Gut Bacterial Communities of Lymantria xylina and Their Associations with Host Development and Diet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Collection
2.2. Effect of Symbiotic Bacteria on L. xylina Development
2.3. DNA Extraction, Bacterial 16s rRNA Gene Amplification, and High-Throughput Sequencing
2.4. Division of Larval Instars
2.5. Bioinformatic Analysis
2.6. Gut Microbiota Composition of L. xylina on Different Diets
2.7. Functional Annotation
2.8. Statistical Analyses
3. Results
3.1. Division of Larval Instars
3.2. Bacterial Community Structures and OTUs
3.3. The Diversity Differences in Bacterial Communities during the Different Development Stages of L. xylina
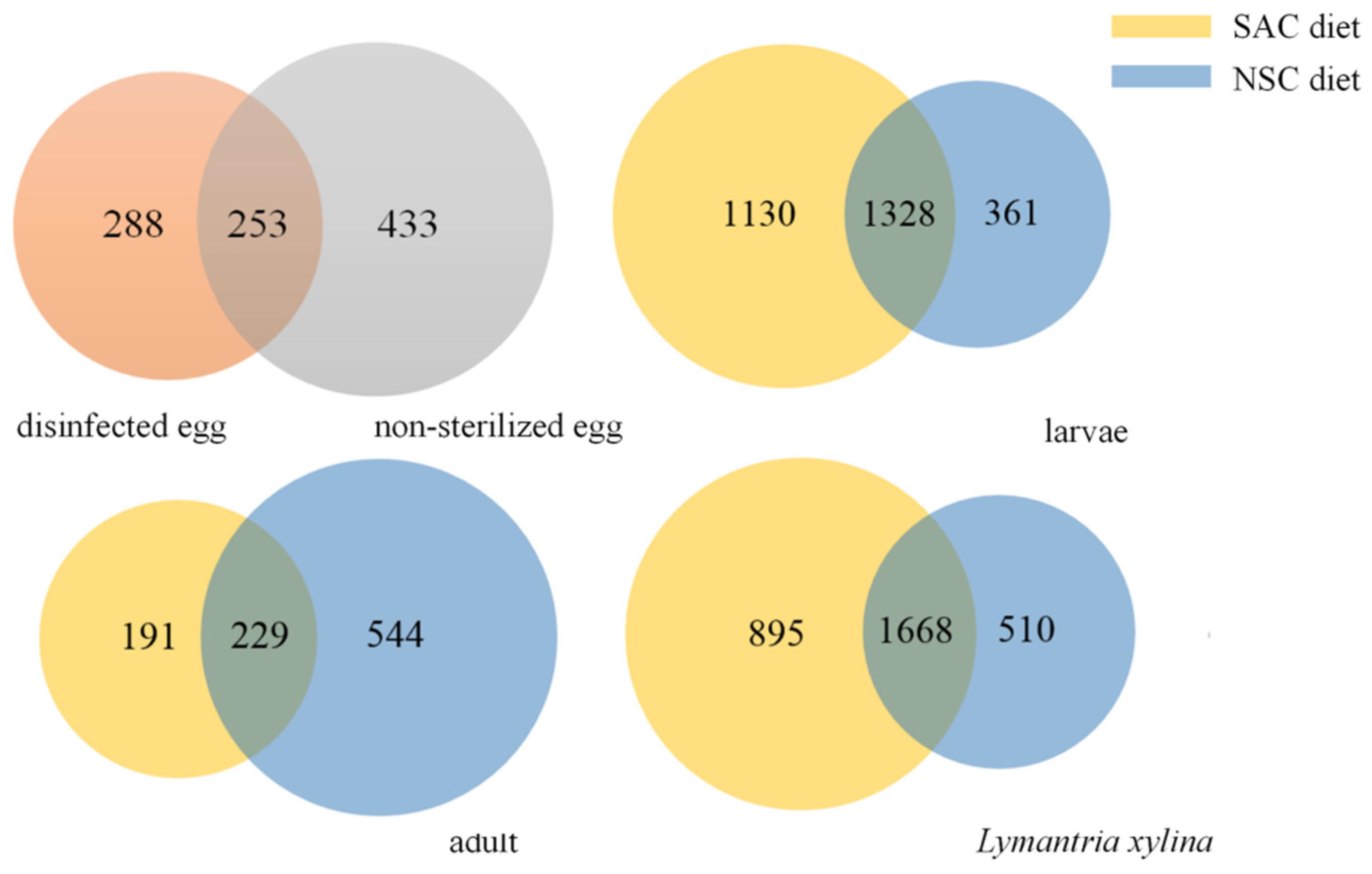
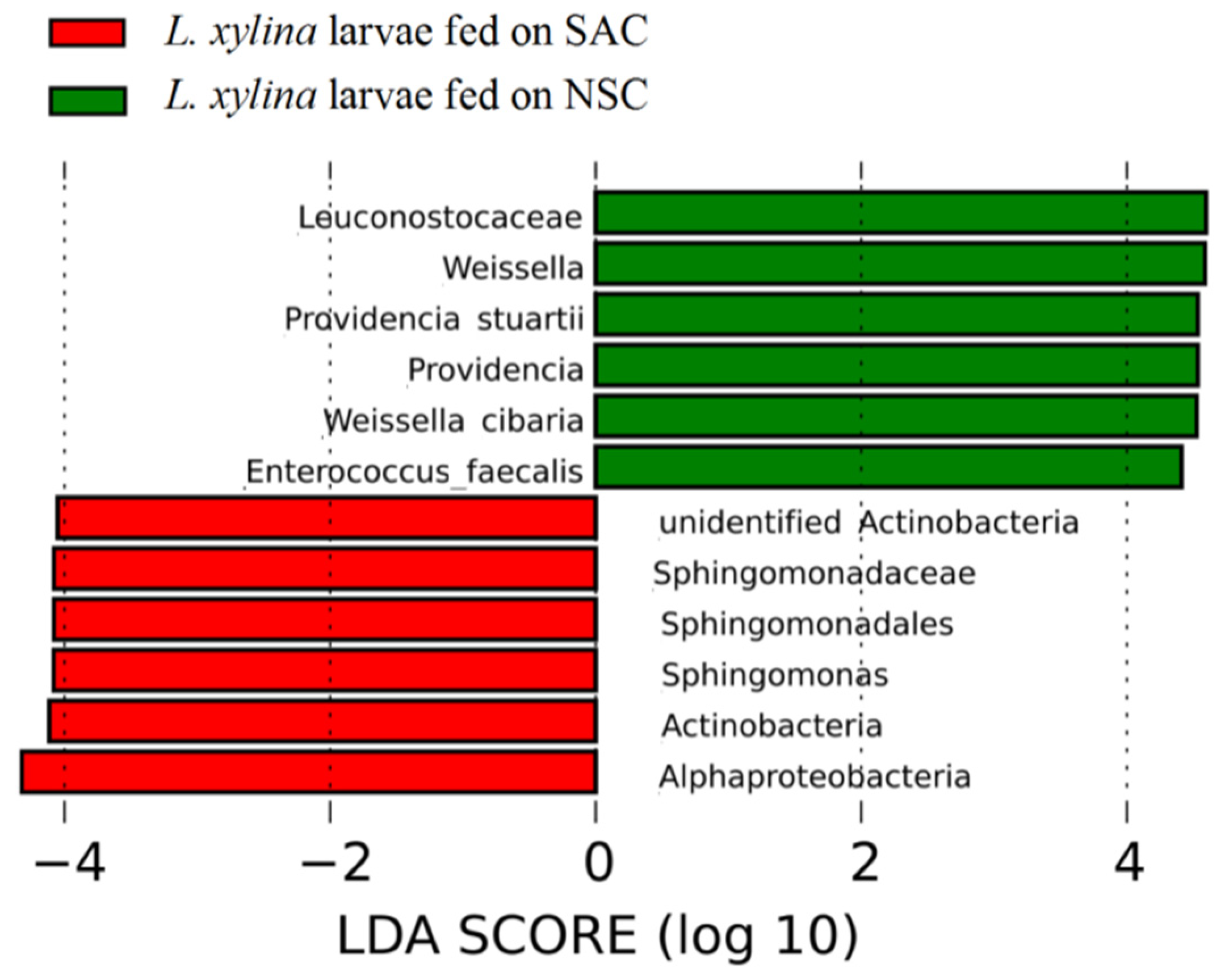
3.4. The Diversity Differences for Bacterial Communities with Different Development Diets for L. xylina
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample | Body Length (mm) | Coefficient Variation | Brooks’ Ratio | Crosby’s Ratio | p Value |
|---|---|---|---|---|---|
| A1 | 3.61 ± 0.195 | 14.2% | - | - | 0.048 * |
| N1 | 5.61 ± 0.768 | 13.7% | - | - | |
| A2 | 5.38 ± 0.187 | 11.1% | 1.489 | - | 0.006 ** |
| N2 | 7.67 ± 3.333 | 7.5% | 1.366 | - | |
| A3 | 7.64 ± 0.188 | 12.1% | 1.420 | −4.6% | 0.046 * |
| N3 | 10.00 ± 0.837 | 18.7% | 1.304 | −4.5% | |
| A4 | 11.11 ± 0.462 | 9.4% | 1.455 | 2.5% | 0.000 *** |
| N4 | 14.71 ± 0.610 | 19.0% | 1.471 | 9.9% | |
| A5 | 19.96 ± 0.334 | 10.1% | 1.796 | 23.4% | 0.324 |
| N5 | 18.84 ± 1.055 | 24.4% | 1.281 | −13.0% | |
| A6 | 31.38 ± 0.507 | 12.5% | 1.572 | −12.5% | 0.000 *** |
| N6 | 21.97 + 0.613 | 15.5% | 1.166 | −9.0% | |
| A7 | 43.43 ± 0.717 | 15.5% | 1.384 | −12.0% | 0.000 *** |
| N7 | 29.74 ± 1.395 | 22.5% | 1.354 | 1.6% |
| Development | OTUs | Chao1 | Shannon-Wiener Index | Simpson Index | Goods Coverage |
|---|---|---|---|---|---|
| Egg | 974 | 334.744 | 3.494 | 0.733 | 0.999 |
| 1st instar larvae | 1245 | 418.548 | 3.062 | 0.664 | 0.998 |
| 2nd instar larvae | 1140 | 402.394 | 3.189 | 0.706 | 0.998 |
| 3rd instar larvae | 1258 | 483.053 | 2.807 | 0.664 | 0.996 |
| 4th instar larvae | 1389 | 531.653 | 4.484 | 0.815 | 0.996 |
| 5th instar larvae | 1250 | 425.975 | 3.392 | 0.665 | 0.998 |
| 6th instar larvae | 619 | 279.847 | 2.101 | 0.586 | 0.998 |
