Titan Cells and Yeast Forms of Cryptococcus neoformans and Cryptococcus gattii Are Recognized by GXMR-CAR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Heat-Killed C. gattii and C. neoformans Cells
2.2. Generation of Cryptococcus spp. Titan Cells
2.3. Construction of CAR Targeting Cryptococcus spp., GXMR-CAR
2.4. Lentiviral Particle Production
2.5. Generation of GXMR-CAR Jurkat Cells by Transduction
2.6. Detection of GXMR-CAR on the Cell Surface by Flow Cytometry
2.7. Interaction of GXMR-CAR+ Cells and Cryptococcus spp. by Fluorescence Microscopy
3. Results
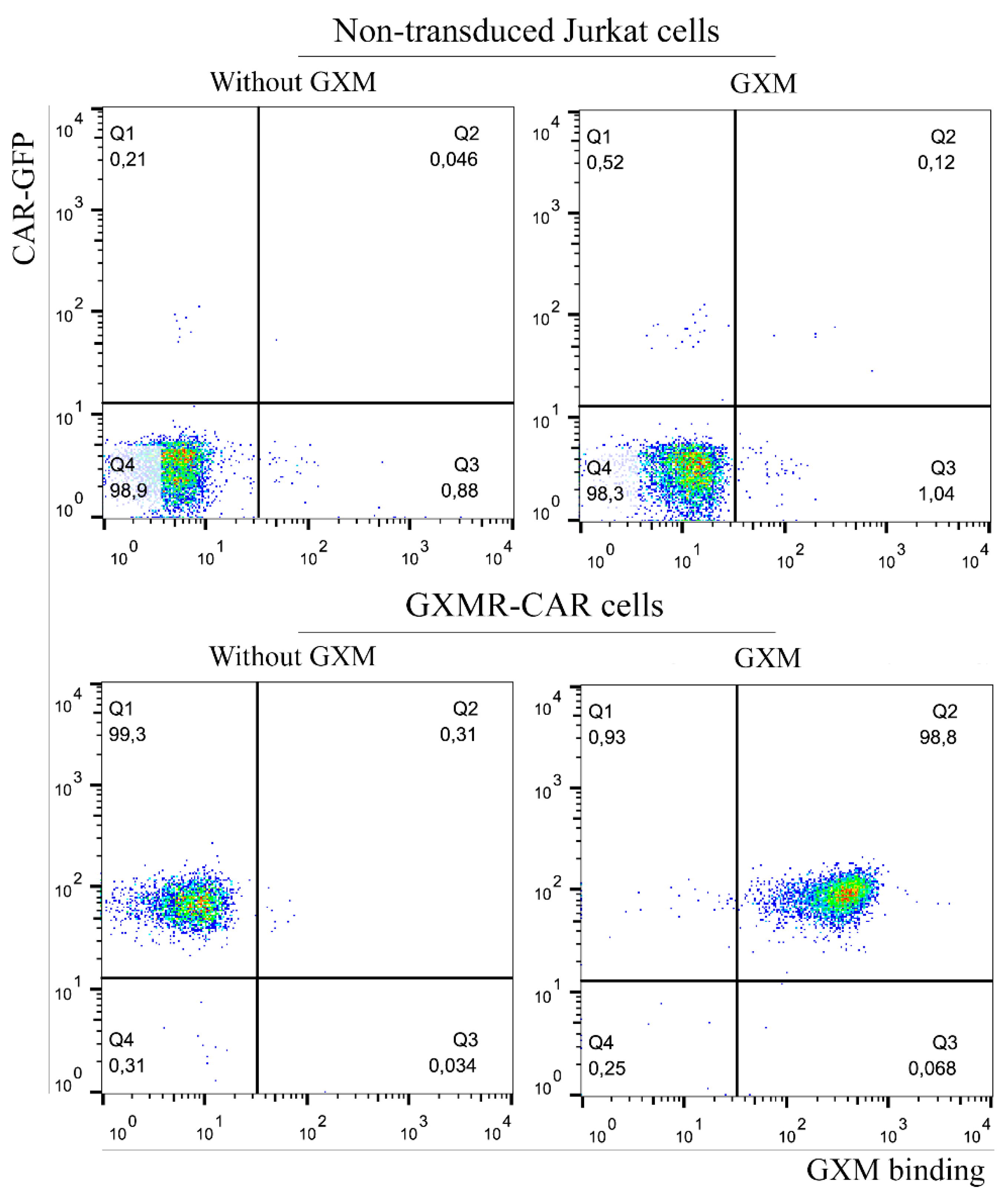
3.1. GXMR-CAR Interacts with Soluble GXM of C. gattii R265
3.2. GXMR-Car Redirects Jurkat Cells to Recognize the Yeast Form of C. neoformans and C. gattii
3.3. Cryptococcus spp. Titan Cells Are Targeted by GXMR-CAR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gullo, F.P.; Rossi, S.A.; Jde, C.S.; Teodoro, V.L.; Mendes-Giannini, M.J.; Fusco-Almeida, A.M. Cryptococcosis: Epidemiology, fungal resistance, and new alternatives for treatment. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1377–1391. [Google Scholar] [CrossRef] [PubMed]
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of hiv-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef] [Green Version]
- Maziarz, E.K.; Perfect, J.R. Cryptococcosis. Infect. Dis. Clin. N. Am. 2016, 30, 179–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Probert, M.; Zhou, X.; Goodall, M.; Johnston, S.A.; Bielska, E.; Ballou, E.R.; May, R.C. A glucuronoxylomannan epitope exhibits serotype-specific accessibility and redistributes towards the capsule surface during titanization of the fungal pathogen cryptococcus neoformans. Infect. Immun. 2019, 87, e00731-18. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.W.; Jiao, A.X.; Wu, X.R.; Zhao, S.Y.; Ma, Y.; Liu, G.; Yin, J.; Xu, B.P.; Shen, K.L. Clinical characteristics of disseminated cryptococcosis in previously healthy children in china. BMC Infect. Dis. 2017, 17, 359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherniak, R.; Sundstrom, J.B. Polysaccharide antigens of the capsule of cryptococcus neoformans. Infect. Immun. 1994, 62, 1507–1512. [Google Scholar] [CrossRef] [Green Version]
- Zaragoza, O. Basic principles of the virulence of cryptococcus. Virulence 2019, 10, 490–501. [Google Scholar] [CrossRef] [Green Version]
- Zaragoza, O.; Rodrigues, M.L.; De Jesus, M.; Frases, S.; Dadachova, E.; Casadevall, A. The capsule of the fungal pathogen cryptococcus neoformans. Adv. Appl. Microbiol. 2009, 68, 133–216. [Google Scholar] [PubMed] [Green Version]
- Wang, Z.A.; Li, L.X.; Doering, T.L. Unraveling synthesis of the cryptococcal cell wall and capsule. Glycobiology 2018, 28, 719–730. [Google Scholar] [CrossRef]
- Urai, M.; Kaneko, Y.; Ueno, K.; Okubo, Y.; Aizawa, T.; Fukazawa, H.; Sugita, T.; Ohno, H.; Shibuya, K.; Kinjo, Y.; et al. Evasion of innate immune responses by the highly virulent cryptococcus gattii by altering capsule glucuronoxylomannan structure. Front. Cell Infect. Microbiol. 2015, 5, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Meara, T.R.; Alspaugh, J.A. The cryptococcus neoformans capsule: A sword and a shield. Clin. Microbiol. Rev. 2012, 25, 387–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaragoza, O.; Casadevall, A. Experimental modulation of capsule size in cryptococcus neoformans. Biol. Proced. Online 2004, 6, 10–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivera, J.; Feldmesser, M.; Cammer, M.; Casadevall, A. Organ-dependent variation of capsule thickness in cryptococcus neoformans during experimental murine infection. Infect. Immun. 1998, 66, 5027–5030. [Google Scholar] [CrossRef] [Green Version]
