Abstract
Klebsiella pneumoniae is an important human pathogen in both developing and industrialised countries that can causes a variety of human infections, such as pneumonia, urinary tract infections and bacteremia. Like many Gram-negative bacteria, it is becoming resistant to many frontline antibiotics, such as carbapenem and cephalosporin antibiotics. In Egypt, K. pneumoniae is increasingly recognised as an emerging pathogen, with high levels of antibiotic resistance. However, few Egyptian K. pneumoniae strains have been sequenced and characterised. Hence, here, we present the genome sequence of a multidrug resistant K. pneumoniae strain, KPE16, which was isolated from a child in Assiut, Egypt. We report that it carries multiple antimicrobial resistance genes, including a blaNDM-1 carbapenemase and extended spectrum β-lactamase genes (i.e., blaSHV-40, blaTEM-1B, blaOXA-9 and blaCTX-M-15). By comparing this strain with other Egyptian isolates, we identified common plasmids, resistance genes and virulence determinants. Our analysis suggests that some of the resistance plasmids that we have identified are circulating in K. pneumoniae strains in Egypt, and are likely a source of antibiotic resistance throughout the world.
1. Introduction
Klebsiella pneumoniae is a Gram-negative, encapsulated, non-motile bacterium that can cause a variety of human infections, including pneumonia, urinary tract infections, bacteremia, and liver abscesses [1]. It readily colonises human mucosal surfaces, such as the gastrointestinal tract and oropharynx, where it is viewed as a commensal, but it is from these sites that it gains entry to other tissues to cause severe disease. In the past, K. pneumoniae infections have mostly targeted immunocompromised patients in hospital settings, but the emergence of hypervirulent (hv) K. pneumoniae strains has seen an increase of community-acquired infection in healthy individuals [1,2,3].
In K. pneumoniae, a number of virulence factors are important for human infection. These include the production of capsule and lipopolysaccharide (LPS), and the expression of fimbriae (both types I and III) and siderophore transport systems. For hv K. pneumoniae strains, the overproduction of capsule is important, due to the action of the plasmid encoded RmpA and/or RmpA2 transcription factors, and such stains usually possess multiple siderophore systems (e.g., aerobactin and yersiniabactin) as well as other iron transport systems (e.g., kfuABC) [1,2,3,4].
Like many bacteria, K. pneumoniae is now becoming resistant to many frontline antibiotics, limiting the treatment of both nosocomial and community acquired infections [1,2,3]. Two types of antibiotic resistance are particularly important, the acquisition of extended spectrum β-lactamase (ESBL) genes (e.g., blaTEM, blaCTX-M and blaOXA), which provide resistance to cephalosporin and monobactam antibiotics, and genes encoding carbapenemases (e.g., blaNDM, blaVIM and blaKPC), providing resistance to carbapenems [1,2,3]. In particular, carbapenemase producing K. pneumoniae have a higher incidence of mortality due to lack of treatment options [5]. Both ESBL and carbapenemases genes are often located on self-transmissible plasmids, contributing to their ease of spread [1,2,3].
In Egypt, K. pneumoniae is increasingly recognised as an emerging pathogen, showing high levels of antibiotic resistance [6,7,8,9,10,11,12,13]. Furthermore, documented transmission of carbapenem resistant K. pneumoniae from Egypt to other countries has been observed [14,15,16,17,18]. Despite this, the genomes of very few K. pneumoniae strains, isolated in Egypt, have been sequenced and fully characterized [19,20]. During a recent screen in Assiut, Egypt [21], we isolated a carbapenem resistant K. pneumoniae strain from a child with diarrhoea. Here we present the antimicrobial resistance profile and genome sequence of this strain, K. pneumoniae KPE16, and compare it with other Egyptian isolates, identifying common plasmids, resistance genes and virulence determinants. We demonstrate that Egyptian K. pneumoniae strains carry specific antimicrobial resistance plasmids, which are similar to those isolated from travellers who previously visited Egypt. We suggest that these plasmids are circulating in K. pneumoniae strains in Egypt and are possibly a source of worldwide antibiotic resistance transmission.
2. Materials and Methods
2.1. Isolation and Characterisation of Klebsiella pneumoniae Strain KPE16
Klebsiella pneumoniae strain KPE16 was isolated from a child, presenting with diarrhoea to the outpatients clinic of Assiut University Children’s Hospital in 2016 [21]. Isolation and original identification were carried out at the Medical Research Center, Faculty of Medicine, Assiut University with growth on MacConkey agar, Eosin Methylene Blue (EMB) agar and IMViC testing, suggesting that KPE16 was an Escherichia coli isolate. However, analysis of the draft genome sequence of KPE16 (see below) with SpeciesFinder 2.0 and KmerFinder 3.2 software at the Center for Genomic Epidemiology (CGE) (http://www.genomicepidemiology.org/, accessed on 9 April 2021) clearly indicated that KPE16 had been misclassified and was K. pneumoniae [22,23,24]. K. pneumoniae KPE16 was tested for susceptibility to a range of antimicrobial agents, using the Kirby-Bauer disc diffusion method [25], which was interpreted according to the CLSI 2014 [26]. The antimicrobial discs (Hi-Media, India) contained the following antibiotics: imipenem (10 µg), meropenem (10 µg), trimethoprim/sulfamethoxazole (5 µg), cefaclor (30 µg), ceftriaxone (30 µg), ampicillin (10 µg), ciprofloxacin (5 µg), oxytetracycline (30 µg), amoxicillin (25 µg), norfloxacin (10 µg), tobramycin (10 µg), and amikacin (30 µg).