| 7th instar larvae | 952 | 318.271 | 2.586 | 0.666 | 0.998 |
| Adult | 964 | 333.503 | 2.096 | 0.470 | 0.998 |
| Phylum Level | Egg | 1st Instar Larvae | 2nd Instar Larvae | 3rd Instar Larvae | 4th Instar Larvae | 5th Instar Larvae | 6th Instar Larvae | 7th Instar Larvae | Pupa | Adult |
|---|---|---|---|---|---|---|---|---|---|---|
| Firmicutes | 44.33 | 55.20 | 80.01 | 68.03 | 42.97 | 74.88 | 90.29 | 79.30 | 63.94 | 86.09 |
| Proteobacteria | 47.23 | 33.95 | 13.40 | 27.64 | 39.09 | 14.04 | 7.29 | 11.43 | 24.95 | 9.92 |
| Actinobacteria | 3.73 | 6.58 | 2.51 | 1.25 | 2.63 | 1.06 | 0.10 | 0.50 | 0.46 | 1.17 |
| Cyanobacteria | 0.27 | 0.20 | 0.35 | 0.21 | 4.67 | 1.76 | 1.44 | 4.82 | 0.75 | 0.28 |
| Bacteroidetes | 2.74 | 1.12 | 2.14 | 1.82 | 6.18 | 5.45 | 0.18 | 2.97 | 8.78 | 2.09 |
| Verrucomicrobia | 0.05 | 0.03 | 0.07 | 0.05 | 1.13 | 0.09 | 0.00 | 0.01 | 0.01 | 0.04 |
| Chloroflexi | 0.02 | 0.97 | 0.06 | 0.01 | 0.12 | 0.13 | 0.00 | 0.04 | 0.03 | 0.01 |
| Acidobacteria | 0.10 | 0.71 | 0.12 | 0.04 | 0.34 | 0.36 | 0.01 | 0.04 | 0.02 | 0.02 |
| Gemmatimonadetes | 0.02 | 0.36 | 0.05 | 0.01 | 0.02 | 0.08 | 0.00 | 0.01 | 0.01 | 0.02 |
| Euryarchaeota | 0.00 | 0.00 | 0.02 | 0.00 | 0.36 | 0.07 | 0.00 | 0.01 | 0.00 | 0.00 |
| Others | 1.51 | 0.88 | 1.27 | 0.92 | 2.50 | 2.10 | 0.68 | 0.88 | 1.04 | 0.36 |
| Phylum Level | Egg | 1st Instar Larvae | 2nd Instar Larvae | 3rd Instar Larvae | 4th Instar Larvae | 5th Instar Larvae | 6th Instar Larvae | 7th Instar Larvae | Pupa | Adult |
|---|---|---|---|---|---|---|---|---|---|---|
| Enterococcus | 23.98 | 34.35 | 57.67 | 56.49 | 28.89 | 63.62 | 89.58 | 75.75 | 61.02 | 79.91 |
| Providencia | 0.00 | 0.01 | 0.01 | 21.31 | 0.02 | 0.02 | 0.01 | 0.01 | 0.00 | 0.01 |
| Weissella | 17.48 | 14.94 | 16.30 | 1.40 | 0.05 | 5.08 | 0.11 | 0.09 | 0.00 | 0.05 |
| Acinetobacter | 19.61 | 16.77 | 3.94 | 1.22 | 2.14 | 5.42 | 5.33 | 8.24 | 20.64 | 2.46 |
| unidentified Cyanobacteria | 0.25 | 0.20 | 0.34 | 0.20 | 4.66 | 1.76 | 1.44 | 4.81 | 0.74 | 0.28 |
| unidentified Burkholderiaceae | 4.87 | 0.00 | 0.00 | 0.06 | 0.12 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Staphylococcus | 0.03 | 1.45 | 2.57 | 6.90 | 0.16 | 0.12 | 0.03 | 0.02 | 0.06 | 0.24 |
| Empedobacter | 1.50 | 0.03 | 0.01 | 0.02 | 0.01 | 0.00 | 0.00 | 2.23 | 6.97 | 0.01 |
