In Vitro and In Vivo Assessment of the Potential of Escherichia coli Phages to Treat Infections and Survive Gastric Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Phages, and Growth Conditions
2.2. Host Range Determination
2.3. Phage DNA Isolation, Sequencing, and Analysis of the Genome of Phage JK1
2.4. The Effect of Various pH and Simulated Gastric Fluid
2.5. In Vivo Phage Therapy Test Using the Galleria Mellonella Model
2.5.1. Bacterial Inoculum and Phage Cocktail
2.5.2. Testing of Phage Cocktail against a Single Strain and against a Pool of the Strains
3. Results
3.1. Characterization of the Phages Comprising the Cocktail
3.2. Evaluation of O-Antigens as Phage Receptors
3.3. Stability Is Highly Phage-Specific
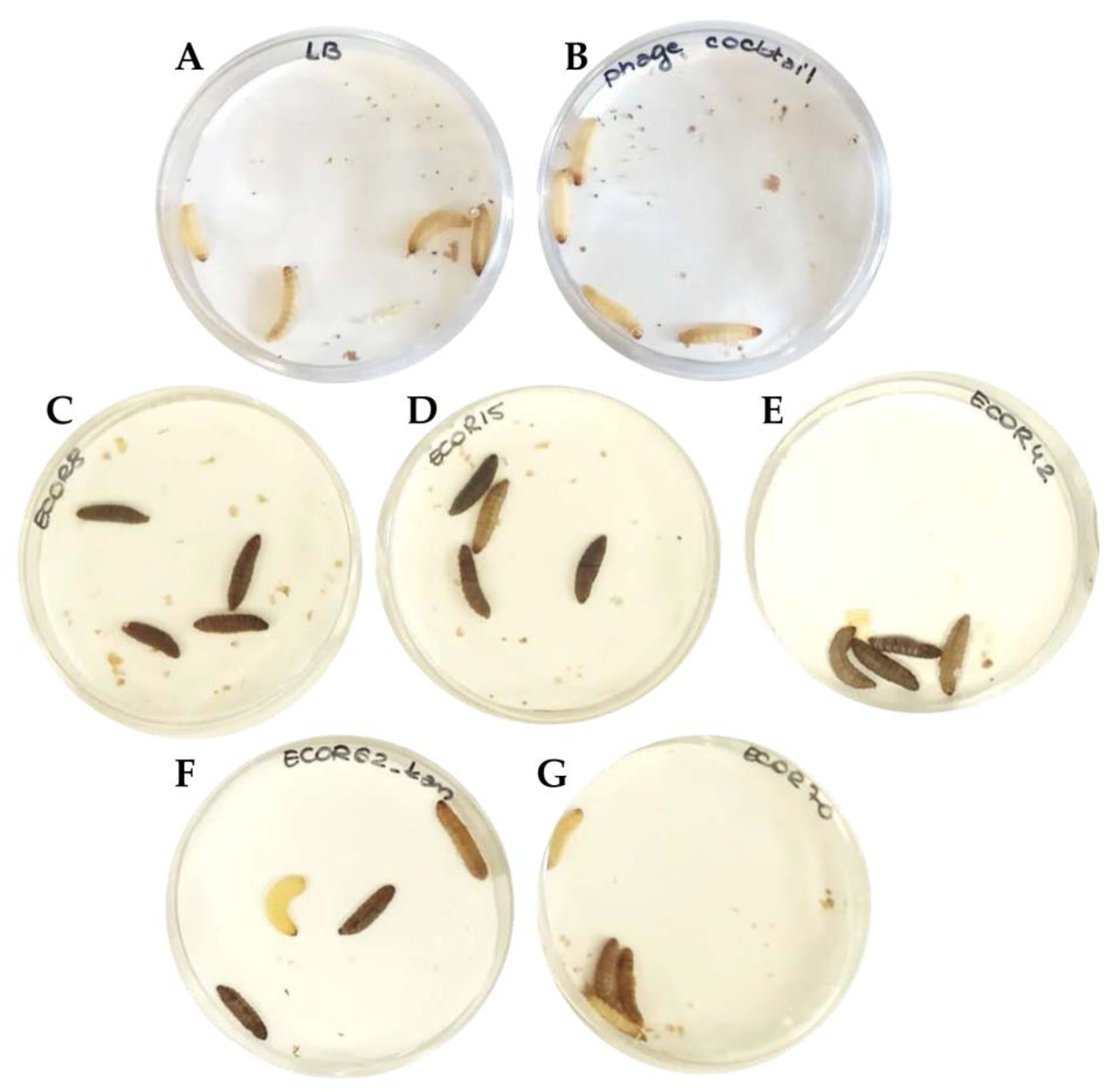
3.4. In Vivo Testing Reveals the Potential of Prophylactic Treatment of Multi-Strain E. coli Infections
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Troeger, C.; Forouzanfar, M.; Rao, P.C.; Khalil, I.; Brown, A.; Reiner, R.C.; Fullman, N.; Thompson, R.L.; Abajobir, A.; Ahmed, M.; et al. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017, 17, 909–948. [Google Scholar] [CrossRef] [Green Version]
- WHO. Diarrhoeal Disease—A Fact Sheet. 2017. Available online: http://www.who.int/mediacentre/factsheets/fs330/en/ (accessed on 15 April 2021).