2.2. Genome Sequencing of K. pneumoniae Strain KPE16
Complete genome sequencing of K. pneumoniae strain KPE16 was carried out using Illumina sequencing by Microbes NG (https://microbesng.com/, accessed on 9 April 2021). Plated cultures were inoculated into a cryopreservative (Microbank™, Pro-Lab Diagnostics, Birkenhead, UK). Ten to 20 µL of this suspension were lysed with 120 µL of TE (10 mM Tris, 1 mM EDTA, pH 7.5) buffer containing lysozyme (final concentration 0.1 mg mL−1) and RNase A (ITW Reagents, Barcelona, Spain) (final concentration 0.1 mg mL−1), incubated for 25 min at 37 °C. Proteinase K (VWR Chemicals, Cleveland, OH, USA) (final concentration 0.1 mg mL−1) and SDS (Sigma-Aldrich, St. Louis, MO, USA) (final concentration 0.5% v/v) were added and incubated for 5 min at 65 °C. Genomic DNA was purified using an equal volume of SPRI beads and resuspended in EB buffer (Qiagen, Hilden, Germany). DNA was quantified with the Quant-iT dsDNA HS kit (ThermoFisher Scientific, Rockford, IL, USA) assay in an Eppendorf AF2200 plate reader (Eppendof UK Ltd., Stevenage, UK). Genomic DNA libraries were prepared using the Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA), following the manufacturer’s protocol with the following modifications: 2 ng of DNA were used as input, and PCR elongation time was increased to 1 min from 30 s. DNA quantification and library preparations were carried out on a Hamilton Microlab STAR automated liquid handling system (Hamilton Bonaduz AG, Bonaduz, Switzerland). Pooled libraries were quantified using the Kapa Biosystems Library Quantification Kit for Illumina on a Roche light cycler 96 qPCR machine. Libraries were sequenced with the Illumina HiSeq using a 250 bp paired end protocol. Illumina reads were adapter trimmed using Trimmomatic 0.30 with a sliding window quality cutoff of Q15 [27]. Genome assembly was performed using Unicycler v0.4.0 [28] and contigs were annotated using Prokka 1.11 [29]. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank with the sequence data for K. pneumoniae KPE16 under the accession number JAGFBT000000000. The genome sequence (chromosome and plasmids) of K. pneumoniae reference strain MGH 78578 was obtained from BioProject PRJNA31 [30], whilst the GenBank accession numbers for the genomes of Egyptian K. pneumoniae strains SF, SK, HM, and SP were RXLV00000000, RXLX00000000, RXLW00000000, and RXLY00000000, respectively [19].
2.3. Bioinformatic Analysis of K. pneumoniae Genome Sequences
Draft genomes were visualised using Artemis [31], comparisons between K. pneumoniae genomes were examined using the CGView Server (http://stothard.afns.ualberta.ca/cgview_server/, accessed on 9 April 2021) [32], the Basic Local Alignment Search Tool (BLAST) at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 9 April 2021) and the Artemis Comparison Tool (ACT) [33]. Representations of genome organisation were drawn using the CGView Server [32] and ACT [33]. K. pneumoniae sequence types were determined using MLST 2.0 [34], plasmid replicons were detected using PlasmidFinder 2.1 [35], antibiotic resistance gene analysis used ResFinder 3.2 [36], and virulence gene analysis was performed using VirulenceFinder 2.0 [37] with the online software from CGE (http://www.genomicepidemiology.org/, accessed on 9 April 2021). In addition, capsule typing, heavy metal resistance genes, and virulence genes were identified using the BIGSdb-Kp database at the Institut Pasteur (https://bigsdb.web.pasteur.fr/index.html, accessed on 9 April 2021) and the Virulence Factor Database (http://www.mgc.ac.cn/VFs/main.htm, accessed on 9 April 2021) [38]. Capsule typing and O-antigen locus type were also determined using Kaptive Web (https://kaptive-web.erc.monash.edu/, accessed on 9 April 2021) [39], insertion sequences were identified using ISfinder (https://www-is.biotoul.fr/blast/resultat.php, accessed on 9 April 2021) [40], and bacteriophage were determined using PHASTER (https://phaster.ca/, accessed on 9 April 2021) [41].
3. Results
3.1. Antimicrobial Resistance and Genomic Characterisation of K. pneumoniae Strain KPE16
K. pneumoniae strain KPE16 was isolated from a child, at the outpatient clinic of Assiut University Children’s Hospital in 2016 [21]. As multidrug resistant K. pneumoniae are a major concern in Egypt [6,7,8,9,10,11,12,13], we examined its susceptibility to various antimicrobials. Results detailed in Table 1 show that K. pneumoniae strain KPE16 was resistant to all antibiotics tested, including the carbapenem antibiotics, imipenem and meropenem, and the cephalosporin antibiotics, cefaclor and ceftriaxone, suggesting that it possesses carbapenemase and ESBL genes.

Table 1.
Antimicrobial resistance profile of K. pneumoniae strain KPE16.
To understand more about the antibiotic resistance and virulence genes that KPE16 carries, we sequenced its genome and compared it to the K. pneumoniae reference strain MGH 78578, and four other recently sequenced Egyptian isolates (K. pneumoniae strains SF, SK, HM and SP) which have not been fully characterised (Table 2) [19,30]. Our analysis indicated that out of all the K. pneumoniae strains examined, SP possessed the smallest genome, containing the fewest coding sequences, whilst the genomes of other strains were relatively similar in size and gene content (Table 2). Comparison of each strain’s genome with the MGH 78578 chromosome indicated that there were a number of common regions of difference (RODs) between the Egyptian strains and MGH 78578 (Figure 1). Although many RODs locate to genes that encode proteins of undefined function, differences between genes responsible for surface structures (e.g., fimbriae, an outer membrane porin, a type I secretion system (TISS), and capsule and O-antigen LPS biosynthetic operons) and prophage were detected. Consistent with this, each Egyptian strain differed from MGH 78578 in their capsule K type and O-antigen locus type: KPE16 was predicted as K43 O2v1, strains SF, SK, and HM were K17 O1v1, and SP was K36 O4 (Table 2). Note that none of the strains were capsule type K1 or K2, which is associated with hv K. pneumoniae [1,2,3]. The Egyptian K. pneumoniae strains also belong to diverse sequence types, with KPE16 belonging to sequence type ST1399, SF, SK, and HM were all ST101, and SP was ST3050 (Table 2). Norsigian et al. [20] suggested that K. pneumoniae strains SF, SK, and HM are very similar. This was confirmed by comparing the draft genome assemblies of each isolate, which showed that SK and HM were almost identical, and SF only differed from SK and HM due to the absence of a number of prophage genes and an IncFIB(pQil) plasmid (Table 2 and Supplementary Figure S1).

Table 2.
Genome analysis of Klebsiella pneumoniae strains isolated in Egypt.