| Acidovorax | 1.10 | 6.04 | 0.79 | 0.28 | 0.82 | 0.34 | 0.35 | 0.46 | 0.69 | 0.83 |
| Sphingomonas | 1.10 | 6.30 | 0.35 | 0.86 | 2.18 | 1.05 | 0.48 | 0.31 | 0.15 | 0.24 |
| Others | 30.07 | 19.91 | 18.01 | 11.27 | 60.95 | 22.60 | 2.66 | 8.07 | 9.72 | 15.97 |
References
- Liu, Y.; Shen, Z.; Yu, J.; Li, Z.; Liu, X.; Xu, H. Comparison of gut bacterial communities and their associations with host diets in four fruit borers. Pest Manag. Sci. 2020, 76, 1353–1362. [Google Scholar] [CrossRef]
- Zhang, J.H.; Yu, N.; Xu, X.X.; Liu, Z.W. Community structure, dispersal ability and functional profiling of microbiome existing in fat body and ovary of the brown planthopper, Nilaparvata lugens. Insect Sci. 2018, 26, 683–694. [Google Scholar] [CrossRef]
- Philipp, E.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 5, 699–735. [Google Scholar]
- Hosokawa, T.; Kikuchi, Y.; Meng, X.Y.; Fukatsu, T. The making of symbiont capsule in the plataspid stinkbug Megacopta punctatissima. FEMS Microbiol. Ecol. 2005, 54, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Guo, Z.; Riegler, M.; Xi, Z.; Liang, G.; Xu, Y. Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 2017, 5, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.S.; Du, K.Q.; Sun, C.; Arunprasanna, V.; Liang, X.; Li, Y.; Wang, B.H.; Lu, X.M.; Li, L.J.; Shao, Y.Q. Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 2018, 52, 1085–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiko, S.; Dubreuil, G.; David, G.; Jean, C.S. Plant–insect interactions under bacterial influence: Ecological implications and underlying mechanisms. J. Exp. Bot. 2014, 2, 2–12. [Google Scholar]
- Su, L.J.; Yang, L.L.; Huang, S.; Li, Y.; Su, X.Q.; Wang, F.Q.; Bo, C.P.; Wang, E.T.; Song, A.D. Variation in the gut microbiota of termites (Tsaitermes ampliceps) against different diets. Appl. Biochem. Biotechnol. 2016, 181, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.Y.; Shi, J.H.; Guo, J.X.; Shi, Z.B.; Zhang, G.C.; Zhang, J. Variation in the pH of experimental diets affects the performance of Lymantria dispar asiatica larvae and its gut microbiota. Arch. Insect Biochem. 2020, 103, e21654. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.C.; Tseng, C.M.; Guan, L.C.; Hwang, S.Y. Performance of Lymantria xylina (Lepidoptera: Lymantriidae) on artificial and host plant diets. J. Econ. Entomol. 2006, 3, 714–721. [Google Scholar] [CrossRef]