- Gerstein, A.C.; Fu, M.S.; Mukaremera, L.; Li, Z.; Ormerod, K.L.; Fraser, J.A.; Berman, J.; Nielsen, K. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio 2015, 6, e01340-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trevijano-Contador, N.; de Oliveira, H.C.; Garcia-Rodas, R.; Rossi, S.A.; Llorente, I.; Zaballos, A.; Janbon, G.; Arino, J.; Zaragoza, O. Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Pathog. 2018, 14, e1007007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dylag, M.; Colon-Reyes, R.J.; Kozubowski, L. Titan cell formation is unique to cryptococcus species complex. Virulence 2020, 11, 719–729. [Google Scholar] [CrossRef]
- da Silva, T.A.; Hauser, P.J.; Bandey, I.; Laskowski, T.; Wang, Q.; Najjar, A.M.; Kumaresan, P.R. Glucuronoxylomannan in the cryptococcus species capsule as a target for chimeric antigen receptor t-cell therapy. Cytotherapy 2021, 23, 119–130. [Google Scholar] [CrossRef]
- Trevijano-Contador, N.; Pianalto, K.M.; Nichols, C.B.; Zaragoza, O.; Alspaugh, J.A.; Pirofski, L.A. Human igm inhibits the formation of titan-like cells in cryptococcus neoformans. Infect. Immun. 2020, 88, e00046-20. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Cleare, W.; Feldmesser, M.; Glatman-Freedman, A.; Goldman, D.L.; Kozel, T.R.; Lendvai, N.; Mukherjee, J.; Pirofski, L.A.; Rivera, J.; et al. Characterization of a murine monoclonal antibody to cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 1998, 42, 1437–1446. [Google Scholar] [CrossRef] [Green Version]
- Greenfield, E.A. Single-cell cloning of hybridoma cells by limiting dilution. Cold Spring Harb. Protoc. 2019, 2019, 726–728. [Google Scholar] [CrossRef]
- Wozniak, K.L.; Levitz, S.M. Isolation and purification of antigenic components of cryptococcus. Methods Mol. Biol. 2009, 470, 71–83. [Google Scholar] [PubMed] [Green Version]
- Kumaresan, P.R.; Manuri, P.R.; Albert, N.D.; Maiti, S.; Singh, H.; Mi, T.; Roszik, J.; Rabinovich, B.; Olivares, S.; Krishnamurthy, J.; et al. Bioengineering t cells to target carbohydrate to treat opportunistic fungal infection. Proc. Natl. Acad. Sci. USA 2014, 111, 10660–10665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohne, F.; Chmielewski, M.; Ebert, G.; Wiegmann, K.; Kurschner, T.; Schulze, A.; Urban, S.; Kronke, M.; Abken, H.; Protzer, U. T cells redirected against hepatitis b virus surface proteins eliminate infected hepatocytes. Gastroenterology 2008, 134, 239–247. [Google Scholar] [CrossRef]
- da Silva, T.A. Car t cell-therapy for infectious diseases with emphasis on invasive fungal infections. Ther. Deliv. 2021, 12, 627–630. [Google Scholar] [CrossRef]