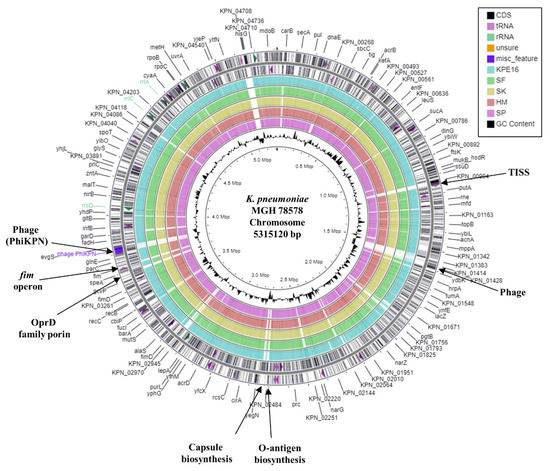
Figure 1.
Comparison of the K. pneumoniae MGH 78578 chromosome with the draft genomes of K. pneumoniae strains KPE16, SF, SK, HM, and SP. The figure shows the comparison of the K. pneumoniae MGH 78578 chromosome with the genomes of K. pneumoniae strains KPE16, SF, SK, HM, and SP using GCview [32]. The outer two rings display the genes (CDS) and features of the K. pneumoniae MGH 78578 chromosome (CP000647.1) on both strands, with selected features labelled [30]. The blue, green, gold, red, and pink rings illustrate the BLAST results when the genome sequences of KPE16, SF, SK, HM, and SP, respectively, are compared to the MGH 78578 chromosome, with shaded regions indicating homology. The inner ring displays the GC content (black) for the K. pneumoniae MGH 78578 chromosome.
3.2. Characterisation of Plasmids Carried by Egyptian K. pneumoniae Strains
Many K. pneumoniae strains carry multiple plasmids, for example, MGH 78578 carries five plasmids (Table 2) [2,3,30]. Consistent with this, analysis of each Egyptian strain identified a number of plasmid replicons (Table 2) [35]. The genome of K. pneumoniae strain KPE16 contains four IncF replicons and one IncQ1 replicon. Due to the draft nature of the KPE16 genome assembly, each plasmid replicon was on a separate contig and it is not clear how many plasmids this strain possesses. It is of note that MGH 78578 plasmids pKPN3 and pKPN4 both carry IncFIB and IncFII replicons (Table 2). This arrangement occurs on other IncF plasmids and, in spite of replicons belonging to the same plasmid incompatibility group, subtle differences in copy number control regions mean that plasmids can be still maintained in the same cell [21,35,42,43]. Analysis of the IncFII(K) replicon (as well as other contigs from the KPE16 genome assembly) indicated that K. pneumoniae strain KPE16 carries a plasmid very similar to plasmid p2 (Figure 2A and Supplementary Figure S2). This plasmid was found in a K. pneumoniae strain that was isolated from the urine of an Egyptian man in the USA in 2012 [14]. Significantly, this plasmid carries multiple antibiotic resistance determinants, including a blaNDM-1 carbapenemase and ESBL genes, amongst others (e.g., blaNDM-1, blaCTX-M-15, blaTEM-1A, blaOXA-9, aac(6′)-Ib-cr, aadA1, aph(3′)-VI, qnrS1) (Figure 2) [14]. Comparison of the KPE16 genome with the Egyptian strains SF, SK, HM, and SP identified one relatively large contig (contig 19) that was only present in KPE16 (Supplementary Figure S3). BLAST analysis indicated that this region was carried by plasmid pKP1–19, which was isolated from an environmental K. pneumoniae strain KP-1 in Australia and carries heavy metal resistance genes for arsenic, silver, and copper (Figure 2B and Supplementary Figure S4) [44]. The organisation of the KPE16 IncQ1 replicon was also found to be similar to the small broad-host range plasmid pRSF1010, which carries sul2, aph(3″)-Ib and aph(6)-Id resistance genes [35,45] (Supplementary Figure S5). Thus, it likely that K. pneumoniae strain KPE16 carries three plasmids, a small IncQ1 plasmid, and two larger plasmids, each of which carry two IncF replicons.
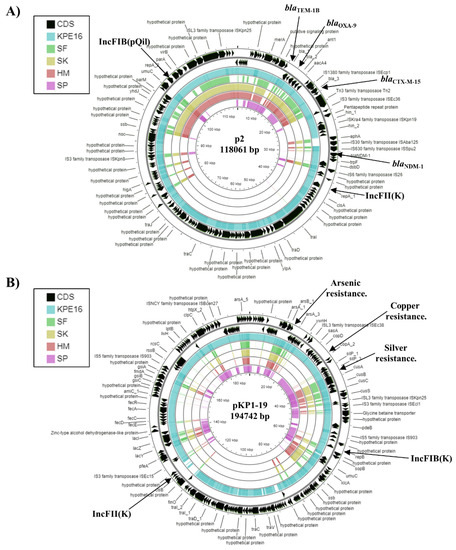
Figure 2.
Analysis of plasmids carried by K. pneumoniae strain KPE16. The figure shows the comparison of the K. pneumoniae plasmids (A) p2 (CP009115.1) [14] and (B) pKP1–19 (CP012884.1) [44] with the genomes of K. pneumoniae strains KPE16, SF, SK, HM, and SP using GCview [32]. In both panels, the outer two rings display the genes (CDS) of each plasmid on both strands, with plasmid replicons and selected features labelled. The blue, green, gold, red, and pink shading illustrates the BLAST results when the genome sequences of KPE16, SF, SK, HM, and SP, respectively, are compared with each plasmid. Shaded regions indicate homology.