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Torres, A.G.; Zhou, X.; Kaper, J.B. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 2005, 73, 18–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robins-Browne, R.M.; Hartland, E.L. Escherichia coli as a cause of diarrhea. J. Gastroenterol. Hepatol. 2002, 17, 467–475. [Google Scholar] [CrossRef]
- Meza-Segura, M.; Zaidi, M.B.; Vera-Ponce de León, A.; Moran-Garcia, N.; Martinez-Romero, E.; Nataro, J.P.; Estrada-Garcia, T. New insights into DAEC and EAEC pathogenesis and phylogeny. Front. Cell. Infect. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Sarantuya, J.; Nishi, J.; Wakimoto, N.; Erdene, S.; Nataro, J.P.; Sheikh, J.; Iwashita, M.; Manago, K.; Tokuda, K.; Yoshinaga, M.; et al. Typical Enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J. Clin. Microbiol. 2004, 42, 133–139. [Google Scholar] [CrossRef] [Green Version]
- Isidean, S.D.; Riddle, M.S.; Savarino, S.J.; Porter, C.K. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 2011, 29, 6167–6178. [Google Scholar] [CrossRef]
- Anderson, M.; Sansonetti, P.J.; Marteyn, B.S. Shigella Diversity and Changing Landscape: Insights for the Twenty-First Century. Front. Cell. Infect. Microbiol. 2016, 6, 45. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, X.; Hou, H.; Lu, Y.; Yu, J.; Mao, L.; Mao, L.; Sun, Z. Characteristics of diarrheagenic Escherichia coli among children under 5 years of age with acute diarrhea: A hospital based study. BMC Infect. Dis. 2018, 18, 63. [Google Scholar] [CrossRef] [Green Version]
- WHO. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Sack, R.B.; Rahman, M.; Yunus, M.; Khan, E.H. Antimicrobial resistance in organisms causing diarrheal disease. Clin. Infect. Dis. 1997, 24, S102–S105. [Google Scholar] [CrossRef]
- Mühlen, S.; Dersch, P. Treatment strategies for infections with Shiga toxin-producing Escherichia coli. Front. Cell. Infect. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Levine, M.M.; Kotloff, K.L.; Barry, E.M.; Pasetti, M.F.; Sztein, M.B. Clinical trials of Shigella vaccines: Two steps forward and one step back on a long, hard road. Nat. Rev. Microbiol. 2007, 5, 540–553. [Google Scholar] [CrossRef] [Green Version]
- Mai, V.; Ukhanova, M.; Reinhard, M.K.; Li, M.; Sulakvelidze, A. Bacteriophage administration significantly reduces Shigella colonization and shedding by Shigella-challenged mice without deleterious side effects and distortions in the gut microbiota. Bacteriophage 2015, 5, e1088124. [Google Scholar] [CrossRef] [Green Version]