- Deeks, S.G.; Wagner, B.; Anton, P.A.; Mitsuyasu, R.T.; Scadden, D.T.; Huang, C.; Macken, C.; Richman, D.D.; Christopherson, C.; June, C.H.; et al. A phase ii randomized study of hiv-specific t-cell gene therapy in subjects with undetectable plasma viremia on combination antiretroviral therapy. Mol. Ther. 2002, 5, 788–797. [Google Scholar] [CrossRef]
- Sautto, G.A.; Wisskirchen, K.; Clementi, N.; Castelli, M.; Diotti, R.A.; Graf, J.; Clementi, M.; Burioni, R.; Protzer, U.; Mancini, N. Chimeric antigen receptor (car)-engineered t cells redirected against hepatitis c virus (hcv) e2 glycoprotein. Gut 2016, 65, 512–523. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Rodas, R.; Cordero, R.J.; Trevijano-Contador, N.; Janbon, G.; Moyrand, F.; Casadevall, A.; Zaragoza, O. Capsule growth in cryptococcus neoformans is coordinated with cell cycle progression. mBio 2014, 5, e00945-14. [Google Scholar] [CrossRef] [Green Version]
- Kuttel, M.M.; Casadevall, A.; Oscarson, S. Cryptococcus neoformans capsular gxm conformation and epitope presentation: A molecular modelling study. Molecules 2020, 25, 2651. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Coelho, C.; Cordero, R.J.B.; Dragotakes, Q.; Jung, E.; Vij, R.; Wear, M.P. The capsule of cryptococcus neoformans. Virulence 2019, 10, 822–831. [Google Scholar] [CrossRef] [Green Version]
- Hommel, B.; Mukaremera, L.; Cordero, R.J.B.; Coelho, C.; Desjardins, C.A.; Sturny-Leclere, A.; Janbon, G.; Perfect, J.R.; Fraser, J.A.; Casadevall, A.; et al. Titan cells formation in cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Pathog. 2018, 14, e1006982. [Google Scholar] [CrossRef] [Green Version]
- Mukaremera, L.; Lee, K.K.; Wagener, J.; Wiesner, D.L.; Gow, N.A.R.; Nielsen, K. Titan cell production in cryptococcus neoformans reshapes the cell wall and capsule composition during infection. Cell Surf. 2018, 1, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Okagaki, L.H.; Strain, A.K.; Nielsen, J.N.; Charlier, C.; Baltes, N.J.; Chretien, F.; Heitman, J.; Dromer, F.; Nielsen, K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Zaragoza, O.; Garcia-Rodas, R.; Nosanchuk, J.D.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L.; Casadevall, A. Fungal cell gigantism during mammalian infection. PLoS Pathog. 2010, 6, e1000945. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, M.H.; Machado, M.P.; Kumaresan, P.R.; da Silva, T.A. Titan Cells and Yeast Forms of Cryptococcus neoformans and Cryptococcus gattii Are Recognized by GXMR-CAR. Microorganisms 2021, 9, 1886. https://doi.org/10.3390/microorganisms9091886
dos Santos MH, Machado MP, Kumaresan PR, da Silva TA. Titan Cells and Yeast Forms of Cryptococcus neoformans and Cryptococcus gattii Are Recognized by GXMR-CAR. Microorganisms. 2021; 9(9):1886. https://doi.org/10.3390/microorganisms9091886
Chicago/Turabian Styledos Santos, Matheus Henrique, Michele Procópio Machado, Pappanaicken R. Kumaresan, and Thiago Aparecido da Silva. 2021. "Titan Cells and Yeast Forms of Cryptococcus neoformans and Cryptococcus gattii Are Recognized by GXMR-CAR" Microorganisms 9, no. 9: 1886. https://doi.org/10.3390/microorganisms9091886
APA Styledos Santos, M. H., Machado, M. P., Kumaresan, P. R., & da Silva, T. A. (2021). Titan Cells and Yeast Forms of Cryptococcus neoformans and Cryptococcus gattii Are Recognized by GXMR-CAR. Microorganisms, 9(9), 1886. https://doi.org/10.3390/microorganisms9091886