The K. pneumoniae strains SK and HM carry three distinct plasmid replicons, whilst strain SF carries two, lacking the InFI(pQil) replicon (Table 2 and Supplementary Figure S1). Analysis of the IncC and IncM1 replicons carried by SF, SK, and HM indicated that they were identical to those carried by plasmids pSH111_166 and pKpn14-4, respectively (Figure 3A,B). Plasmid pSH111_166 was found in a multi drug resistant Salmonella enterica subsp. enterica serovar Heidelberg strain that was isolated from a bovine diagnostic specimen in Ohio, USA in 2001 and carries a number of antibiotic resistance genes (i.e., aac(3)-IV, blaCMY-2, aph(4)-Ia, aph(6)-Id, aph(3″)-Ib, floR, sul2, tetA) (Figure 3A) [46]. Plasmid pKpn14-4 was carried by K. pneumoniae strain Kpn-14 isolated from a patient in Ontario, Canada in 2014, who had previously been hospitalised in Egypt and also possesses multiple antibiotic resistance genes (i.e., aph(3′)-VIb, aph(3″)-Ib, aph(6)-Id, blaCTX-M-14b, qnrS1) (Figure 3B) [15]. Analysis of the InFI(pQil) replicon from both SK and HM, indicated that they were identical to that of MGH 78578 plasmid pKPN4. Genome subtraction, as well as comparison of pKPN4 with the complete genomes of SK and HM, indicated that each strain carries a similar plasmid, though it lacks the tra genes of pKPN4 and is likely much smaller (Figure 3C). It is of note that pKPN4 also carries a multiple antimicrobial resistance genes (aac(6′)-Ib, aac(6′)-Ib-cr, aadA1, blaOXA-9ψ, blaSHV-12, blaTEM-1A) (Table 3 and Figure 3C).
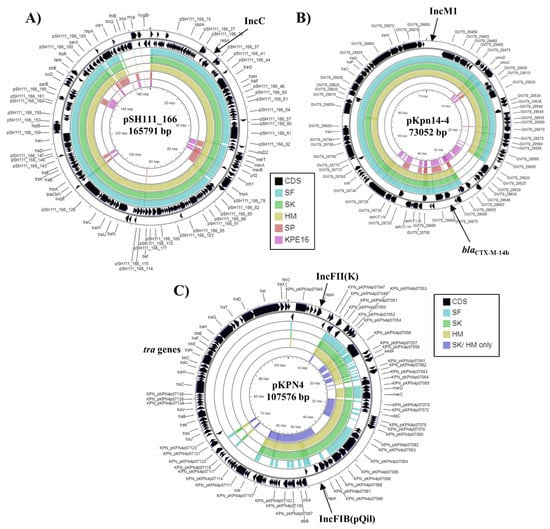
Figure 3.
Analysis of plasmids carried by K. pneumoniae strains SF, SK, and HM. The figure shows the comparison of (A) Salmonella enterica subsp. enterica serovar Heidelberg plasmid pSH111_166 (JN983043.1) [46], (B) K. pneumoniae plasmid pKpn14-4 (CP047704.1) [15] and (C) K. pneumoniae MGH 78578 plasmid pKPN4 (CP000649.1) [30] with the genomes of K. pneumoniae strains SF, SK, HM, SP and KPE16, using GCview [32]. In all panels, the outer two rings display the genes (CDS) of each plasmid on both strands, with plasmid replicons labelled. In panels (A,B) the blue, green, gold, red, and pink shading illustrates the BLAST results when the genome sequences of SF, SK, HM, SP, and KPE16, respectively, are compared with each plasmid. In panel (C), the light blue, green, gold, and dark blue shading illustrates the BLAST results when the genome sequences of SF, SK, HM and the contigs that are specific to SK/HM only (after genome subtraction) are compared to pKPN4. Shaded regions indicate homology.

Table 3.
Analysis of antimicrobial resistance, heavy metal resistance, and virulence genes carried by Egyptian K. pneumoniae strains.
The K. pneumoniae isolate SP also contained three different plasmid replicons (Table 2), suggesting that it possesses three plasmids. Analysis of the IncFIB(K) replicon indicated it was 99.8% identical to plasmid pTHO-004-1, which was isolated from K. pneumoniae strain THO-004, infecting a patient in Tokyo, Japan in 2018 (Figure 4). Plasmid pTHO-004-1 carries many gene associated with heavy metal resistance, such as arsenic, copper, and silver (Figure 4). The contig carrying the IncR replicon was similar to that carried by a number of IncR plasmids, though similarity between these plasmids was limited to the replicon region only (not shown). Interestingly, the IncQ1 replicon in K. pneumoniae strain SP is also very similar to that carried by K. pneumoniae strain KPE16 and pRSF1010 [35,45] (Supplementary Figure S5).
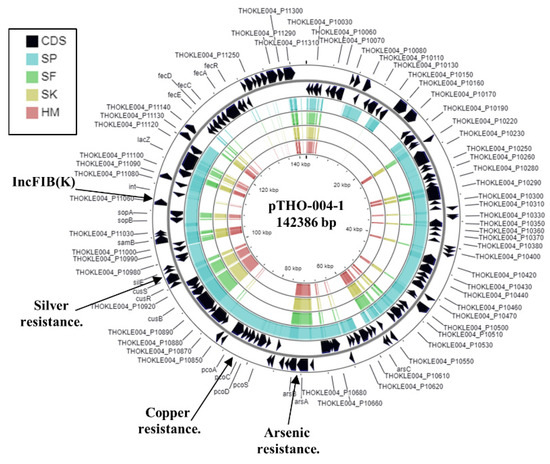
Figure 4.
Analysis of the IncFIB(K) replicon carried by K. pneumoniae strain SP. The figure shows the comparison of K. pneumonia plasmid pTHO-004-1 (AP022528.1) with the genomes of K. pneumoniae strains SP, SF, SK, and HM, using GCview [32]. The outer two rings display the genes (CDS) on both strands, with the IncFIB(K) plasmid replicon and selected features labelled. The blue, green, gold, and red shading illustrates the BLAST results when the genome sequences of SP, SF, SK, and HM, respectively, are compared with pTHO-004-1. Shaded regions indicate homology.