- Sree, K.S.; Varma, A. Biocontrol of Lepidopteran Pests; Springer International Publishing: Berlin, Switzerland, 2015; p. 43. [Google Scholar]
- Chao, J.T.; Paul, W.S.; Fan, Y.B.; Lu, S.S. Host plants and infestation of casuarina moth Lymantria xylina in Taiwan. Taiwan For. Sci. 1996, 11, 23–28. [Google Scholar]
- Wang, R.; Zhang, Z.H.; Hu, X.; Wu, S.Q.; Wang, J.D.; Zhang, F.P. Molecular detection and genetic diversity of casuarina moth, Lymantria xylina (Lepidoptera: Erebidae). J. Insect Sci. 2018, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mitter, C.; Davis, D.R.; Cummings, M.P. Phylogeny and evolution of Lepidoptera. Annu. Rev. Entomol. 2017, 62, 265–283. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, C.; Chen, H.; Ma, J. Differences in the structure of the gut bacteria communities in development stages of the Chinese white pine beetle (Dendroctonus armandi). Int. J. Mol. Sci. 2013, 14, 21006–21020. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Zhang, J.T.; Zong, S.X.; Luo, Y.Q.; Niu, H.L.; Zhang, B. Determination of the larval instar number of the carpenter moth Holcocerus vicarius (Lepidoptera: Cossidae). Acta Entomol. Sin. 2012, 55, 710–718. [Google Scholar]
- Loerch, C.R.; Cameron, E.A. Determination of larval instars of the bronze birch borer, Agrilus anxius (Coleoptera: Buprestidae). Ann. Entomol. Soc. Am. 1983, 76, 948–952. [Google Scholar] [CrossRef]
- Ablhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Torbjørn, R.; Tomáš, F.; Nichols, B.; Quince, C.; Frédéric, M. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar]
- Brian, J.H.; Dirk, G.; Ashlee, M.E.; Mike, F.; Doyle, V.W.; Georgia, G.; Dawn, C.; Diana, T.; Sarah, K.H.; Erica, S.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.G.; Liu, X.G.; Xu, H.X.; Liu, Y.H.; Panna, A.; Muhammad, A.B.; Lu, Z.X. The abundance and diversity of gut bacteria of rice leaffolder Cnaphalocrocis medinalis (Guenée) across life stages. J. Asia-Pac. Entomol. 2020, 23, 430–438. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Luis, R.P.V.; Enric, F.; Martin, K.; Monika, H.; Nina, E.F. Bacterial symbionts in Lepidoptera: Their diversity, transmission, and impact on the host. Front. Microbiol. 