- Mirhoseini, A.; Amani, J.; Nazarian, S. Review on pathogenicity mechanism of enterotoxigenic Escherichia coli and vaccines against it. Microb. Pathog. 2018, 117, 162–169. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Platts-Mills, J.A.; Nasrin, D.; Roose, A.; Blackwelder, W.C.; Levine, M.M. Global burden of diarrheal diseases among children in developing countries: Incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine 2017, 35, 6783–6789. [Google Scholar] [CrossRef]
- Hamdi, S.; Rousseau, G.M.; Labrie, S.J.; Tremblay, D.M.; Kourda, R.S.; Ben Slama, K.; Moineau, S. Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci. Rep. 2017, 7, 40349. [Google Scholar] [CrossRef]
- Brussow, H. Bacteriophage-host interaction: From splendid isolation into a messy reality. Curr. Opin. Microbiol. 2013, 16, 500–506. [Google Scholar] [CrossRef]
- Lai, X.H.; Wang, S.Y.; Uhlin, B.E. Expression of cytotoxicity by potential pathogens in the standard Escherichia coli collection of reference (ECOR) strains. Microbiology 1999, 145, 3295–3303. [Google Scholar] [CrossRef] [Green Version]
- Barnoy, S.; Gancz, H.; Zhu, Y.; Honnold, C.L.; Zurawski, D.V.; Venkatesan, M.M. The Galleria mellonella larvae as an in vivo model for evaluation of Shigella virulence. Gut Microbes 2017, 8, 335–350. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.J.; Loh, J.M.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [Green Version]
- Nale, J.Y.; Chutia, M.; Carr, P.; Hickenbotham, P.T.; Clokie, M.R. ‘Get in Early’; Biofilm and Wax Moth (Galleria mellonella) Models Reveal New Insights into the Therapeutic Potential of Clostridium difficile Bacteriophages. Front. Microbiol. 2016, 7, 1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedhila, S.; Buisson, C.; Dussurget, O.; Serror, P.; Glomski, I.J.; Liehl, P.; Lereclus, D.; Nielsen-LeRoux, C. Comparative analysis of the virulence of invertebrate and mammalian pathogenic bacteria in the oral insect infection model Galleria mellonella. J. Invertebr. Pathol. 2010, 103, 24–29. [Google Scholar] [CrossRef]
- Ramarao, N.; Nielsen-Leroux, C.; Lereclus, D. The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J. Vis. Exp. JoVE 2012, 70, e4392. [Google Scholar] [CrossRef] [Green Version]
- Heitmueller, M.; Billion, A.; Dobrindt, U.; Vilcinskas, A.; Mukherjee, K. Epigenetic Mechanisms Regulate Innate Immunity against Uropathogenic and Commensal-Like Escherichia coli in the Surrogate Insect Model Galleria mellonella. Infect. Immun. 2017, 85, e00336-17. [Google Scholar] [CrossRef] [Green Version]
- Kaczorowska, J.; Casey, E.; Neve, H.; Franz, C.; Noben, J.P.; Lugli, G.A.; Ventura, M.; Sinderen, D.V.; Mahony, J. A quest of great importance-Developing a broad spectrum Escherichia coli phage collection. Viruses 2019, 11, 899. [Google Scholar] [CrossRef] [Green Version]