3.3. Antibiotic- and Heavy Metal-Resistance Genes Carried by Egyptian K. pneumoniae Isolates
In the genome of K. pneumoniae KPE16, we detected various antibiotic resistance genes (Table 3). These can result in aminoglycoside resistance (aac(6′)-Ib-cr, aac(6′)-Ib, aadA1, aph(6)-Id, aph(3″)-Ib, aph(3′)-Ia), β-lactam resistance (blaSHV-40, blaTEM-1B, blaOXA-9, blaNDM-1, blaCTX-M-15), fosfomycin resistance (fosA5), quinolone/fluoroquinolone resistance (aac(6′)-Ib-cr, oqxA, oqxB, qnrS1), sulphonamide resistance (sul2), and tetracycline resistance (tetA) [36]. This is consistent with the multidrug resistance phenotype of K. pneumoniae KPE16 (Table 1). In particular, its resistance to carbapenem antibiotics is due to the carriage of blaNDM-1 carbapenemase gene. Note that the blaNDM-1 gene is located on same contig as the IncFII(K) replicon (Supplementary Figure S2) and that the sul2, aph(3″)-Ib, aph(6)-Id and aph(3′)-Ia genes co-localise with the IncQ1 replicon (Supplementary Figure S5), indicating that these resistance determinants are plasmid-borne. Furthermore, the aadA1, aac(6′)-Ib, blaCTX-M-15 and qnrS1 resistance genes are all localised on a large contig, which is similar to the sequence of K. pneumoniae plasmid p2, suggesting that these genes are likely plasmid encoded (Figure 2 and Supplementary Figure S2).
Reference strain MGH 78578 also carries multiple antibiotic resistance genes that could offer resistance to aminoglycoside, β-lactam, fluoroquinolone, fosfomycin, phenicol, sulphonamide, and tetracycline antibiotics (Table 3) [36]. Many of these resistance genes are located on plasmids pKPN4 and pKPN5 (Table 3). As pKPN4 carries many tra genes it is potentially conjugative (Figure 3). Norsigian et al. [20] demonstrated that K. pneumoniae strains SF, SK, and HM were resistant to many classes of antibiotics (e.g., aminoglycosides, β-lactams (penicillins, cephalosporins, and carbapenems), fluoroquinolones, phenicols, polymixins, sulphonamides, and tetracyclines), whilst strain SP was generally less resistant. This is consistent with the antibiotic resistance gene profile observed for the four strains (Table 3 and Supplementary Table S1). Strains SF, SK, and HM all contained the blaVIM-29 carbapenemase gene, which explains their resistance to carbapenems, and all four Egyptian strains carry multiple ESBL genes. For SF, SK, and HM the blaCTX-M-14b gene was co-localised with the IncM1 contig, indicating that it is plasmid borne, which is consistent with this plasmid being similar to pKpn14-4 (Figure 3B and Supplementary Figure S6). For K. pneumoniae strain SP the sul2, aph(3″)-Ib, aph(6)-Id resistance genes co-localised with the IncQ1 replicon as previously mentioned (Supplementary Figure S5).
As well as acquired antibiotic resistance genes, all five strains also possess a number of chromosomal point mutations in the genes encoding the outer membrane porins, ompK36 and ompK37, which are associated with cephalosporin and carbapenem resistance, and the regulator acrR, which is associated with fluoroquinolone resistance (Table 3 and Supplementary Table S1) [36]. These, mutations could contribute to the antimicrobial resistance of each strain [36] and may explain why strain SP is resistant to the carbapenem antibiotic imipenem, yet seems to lack carbapenemase genes [3,20,47]. Isolates SF and SK were previously shown to be resistant to the polymixin antibiotic colistin, whilst strains HM and SP were sensitive [19,20]. Genome analysis of SF and SK indicates that products of the phoPQ, pmrAB, mgrB, yciM, and lpxM genes, which have all been implicated in colistin resistance [3,48,49,50,51,52], were either identical to the colistin sensitive reference strain MGH 78578 or differences were also present in HM and/or SP (Supplementary Figure S7). Thus, the mechanism of colistin resistance in strains SF and SK is unclear.
In addition to antibiotic resistance genes, all strains carried loci for heavy metal resistance (Table 3). For example, K. pneumoniae KPE16 carries resistance genes for arsenic (arsRDABC), cobalt (corA, corC, mgtA), copper (pcoABCDERS), mercury (merRTPCADE), and silver (silBCEFGPRS). The arsenic, copper, and silver resistance determinates were all located on contig 19, which is also carried by plasmid pKP1-19 from the environmental K. pneumoniae strain, KP-1, (see above) (Figure 2B and Supplementary Figure S4) [44], indicating that these genes are likely plasmid encoded. Due to the draft nature of the K. pneumoniae SF, SK, HM, and SP genome assemblies, it is unclear if their heavy metal resistance genes are plasmid borne. However, as such genes are often carried by plasmids (as seen for MGH 78578 pKPN3 and pKPN4 (Table 3)), it is likely that this will be the case, especially as we predict that SP contains a similar plasmid to pTHO-004-1 (Figure 4).
3.4. Virulence Genes of K. pneumoniae Isolates
Analysis of potential virulence determinants in each K. pneumoniae isolate (Table 3) indicated that all strains possess genes encoding the E. coli common pilus (ecpRABCDE), mannose-sensitive type I fimbriae (fimABCDFGH), and Klebsiella mannose-insensitive type III fimbriae (mrkABCDF). They also carry genes involved in biofilm (treC and sugE) and capsule (rcsA and rcsB) formation, as well as genes encoding urease (ureDABCEFG) [2,3,53]. With respect to siderophore production, each strain carried the genes for enterobactin production (entAB and fepC) and the aerobactin transporter (iutA) but not the aerobactin gene cluster (iucABCD) [1,2,3]. Only SK, SF, and HM carried the yersiniabactin uptake system (irp1, ipr2, fyuA) and the ferric uptake genes kfuABC, which are both associated with hv K. pneumoniae (Table 3) [1,2,3,4]. Thus, it is clear that each Egyptian isolate possess many genes associated with K. pneumoniae virulence. Note that none of the strains carried the RmpA or RmpA2 transcription factors, often found in hv K. pneumoniae strains [1,2,3].