2018, 9, 556. [Google Scholar] [CrossRef]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malhotra, J.; Dua, A.; Saxena, A.; Sangwan, N.; Lal, R. Genome sequence of Acinetobacter sp. strain HA, isolated from the gut of the polyphagous insect pest Helicoverpa Armigera. J. Bacteriol. 2012, 194, 5156. [Google Scholar] [CrossRef] [Green Version]
- Indiragandhi, P.; Anandham, R.; Madhaiyan, M.; Sa, T.M. Characterization of plant growth–promoting traits of bacteria isolated from larval guts of diamondback moth Plutella xylostella (Lepidoptera: Plutellidae). Curr. Microbiol. 2008, 56, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; Dillon, V.M. The gut bacteria of insects: Nonpathogenic interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef]
- Eberle, M.W.; McLean, D.L. Observation of symbiote migration in human body lice with scanning and transmission electron microscopy. Can. J. Microbiol. 1983, 29, 755–762. [Google Scholar] [CrossRef]
- Salem, H.; Florez, L.; Gerardo, N.; Kaltenpoth, M. An out-of-body experience: The extracellular dimension for the transmission of mutualistic bacteria in insects. Proc. R. Soc. B 2015, 282, 20142957. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, Y.; Hosokawa, T.; Nikoh, N.; Meng, X.Y.; Kamagata, Y.; Fukatsu, T. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 2009, 7, 2. [Google Scholar] [CrossRef] [Green Version]
- Salem, H.; Kreutzer, E.; Sudakaran, S.; Kaltenpoth, M. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae). Environ. Microbiol. 2013, 15, 1956–1968. [Google Scholar] [CrossRef]
- Chu, C.C.; Joseph, L.S.; Matías, J.C.; Jorge, A.Z.; Manfredo, J.S. Gut bacteria facilitate adaptation to crop rotation in the western corn rootworm. Proc. Natl. Acad. Sci. USA 2013, 110, 11917–11922. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhu, J.Q.; Fang, J.; Shen, L.; Ma, S.J.; Zhao, Z.M.; Yu, W.D.; Jiang, W.B. Diversity, composition and functional inference of gut microbiota in indian cabbage white Pieris canidia (Lepidoptera: Pieridae). Life 2020, 10, 254. [Google Scholar] [CrossRef] [PubMed]