- Haines, M.E.K.; Hodges, F.E.; Nale, J.Y.; Mahony, J.; van Sinderen, D.; Kaczorowska, J.; Alrashid, B.; Akter, M.; Brown, N.; Sauvageau, D.; et al. Analysis of selection methods to develop novel phage therapy cocktails against antimicrobial resistant clinical isolates of bacteria. Front. Microbiol. 2021, 12, 613529. [Google Scholar] [CrossRef]
- Jeong, H.; Barbe, V.; Lee, C.H.; Vallenet, D.; Yu, D.S.; Choi, S.H.; Couloux, A.; Lee, S.W.; Yoon, S.H.; Cattolico, L.; et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 2009, 394, 644–652. [Google Scholar] [CrossRef]
- Ochman, H.; Selander, R.K. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 1984, 157, 690–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pacan, J.C.; Wang, Q.; Xu, Y.; Huang, X.; Korenevsky, A.; Sabour, P.M. Microencapsulation of bacteriophage felix O1 into chitosan-alginate microspheres for oral delivery. Appl. Environ. Microbiol. 2008, 74, 4799–4805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Los, J.M.; Golec, P.; Wegrzyn, G.; Wegrzyn, A.; Los, M. Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Appl. Environ. Microbiol. 2008, 74, 5113–5120. [Google Scholar] [CrossRef] [Green Version]
- Zrelovs, N.; Dislers, A.; Kazaks, A. Motley crew: Overview of the currently available phage diversity. Front. Microbiol. 2021, 11. [Google Scholar] [CrossRef]
- Olsen, N.S.; Forero-Junco, L.; Kot, W.; Hansen, L.H. Exploring the remarkable diversity of culturable Escherichia coli phages in the Danish wastewater environment. Viruses 2020, 12, 986. [Google Scholar] [CrossRef]
- Heller, K.; Braun, V. Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J. Virol. 1982, 41, 222–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandulache, R.; Prehm, P.; Kamp, D. Cell wall receptor for bacteriophage Mu G(+). J. Bacteriol. 1984, 160, 299–303. [Google Scholar] [CrossRef] [Green Version]
- Prehm, P.; Jann, B.; Jann, K.; Schmidt, G.; Stirm, S. On a bacteriophage T3 and T4 receptor region within the cell wall lipopolysaccharide of Escherichia coli B. J. Mol. Biol. 1976, 101, 277–281. [Google Scholar] [CrossRef]
- Amor, K.; Heinrichs, D.E.; Frirdich, E.; Ziebell, K.; Johnson, R.P.; Whitfield, C. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 2000, 68, 1116–1124. [Google Scholar] [CrossRef] [Green Version]
- Broeker, N.K.; Barbirz, S. Not a barrier but a key: How bacteriophages exploit host’s O-antigen as an essential receptor to initiate infection. Mol. Microbiol. 2017, 105, 353–357. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.J.; Lee, H.; Lee, J.H.; Ryu, S.; Park, J.H. Morphological features and lipopolysaccharide attachment of coliphages specific to Escherichia coli O157:H7 and to a broad range of E. coli hosts. Appl. Biol. Chem. 2016, 59, 109–116. [Google Scholar] [CrossRef]
- Johnson, J.R.; Delavari, P.; Stell, A.L.; Prats, G.; Carlino, U.; Russo, T.A. Integrity of archival strain collections: The ECOR collection. Asm News 2001, 67, 288–289. [Google Scholar]