4. Discussion
Here, we have reported the genome sequence of a multidrug-resistant K. pneumoniae strain, KPE16, isolated from an Egyptian child with diarrhoea. By comparing its genome with other Egyptian isolates, we have gained important insights into the distribution and mobility of virulence and resistance determinants, especially those that are plasmid-associated. As K. pneumoniae is a normal resident in the human gastrointestinal tract, we think it unlikely that strain KPE16 was the cause of diarrhoea [2,3]. However, irrespective of this, KPE16, alongside the other Egyptian K. pneumoniae isolates, possesses many genes associated with virulence, including those involved in capsule, siderophore, and fimbriae biosynthesis [1,2,3]. Many of these genes are shared with the archetype strain, MGH 78578, which is considered to be a low virulence strain (Table 3) [54]. Usually, hv K. pneumoniae strains have capsule type K1 or K2, carry the regulator RmpA/RmpA2 and have multiple siderophore genes and iron acquisition systems. None of the Egyptian strains fit this profile (Table 2 and Table 3), though strains SF, SK, and HM do possess an additional yersiniabactin uptake system and the kfuABC ferric transport genes [1,2,3,4,55,56]. Although, this may make SF, SK, and HM more virulent, as iron scavenging in important during infection [1,2,3,56], it is unlikely that any of these strains are hypervirulent in character.
KPE16 was resistant to all antibiotics tested (Table 1), highlighting the possibility that, if treatment were required with frontline antibiotics, it would likely fail to resolve an infection. This is consistent with the antibiotic resistance genes carried by this strain (Table 3). Of note is the presence of the blaNDM-1 carbapenemase and multiple ESBL genes (blaSHV-40, blaTEM-1B, blaOXA-9 and blaCTX-M-15), which likely account for this strain’s resistance to carbapenem and cephalosporin antibiotics (Table 1 and Table 3). K. pneumoniae strain KPE16 is sequence type ST1399, which has previously been detected in Egypt and was found, in each case, to be multidrug resistant [9,13]. Similarly, K. pneumoniae strains SF, SK, and HM were all multidrug resistant, possessing a plethora of antibiotic resistance genes, in particular, the blaVIM-29 carbapenemase gene and multiple ESBL genes. All three isolates were ST101, which is considered to be the most prevalent sequence type in Egypt and a major MDR sequence type throughout the world [3,9,13,57].
Like many K. pneumoniae strains, the Egyptian isolates presented here all carry plasmids, which encode antibiotic and/or heavy metal resistance determinants [3]. Our analysis indicates that K. pneumoniae strains KPE16 and SP carry an IncQ1 replicon, which is similar to that of pRSF100 [35,45] (Supplementary Figure S5). This plasmid was also detected in Enteroaggregative E. coli strain E36, which we isolated in Assuit, Egypt in 2016 [21] and this suggests that similar IncQ1 plasmids are likely endemic in Egyptian Enterobacteriaceae (Supplementary Figure S5). Furthermore, strain KPE16 possessed a plasmid very similar to K. pneumoniae plasmid p2, which was isolated in the USA from a man from Egypt [14] (Figure 2a). For K. pneumoniae strains SF, SK, and HM, we detected a plasmid similar to pKpn14-4 (Figure 2), which was isolated from a patient in Ontario, Canada, who had been previously hospitalised in Egypt. As such plasmids have been detected in Egyptian K. pneumoniae strains, as well as being isolated from patients that have visited Egypt, we suggest that these plasmids may be a source of antimicrobial resistance transmission worldwide. These data are consistent with the view that strains carrying high levels of resistance to antibiotics are not hypervirulent, and vice versa, presumably, because there is a trade-off, due the burden of carrying different genetic determinants. Notwithstanding this, the emerging acquisition of multidrug resistant by hv K. pneumoniae is particularly concerning [1,2,3,55,58,59,60].
5. Conclusions
In this study we used whole genome sequencing to determine the antimicrobial resistance genes and virulence determinants carried by a multi-drug resistant K. pneumoniae strain isolated in Assiut, Egypt, comparing it to other sequenced Egyptian isolates. We show that all K. pneumoniae strains examined carry multiple antimicrobial resistance genes, which include carbapenemase and/or ESBL genes, and that these determinants are likely plasmid encoded. Our analysis also identifies similar resistance plasmids carried by K. pneumoniae strains isolated from individuals who had visited Egypt.
K. pneumoniae has been defined as an ESKAPE pathogen (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Enterobacter species) and a particular global threat due to its increased resistance to antimicrobial agents [61]. It is clear that the success of K. pneumoniae as a human pathogen, in both healthcare and community settings, is due to the acquisition of antimicrobial resistance genes and specific virulence determinates on transmissible plasmids [1,2,3]. Thus, understanding the transmission of these plasmids and the population dynamics of K. pneumoniae is key in devising future strategies to intervene and disarm this important human pathogen.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9091880/s1, Table S1: Antibiotic resistance gene analysis of Egyptian Klebsiella pneumoniae strains SF, SK, and HM. Figure S1: Comparison of draft genome of Klebsiella pneumoniae strain SK with those of K. pneumoniae strains HM and SF. Figure S2: Analysis of the IncFII(K) plasmid carried by K. pneumoniae strain KPE16. Figure S3: Comparison of the K. pneumoniae KPE16 draft genome with those of K. pneumoniae SF, SK, HM, and SP. Figure S4: Comparison of plasmid pKP1-19 with the K. pneumoniae KPE16 draft genome. Figure S5: Analysis of IncQ1 replicons carried by Egyptian K. pneumoniae and EAEC strains. Figure S6: Analysis of the IncM replicon carried by K. pneumoniae strain SK. Figure S7: Analysis of proteins implicated in colistin resistance in K. pneumoniae.
Author Contributions
Conceptualization, E.A.H., E.H.A., N.H.A.-F., E.A.D., S.J.W.B., and D.F.B.; formal analysis, R.A., S.J.W.B., and D.F.B.; funding acquisition, S.J.W.B., and D.F.B.; investigation, R.A., M.M.A., N.H.A.-F., and D.F.B.; supervision, E.A.D., S.J.W.B., and D.F.B.; writing—original draft, R.A., S.J.W.B., and D.F.B.; writing—review and editing, R.A., M.M.A., E.A.H., E.H.A., N.H.A.-F., E.A.D., S.J.W.B., and D.F.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was generously supported by a studentship from the Egyptian Ministry of Higher Education (Cultural Affairs and Missions Sector) and the Grant Office from the Medical School, Assiut University to R.A., and BBSRC research grant BB/R017689/1 to D.F.B. and S.J.W.B. The authors extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for funding Munirah Alhammadi.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank with the sequence data for K. pneumoniae KPE16 under the accession number JAGFBT000000000.