- Staudacher, H.; Kaltenpoth, M.; Breeuwer, J.A.; Menken, S.B.; Heckel, D.G.; Groot, A.T. Variability of bacterial communities in the moth Heliothis virescens indicates transient association with the host. PLoS ONE 2016, 11, e0154514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awmack, C.S.; Leather, S.R. Host plant quality and fecundity in herbivorous insects. Annu. Rev. Entomol. 2002, 47, 817–844. [Google Scholar] [CrossRef]
- Xia, X.; Gurr, G.M.; Vasseur, L.; Zheng, D.; Zhong, H.; Qin, B.; Lin, J.H.; Wang, Y.; Song, F.Q.; Li, Y.; et al. Metagenomic sequencing of diamondback moth gut microbiome unveils key holobiont adaptations for herbivory. Front. Microbiol. 2017, 8, 663. [Google Scholar] [CrossRef]
- Xiang, H.; Wei, G.F.; Jia, S.H.; Huang, J.H.; Miao, X.X.; Zhou, Z.H.; Zhao, L.P.; Huang, Y.P. Microbial communities in the larval midgut of laboratory and field populations of cotton bollworm (Helicoverpa armigera). Can. J. Microbiol. 2007, 52, 23–24. [Google Scholar] [CrossRef] [PubMed]
- Broderick, N.A.; Raffa, K.F.; Goodman, R.M.; Handelsman, J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 2004, 70, 293–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.H.; Dalial, F.; Heiko, V.; Ping, L.Y.; Shao, Y.Q.; Erika, A.C.; Gary, A.; Martin, W.; David, G.H.; Wilhelm, B. Complexity and variability of gut commensal microbiota in polyphagous Lepidopteran larvae. PLoS ONE 2012, 7, e36978. [Google Scholar] [CrossRef] [Green Version]
- Breznak, J.A. Intestinal microbiota of termites and other xylophagous insects. Annu. Rev. Microbiol. 1982, 36, 323. [Google Scholar] [CrossRef] [PubMed]
- Dow, J.A. pH gradients in lepidopteran midgut. J. Exp. Biol. 1992, 172, 255–375. [Google Scholar] [CrossRef]
- Li, P.H.; Niu, Q.; Wei, Q.T.; Zhang, Y.Q.; Ma, X.; Sung, W.K.; Ling, M.X.; Huang, R.H. Microbial shifts in the porcine distal gut in response to diets supplemented with Enterococcus Faecalis as alternatives to antibiotics. Sci. Rep. 2017, 7, 41395. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Chen, B.; Sun, C.; Ishida, K.; Hertweck, C.; Boland, W. Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Cell Chem. Biol. 2017, 24, 66–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundgren, J.G.; Lehman, R. Bacterial gut symbionts contribute to seed digestion in an omnivorous beetle. PLoS ONE 2010, 5, e10831. [Google Scholar] [CrossRef]