- Patel, I.R.; Gangiredla, J.; Mammel, M.K.; Lampel, K.A.; Elkins, C.A.; Lacher, D.W. Draft genome sequences of the Escherichia coli reference (ECOR) collection. Microbiol. Resour. Announc. 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Clermont, O.; Cordevant, C.; Bonacorsi, S.; Marecat, A.; Lange, M.; Bingen, E. Automated ribotyping provides rapid phylogenetic subgroup affiliation of clinical extraintestinal pathogenic Escherichia coli strains. J. Clin. Microbiol. 2001, 39, 4549–4553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulikov, E.E.; Golomidova, A.K.; Prokhorov, N.S.; Ivanov, P.A.; Letarov, A.V. High-throughput LPS profiling as a tool for revealing of bacteriophage infection strategies. Sci. Rep. 2019, 9, 2958. [Google Scholar] [CrossRef]
- Ryan, E.M.; Gorman, S.P.; Donnelly, R.F.; Gilmore, B.F. Recent advances in bacteriophage therapy: How delivery routes, formulation, concentration and timing influence the success of phage therapy. J. Pharm. Pharmacol. 2011, 63, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Nagel, T.E.; Chan, B.K.; De Vos, D.; El-Shibiny, A.; Kang’ethe, E.K.; Makumi, A.; Pirnay, J.P. The Developing world urgently needs phages to combat pathogenic bacteria. Front. Microbiol. 2016, 7, 882. [Google Scholar] [CrossRef]
- Górski, A.; Borysowski, J.; Miedzybrodzki, R.; Weber-Dabrowska, B. Bacteriophages in Medicine; Caister Academic Press: Norfolk, UK, 2007. [Google Scholar]
- Joerger, R. Alternatives to antibiotics: Bacteriocins, antimicrobial peptides and bacteriophages. Poult. Sci. 2003, 82, 640–647. [Google Scholar] [CrossRef]
- Brussow, H. What is needed for phage therapy to become a reality in Western medicine? Virology 2012, 434, 138–142. [Google Scholar] [CrossRef] [Green Version]
- Jurczak-Kurek, A.; Gasior, T.; Nejman-Falenczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016, 6, 34338. [Google Scholar] [CrossRef]
- Dini, C.; De Urraza, P.J. Isolation and selection of coliphages as potential biocontrol agents of enterohemorrhagic and Shiga toxin-producing E. coli (EHEC and STEC) in cattle. J. Appl. Microbiol. 2010, 109, 873–887. [Google Scholar] [CrossRef] [Green Version]
- Jun, J.W.; Kim, J.H.; Shin, S.P.; Han, J.E.; Chai, J.Y.; Park, S.C. Characterization and complete genome sequence of the Shigella bacteriophage pSf-1. Res. Microbiol. 2013, 164, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Allue-Guardia, A.; Martinez-Castillo, A.; Muniesa, M. Persistence of infectious Shiga toxin-encoding bacteriophages after disinfection treatments. Appl. Environ. Microbiol. 2014, 80, 2142–2149. [Google Scholar] [CrossRef] [Green Version]