Acknowledgments
We thank MicrobesNG for sequencing and genome annotation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.C.; Fang, C.T.; Lee, C.Z.; Shun, C.T.; Wang, J.T. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J. Infect. Dis. 2005, 192, 117–128. [Google Scholar] [CrossRef] [Green Version]
- CDC. Vital signs: Carbapenem-resistant Enterobacteriaceae. MMWR Morb. Mortal. Wkly. Rep. 2013, 62, 165–170. [Google Scholar]
- Abdelaziz, M.O.; Bonura, C.; Aleo, A.; Fasciana, T.; Mammina, C. NDM-1- and OXA-163-producing Klebsiella pneumoniae isolates in Cairo, Egypt, 2012. J. Glob. Antimicrob. Resist. 2013, 1, 213–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamal, D.; Fernández-Martínez, M.; Salem, D.; El-Defrawy, I.; Montes, L.; Ocampo-Sosa, A.A.; Martinez-Martinez, L. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int. J. Infect. Dis. 2016, 43, 17–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaith, D.M.; Zafer, M.M.; Said, H.M.; Elanwary, S.; Elsaban, S.; Al-Agamy, M.H.; Bohol, M.F.F.; Bendary, M.M.; Al-Qahtani, A.; Al-Ahdal, M.N. Genetic diversity of carbapenem-resistant Klebsiella Pneumoniae causing neonatal sepsis in intensive care unit, Cairo, Egypt. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Zafer, M.M.; El-Mahallawy, H.A.; Abdulhak, A.; Amin, M.A.; Al-Agamy, M.H.; Radwan, H.H. Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 40. [Google Scholar] [CrossRef]
- El-Kholy, A.A.; Girgis, S.A.; Shetta, M.A.F.; Abdel-Hamid, D.H.; Elmanakhly, A.R. Molecular characterization of multidrug-resistant Gram-negative pathogens in three tertiary hospitals in Cairo, Egypt. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 987–992. [Google Scholar] [CrossRef] [Green Version]
- Hassuna, N.A.; AbdelAziz, R.A.; Zakaria, A.; Abdelhakeem, M. Extensively-Drug Resistant Klebsiella pneumoniae Recovered From Neonatal Sepsis Cases From a Major NICU in Egypt. Front Microbiol. 2020, 11, 1375. [Google Scholar] [CrossRef] [PubMed]
- Dandachi, I.; Chaddad, A.; Hanna, J.; Matta, J.; Daoud, Z. Understanding the Epidemiology of Multi-Drug Resistant Gram-Negative Bacilli in the Middle East Using a One Health Approach. Front Microbiol. 2019, 10, 1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ElMahallawy, H.; Zafer, M.M.; Al-Agamy, M.; Amin, M.A.; Mersal, M.M.; Booq, R.Y.; Alyamani, E.; Radwan, S. Dissemination of ST101 blaOXA-48 producing Klebsiella pneumoniae at tertiary care setting. J. Infect. Dev. Ctries. 2018, 12, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Van Duin, D.; Perez, F.; Rudin, S.D.; Cober, E.; Hanrahan, J.; Ziegler, J.; Webber, R.; Fox, J.; Mason, P.; Richter, S.S. Surveillance of carbapenem-resistant Klebsiella pneumoniae: Tracking molecular epidemiology and outcomes through a regional network. Antimicrob. Agents Chemother. 2014, 58, 4035–4041. [Google Scholar] [CrossRef] [Green Version]
- Kohler, P.; Tijet, N.; Kim, H.C.; Johnstone, J.; Edge, T.; Patel, S.N.; Seah, C.; Willey, B.; Coleman, B.; Green, K.; et al. Dissemination of Verona Integron-encoded Metallo-β-lactamase among clinical and environmental Enterobacteriaceae isolates in Ontario, Canada. Sci. Rep. 2020, 10, 18580. [Google Scholar] [CrossRef]
- Ludden, C.; Lötsch, F.; Alm, E.; Kumar, N.; Johansson, K.; Albiger, B.; Huang, T.-D.; Denis, O.; Hammerum, A.M.; Hasman, H.; et al. Cross-border spread of bla (NDM-1)- and bla (OXA-48)-positive Klebsiella pneumoniae: A European collaborative analysis of whole genome sequencing and epidemiological data, 2014 to 2019. Euro Surveill. 2020, 25, 2000627. [Google Scholar] [CrossRef] [PubMed]
- Alghoribi, M.F.; Binkhamis, K.; Alswaji, A.A.; Alhijji, A.; Alsharidi, A.; Balkhy, H.H.; Doumith, M.; Somily, A. Genomic analysis of the first KPC-producing Klebsiella pneumoniae isolated from a patient in Riyadh: A new public health concern in Saudi Arabia. J. Infect. Public Health. 2020, 13, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Nucleo, E.; Marchetti, V.M.; Mercato, A.; Quatela, M.; Villa, L.; Migliavacca, R. OXA-48 and NDM-1 Klebsiella pneumoniae of Sequence Type 101 from blood in a patient with travel history abroad, Italy. New Microbiol. 2020, 43, 41–43. [Google Scholar]
- Attia, H.; Szubin, R.; Yassin, A.S.; Monk, J.M.; Aziz, R.K. Draft Genome Sequences of Four Metallo-Beta-Lactamase-Producing Multidrug-Resistant Klebsiella pneumoniae Clinical Isolates, Including Two Colistin-Resistant Strains, from Cairo, Egypt. Microbiol. Resour. Announc. 2019, 8, e01418-18. [Google Scholar] [CrossRef] [Green Version]
- Norsigian, C.J.; Attia, H.; Szubin, R.; Yassin, A.S.; Palsson, B.; Aziz, R.K.; Monk, J.M. Comparative Genome-Scale Metabolic Modeling of Metallo-Beta-Lactamase-Producing Multidrug-Resistant Klebsiella pneumoniae Clinical Isolates. Front Cell Infect. Microbiol. 2019, 9, 161. [Google Scholar] [CrossRef]