- Schmid, R.B.; Michael, L.R.; Brözel, V.S.; Lundgren, J.G. Gut bacterial symbiont diversity within beneficial insects linked to reductions in local biodiversity. Ann. Entomol. Soc. Am. 2015, 6, 993–999. [Google Scholar] [CrossRef] [Green Version]



| Sample | Head Capsule Width (mm) | Coefficient Variation | Brooks Ratio | Crosby Ratio | p Value |
|---|---|---|---|---|---|
| A1 | 0.66 ± 0.175 | 6.6% | - | - | 0.193 |
| N1 | 0.80 ± 0.099 | 14.1% | - | - | |
| A2 | 0.99 ± 0.012 | 6.0% | 1.500 | - | 0.065 |
| N2 | 1.23 ± 0.066 | 9.4% | 1.522 | - | |
| A3 | 1.40 ± 0.029 | 5.4% | 1.406 | −6.3% | 0.001 ** |
| N3 | 1.96 ± 0.105 | 5.8% | 1.590 | 4.3% | |
| A4 | 1.94 ± 0.081 | 9.0% | 1.389 | 1.2% | 0.000 *** |
| N4 | 3.36 ± 0.059 | 5.8% | 1.715 | 7.9% | |
| A5 | 3.08 ± 0.065 | 6.1% | 1.588 | 14.4% | 0.000 *** |
| N5 | 4.26 ± 0.034 | 3.5% | 1.268 | −26.1% | |
| A6 | 4.90 ± 0.265 | 8.8% | 1.591 | 0.1% | 0.605 |
| N6 | 5.17 ± 0.024 | 2.6% | 1.213 | −4.3% | |
| A7 | 6.17 ± 4.941 | 11.2% | 1.259 | −38.4% | 0.005 * |
| N7 | 6.15 ± 0.091 | 7.1% | 1.190 | −1.9% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Q.; Cui, Y.; Chu, X.; Li, G.; Yang, M.; Wang, R.; Liang, G.; Wu, S.; Tigabu, M.; Zhang, F.; et al. Gut Bacterial Communities of Lymantria xylina and Their Associations with Host Development and Diet. Microorganisms 2021, 9, 1860. https://doi.org/10.3390/microorganisms9091860
Ma Q, Cui Y, Chu X, Li G, Yang M, Wang R, Liang G, Wu S, Tigabu M, Zhang F, et al. Gut Bacterial Communities of Lymantria xylina and Their Associations with Host Development and Diet. Microorganisms. 2021; 9(9):1860. https://doi.org/10.3390/microorganisms9091860
Chicago/Turabian StyleMa, Qiuyu, Yonghong Cui, Xu Chu, Guoqiang Li, Meijiao Yang, Rong Wang, Guanghong Liang, Songqing Wu, Mulualem Tigabu, Feiping Zhang, and et al. 2021. "Gut Bacterial Communities of Lymantria xylina and Their Associations with Host Development and Diet" Microorganisms 9, no. 9: 1860. https://doi.org/10.3390/microorganisms9091860
APA StyleMa, Q., Cui, Y., Chu, X., Li, G., Yang, M., Wang, R., Liang, G., Wu, S., Tigabu, M., Zhang, F., & Hu, X. (2021). Gut Bacterial Communities of Lymantria xylina and Their Associations with Host Development and Diet. Microorganisms, 9(9), 1860. https://doi.org/10.3390/microorganisms9091860