- Shende, R.K.; Hirpurkar, S.D.; Sannat, C.; Rawat, N.; Pandey, V. Isolation and characterization of bacteriophages with lytic activity against common bacterial pathogens. Vet. World 2017, 10, 973–978. [Google Scholar] [CrossRef] [Green Version]
- Sarker, S.A.; McCallin, S.; Barretto, C.; Berger, B.; Pittet, A.C.; Sultana, S.; Krause, L.; Huq, S.; Bibiloni, R.; Bruttin, A.; et al. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 2012, 434, 222–232. [Google Scholar] [CrossRef] [Green Version]
- Kutter, E.; De Vos, D.; Gvasalia, G.; Alavidze, Z.; Gogokhia, L.; Kuhl, S.; Abedon, S. Phage therapy in clinical practice: Treatment of human infections. Curr. Pharm. Biotechnol. 2010, 11, 69–86. [Google Scholar] [CrossRef]
- Sarker, S.A.; Sultana, S.; Reuteler, G.; Moine, D.; Descombes, P.; Charton, F.; Bourdin, G.; McCallin, S.; Ngom-Bru, C.; Neville, T.; et al. Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine 2016, 4, 124–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]

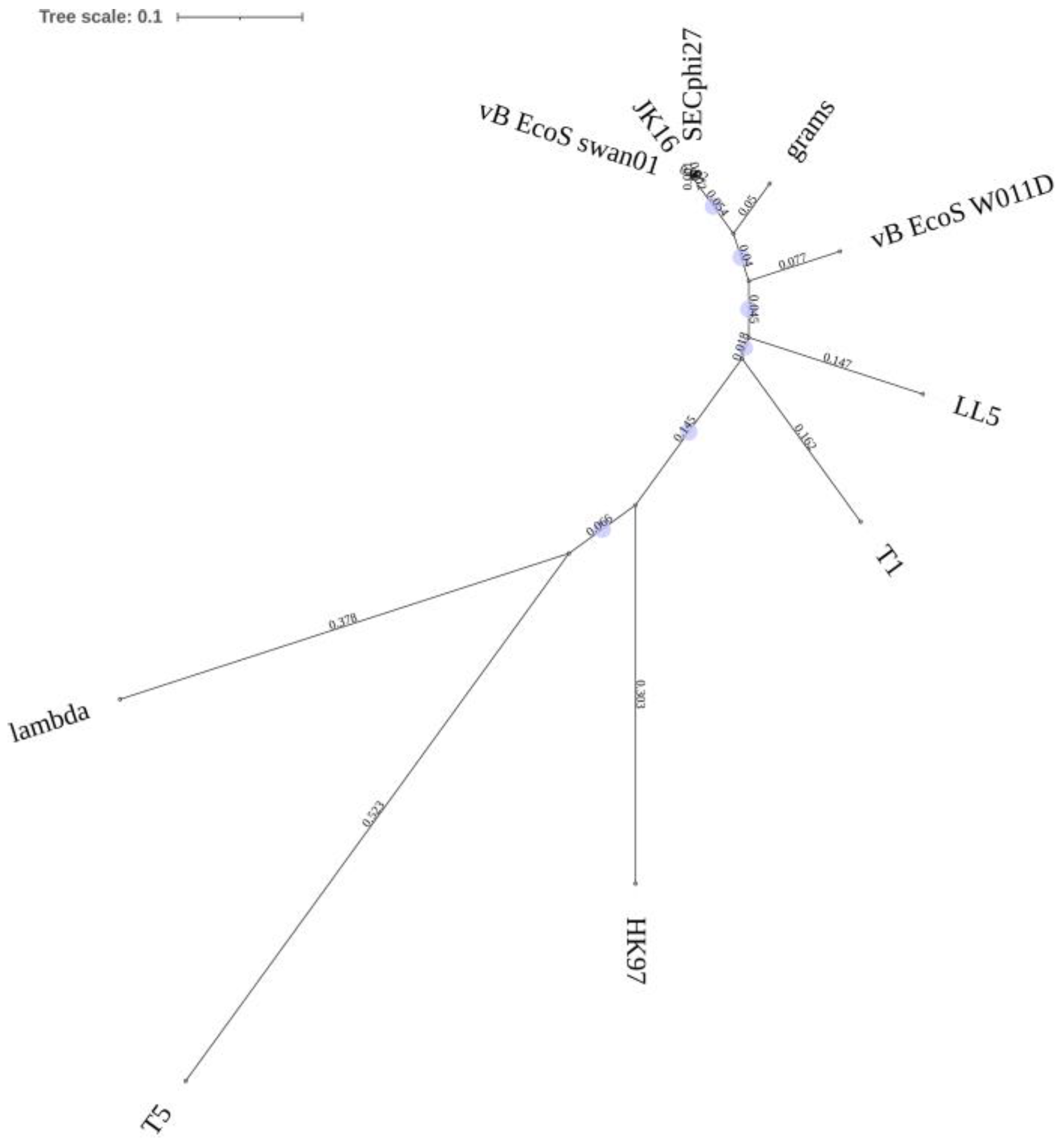


| Phage Name | Source | Propagation Host | Family | Genus | Reference |
|---|---|---|---|---|---|
| JK1 | River Lee, Cork | BL21 | Myoviridae | Tequatrovirus | This study |
| JK08 | Cork City stream | DH5α | Myoviridae | Tequatrovirus | [29] |
| JK16 | Cork City stream | DH5α | Drexlerviridae | Warickvirus | [28] |
| JK36 | Sewage | Top10 | Myoviridae | Mosigvirus | [28] |
| JK38 | Sewage | BL21 | Myoviridae | Tequatrovirus | [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaczorowska, J.; Casey, E.; Lugli, G.A.; Ventura, M.; Clarke, D.J.; van Sinderen, D.; Mahony, J. In Vitro and In Vivo Assessment of the Potential of Escherichia coli Phages to Treat Infections and Survive Gastric Conditions. Microorganisms 2021, 9, 1869. https://doi.org/10.3390/microorganisms9091869
Kaczorowska J, Casey E, Lugli GA, Ventura M, Clarke DJ, van Sinderen D, Mahony J. In Vitro and In Vivo Assessment of the Potential of Escherichia coli Phages to Treat Infections and Survive Gastric Conditions. Microorganisms. 2021; 9(9):1869. https://doi.org/10.3390/microorganisms9091869
Chicago/Turabian StyleKaczorowska, Joanna, Eoghan Casey, Gabriele A. Lugli, Marco Ventura, David J. Clarke, Douwe van Sinderen, and Jennifer Mahony. 2021. "In Vitro and In Vivo Assessment of the Potential of Escherichia coli Phages to Treat Infections and Survive Gastric Conditions" Microorganisms 9, no. 9: 1869. https://doi.org/10.3390/microorganisms9091869
APA StyleKaczorowska, J., Casey, E., Lugli, G. A., Ventura, M., Clarke, D. J., van Sinderen, D., & Mahony, J. (2021). In Vitro and In Vivo Assessment of the Potential of Escherichia coli Phages to Treat Infections and Survive Gastric Conditions. Microorganisms, 9(9), 1869. https://doi.org/10.3390/microorganisms9091869