- Abdelwahab, R.; Yasir, M.; Godfrey, R.E.; Christie, G.S.; Element, S.J.; Saville, F.; Hassan, E.A.; Ahmed, E.H.; Abu-Faddan, N.H.; Daef, E.A.; et al. Antimicrobial resistance and gene regulation in Enteroaggregative Escherichia coli from Egyptian children with diarrhoea: Similarities and differences. Virulence 2021, 12, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Ponten, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasman, H.; Saputra, D.; Sicheritz-Ponten, T.; Lund, O.; Svendsen, C.A.; Frimodt-Møller, N.; Aarestrup, F.M. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 2014, 52, 139–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clausen, P.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; CLSI Document M100-S24; Twenty-Fourth Informational Supplement: Wayne, MI, USA, 2014; Volume 34. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- McClelland, M.; Sanderson, K.E.; Spieth, J.; Clifton, S.W.; Latreille, P.; Courtney, L.; Porvollik, S.; Ali, J.; Dante, M.; Du, F.; et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 2001, 413, 852–856. [Google Scholar] [CrossRef] [Green Version]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef] [Green Version]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis Comparison Tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef] [Green Version]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Ponten, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Wick, R.R.; Heinz, E.; Holt, K.E.; Wyres, K.L. Kaptive Web: User-Friendly Capsule and Lipopolysaccharide Serotype Prediction for Klebsiella Genomes. J. Clin. Microbiol. 2018, 56, e00197-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villa, L.; Garcia-Fernandez, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef] [Green Version]
- Osborn, A.M.; da Silva Tatley, F.M.; Steyn, L.M.; Pickup, R.W.; Saunders, J.R. Mosaic plasmids and mosaic replicons: Evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 2000, 146, 2267–2275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.W.; Arumugam, K.; Purbojati, R.W.; Tay, Q.X.; Williams, R.B.; Kjelleberg, S.; Rice, S.A. Draft Genome Sequence of Klebsiella pneumoniae Strain KP-1. Genome Announc. 2013, 1, e01082-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholz, P.; Haring, V.; Wittmann-Liebold, B.; Ashman, K.; Bagdasarian, M.; Scherzinger, E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Genesis 1989, 75, 271–288. [Google Scholar] [CrossRef]
- Han, J.; Lynne, A.M.; David, D.E.; Tang, H.; Xu, J.; Nayak, R.; Kaldhone, P.; Logue, C.M.; Foley, S.L. DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates. PLoS ONE 2012, 7, e51160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poulou, A.; Voulgari, E.; Vrioni, G.; Koumaki, V.; Xidopoulos, G.; Chatzipantazi, V.; Markou, F.; Tsakris, A. Outbreak caused by an ertapenem-resistant, CTX-M-15-producing Klebsiella pneumoniae sequence type 101 clone carrying an OmpK36 porin variant. J. Clin. Microbiol. 2013, 51, 3176–3182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannatelli, A.; D′Andrea, M.M.; Giani, T.; Di Pilato, V.; Arena, F.; Ambretti, S.; Gaibani, P.; Rossolini, G.M. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 2013, 57, 5521–5526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannatelli, A.; Di Pilato, V.; Giani, T.; Arena, F.; Ambretti, S.; Gaibani, P.; D′Andrea, M.M.; Rossolini, G.M. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob. Agents Chemother. 2014, 58, 4399–4403. [Google Scholar] [CrossRef] [Green Version]
- Jayol, A.; Poirel, L.; Brink, A.; Villegas, M.V.; Yilmaz, M.; Nordmann, P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 2014, 58, 4762–4766. [Google Scholar] [CrossRef] [Green Version]
- Wright, M.S.; Suzuki, Y.; Jones, M.B.; Marshall, S.H.; Rudin, S.D.; van Duin, D.; Kaye, K.; Jacobs, M.R.; Bonomo, R.A.; Adams, M.D. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 2015, 59, 536–543. [Google Scholar] [CrossRef] [Green Version]
- Halaby, T.; Kucukkose, E.; Janssen, A.B.; Rogers, M.R.; Doorduijn, D.J.; van der Zanden, A.G.; al Naiemi, N.; Vandenbroucke-Grauls, C.M.J.E.; van Schaik, W. Genomic Characterization of Colistin Heteroresistance in Klebsiella pneumoniae during a Nosocomial Outbreak. Antimicrob. Agents Chemother. 2016, 60, 6837–6843. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.C.; Lin, T.L.; Hsieh, P.F.; Yang, H.C.; Wang, J.T. Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS ONE 2011, 6, e23500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fodah, R.A.; Scott, J.B.; Tam, H.H.; Yan, P.; Pfeffer, T.L.; Bundschuh, R.; Warawa, J.M. Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS ONE 2014, 9, e107394. [Google Scholar] [CrossRef] [Green Version]
- Fasciana, T.; Gentile, B.; Aquilina, M.; Ciammaruconi, A.; Mascarella, C.; Anselmo, A.; Fortunato, A.; Fillo, S.; Petralito, G.; Lista, F.; et al. Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect. Dis. 2019, 19, 928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fasciana, T.; Ciammaruconi, A.; Gentile, B.; Di Carlo, P.; Virruso, R.; Tricoli, M.R.; Palma, D.M.; Pitarresi, G.L.; Lista, F.; Giammanco, A. Draft Genome Sequence and Biofilm Production of a Carbapenemase-Producing Klebsiella pneumoniae (KpR405) Sequence Type 405 Strain Isolated in Italy. Antibiotics 2021, 10, 560. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Poirel, L.; Rondinaud, E.; Nordmann, P. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill. 2013, 18, 20549. [Google Scholar] [CrossRef]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae-clinical and molecular perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales-León, F.; Opazo-Capurro, A.; Caro, C.; Lincopan, N.; Cardenas-Arias, A.; Esposito, F.; Illesca, V.; Rioseco, M.L.; Dominguez-Yevenes, M.; Lima, C.A.; et al. Hypervirulent and hypermucoviscous extended-spectrum β-lactamase-producing Klebsiella pneumoniae and Klebsiella variicola in Chile. Virulence 2021, 12, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liang, Q.; Liu, W.; Zheng, B.; Liu, L.; Wang, W.; Xu, Z.; Huang, M.; Feng, Y. Convergence of carbapenem resistance and hypervirulence in a highly-transmissible ST11 clone of K. pneumoniae: An epidemiological, genomic and functional study. Virulence 2021, 12, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).