | 5.22 ± 0.02 a | 5.36 ± 0.03 a | 5.29 ± 0.01 b | 6.18 ± 0.08 b | 4.85 ± 0.06 a | 6.59 ± 0.08 b | 4.14 ± 0.02 b | 6.53 ± 0.05 b |
| PT1-8 | 5.84 ± 0.05 a | 4.80 ± 0.02 a | 5.79 ± 0.03 b | 5.54 ± 0.01 a | 5.16 ± 0.04 b | 5.46 ± 0.02 b | 5.04 ± 0.06 b | 6.33 ± 0.03 a | 3.94 ± 0.04 b | 7.75 ± 0.06 a |
| PT3-9 | 5.86 ± 0.01 a | 4.52 ± 0.01 a | 5.63 ± 0.02 b | 5.17 ± 0.04 a | 5.44 ± 0.04 a | 4.99 ± 0.03 b | 4.67 ± 0.06 b | 6.09 ± 0.04 b | 4.05 ± 0.04 b | 6.64 ± 0.08 b |
| PT5-6 | 5.80 ± 0.03 a | 4.55 ± 0.02 a | 5.41 ± 0.05 b | 5.20 ± 0.01 a | 5.20 ± 0.04 a | 5.20 ± 0.02 a | 5.18 ± 0.03 a | 6.45 ± 0.05 a | 4.25 ± 0.03 a | 7.67 ± 0.02 b |
| PT7-3 | 5.74 ± 0.00 a | 4.79 ± 0.00 a | 5.71 ± 0.04 b | 5.44 ± 0.01 a | 5.40 ± 0.01 b | 5.36 ± 0.01 b | 5.09 ± 0.03 a | 8.31 ± 0.02 b | 4.27 ± 0.04 a | 9.44 ± 0.03 b |
| PT9-8 | 5.81 ± 0.03 a | 4.95 ± 0.02 a | 5.71 ± 0.01 a | 5.53 ± 0.01 a | 5.49 ± 0.02 a | 5.46 ± 0.01 b | 5.12 ± 0.06 b | 7.33 ± 0.01 b | 4.32 ± 0.03 a | 8.72 ± 0.01 b |
| PT9-11 | 5.83 ± 0.03 a | 4.82 ± 0.01 a | 5.80 ± 0.00 b | 5.38 ± 0.00 a | 5.30 ± 0.02 b | 5.32 ± 0.01 b | 5.14 ± 0.01 b | 7.67 ± 0.03 b | 4.25 ± 0.01 a | 8.84 ± 0.01 b |
| PT9-3 | 5.86 ± 0.02 a | 4.93 ± 0.03 a | 5.63 ± 0.01 a | 5.64 ± 0.00 a | 5.09 ± 0.01 a | 5.30 ± 0.01 b | 5.08 ± 0.04 a | 7.29 ± 0.00 b | 4.31 ± 0.04 a | 9.58 ± 0.05 b |
| PT13-5 | 5.81 ± 0.02 a | 4.43 ± 0.01 a | 5.47 ± 0.02 b | 5.30 ± 0.03 a | 5.41 ± 0.03 b | 5.26 ± 0.04 a | 4.91 ± 0.01 b | 6.61 ± 0.07 a | 4.12 ± 0.02 a | 7.29 ± 0.04 a |
| PT18-6 | 5.86 ± 0.03 a | 4.87 ± 0.03 a | 5.47 ± 0.03 b | 5.83 ± 0.07 a | 5.31 ± 0.05 a | 5.62 ± 0.03 b | 4.47 ± 0.03 b | 6.90 ± 0.04 b | 4.09 ± 0.02 a | 7.86 ± 0.05 a |
| PT18-8 | 5.85 ± 0.05 a | 4.50 ± 0.03 a | 5.51 ± 0.01 b | 5.32 ± 0.01 a | 5.42 ± 0.08 a | 5.10 ± 0.02 b | 4.87 ± 0.04 b | 6.04 ± 0.10 a | 4.05 ± 0.06 a | 7.07 ± 0.03 b |
| PT23-1 | 5.81 ± 0.06 a | 4.45 ± 0.05 a | 5.49 ± 0.05 a | 5.27 ± 0.02 a | 5.43 ± 0.07 b | 5.12 ± 0.02 b | 4.69 ± 0.08 b | 5.95 ± 0.03 b | 3.82 ± 0.05 b | 6.67 ± 0.04 b |
| L. plantarum strains | Hys | Lys | Arg | Orn | Tyr | Trp | Phe |
|---|---|---|---|---|---|---|---|
| PT0-3 | − | + | − | − | − | − | − |
| PT9-2 | − | + | + | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mangia, N.P.; Cottu, M.; Mura, M.E.; Murgia, M.A.; Blaiotta, G. Technological Parameters, Anti-Listeria Activity, Biogenic Amines Formation and Degradation Ability of L. plantarum Strains Isolated from Sheep-Fermented Sausage. Microorganisms 2021, 9, 1895. https://doi.org/10.3390/microorganisms9091895
Mangia NP, Cottu M, Mura ME, Murgia MA, Blaiotta G. Technological Parameters, Anti-Listeria Activity, Biogenic Amines Formation and Degradation Ability of L. plantarum Strains Isolated from Sheep-Fermented Sausage. Microorganisms. 2021; 9(9):1895. https://doi.org/10.3390/microorganisms9091895
Chicago/Turabian StyleMangia, Nicoletta P., Michele Cottu, Maria E. Mura, Marco A. Murgia, and Giuseppe Blaiotta. 2021. "Technological Parameters, Anti-Listeria Activity, Biogenic Amines Formation and Degradation Ability of L. plantarum Strains Isolated from Sheep-Fermented Sausage" Microorganisms 9, no. 9: 1895. https://doi.org/10.3390/microorganisms9091895
APA StyleMangia, N. P., Cottu, M., Mura, M. E., Murgia, M. A., & Blaiotta, G. (2021). Technological Parameters, Anti-Listeria Activity, Biogenic Amines Formation and Degradation Ability of L. plantarum Strains Isolated from Sheep-Fermented Sausage. Microorganisms, 9(9), 1895. https://doi.org/10.3390/microorganisms9091895






