Lactic Acid Bacteria Exert a Hepatoprotective Effect against Ethanol-Induced Liver Injury in HepG2 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Bacterial Strains and Preparation of Cell-Free Extracts (CFEs)
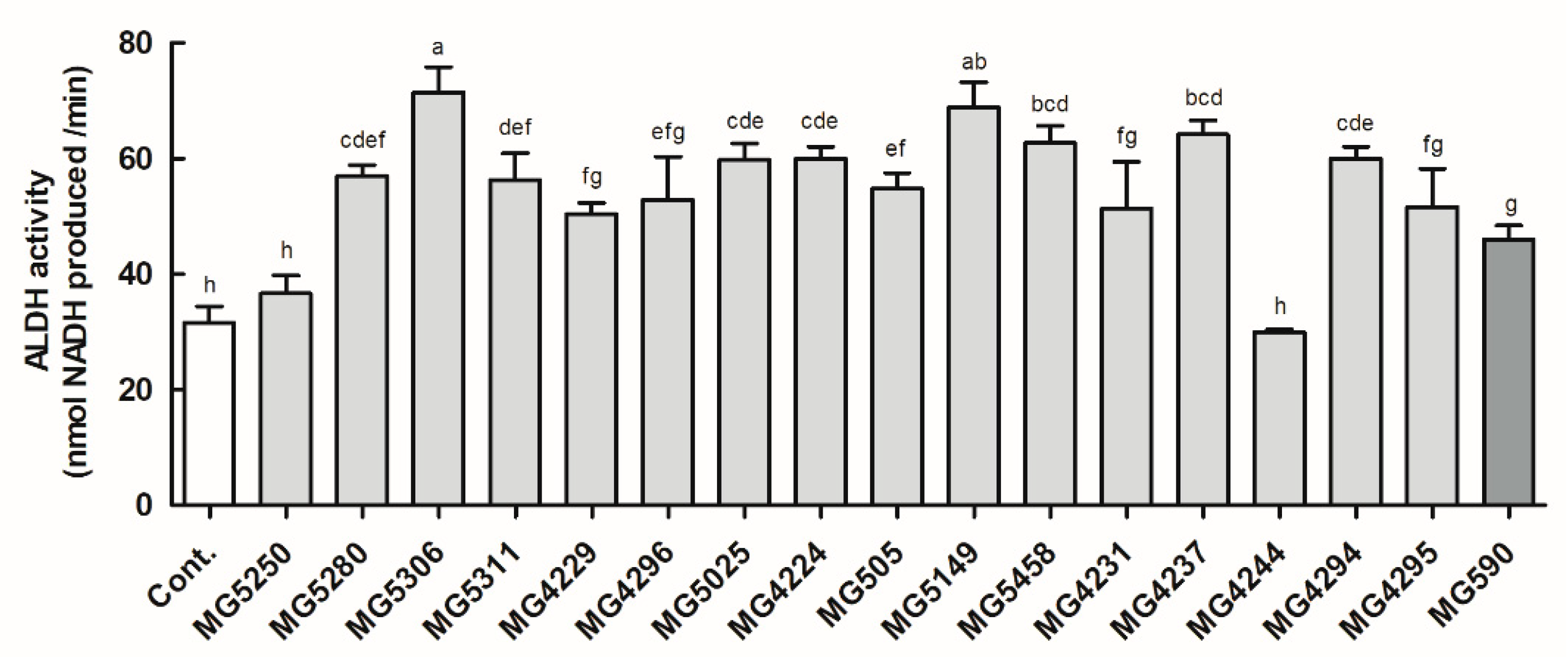
2.2. ALDH Activity
2.3. Cell Culture
2.4. Cell Viability
2.5. Determination of Lipid Peroxidation and Glutathione (GSH) Content
2.6. Measurement of Liver Injury
2.7. mRNA Extraction and Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
2.8. Probiotic Properties
2.8.1. Antibiotic Susceptibility and Hemolysis Assay
2.8.2. Gastrointestinal Tract (GIT) Stability and Adhesion
2.9. Statistical Analysis
3. Results
3.1. ALDH Activity of the LAB Strains
3.2. Protective Effect of LAB Strains on Ethanol-Induced HepG2 Cells
3.3. LAB Strains Regulate Oxidative Stress in Ethanol-Induced HepG2 Cells
3.4. LAB Strains Protect against Liver Injury Induced by Ethanol in HepG2 Cells
3.5. LAB Strains Modulate Ethanol Metabolism by Enhancing mRNA Expression of Antioxidant Enzyme and Lipid Metabolism in Ethanol-Induced HepG2 Cells
3.6. Probiotic Properties of the LAB Strains
3.6.1. Safety of LAB as Probiotics
3.6.2. GIT Stability and Adhesion on HT-29 Colorectal Cells of L. brevis MG5311 and L. fermentum MG4237
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, L.; Yang, C.; Thomes, P.G.; Kharbanda, K.K.; Casey, C.A.; McNiven, M.A.; Donohue, T.M., Jr. Lipophagy and alcohol-induced fatty liver. Front. Pharmacol. 2019, 10, 495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Z.; Liu, Y.; Hu, S.; You, Y.; Wen, J.; Li, W.; Wang, Y. Probiotics for alleviating alcoholic liver injury. Gastroenterol. Res. Pract. 2019, 2019, 9097276. [Google Scholar] [CrossRef] [Green Version]
- Zheng, T.-X.; Pu, S.-L.; Tan, P.; Du, Y.-C.; Qian, B.-L.; Chen, H.; Fu, W.-G.; Huang, M.-Z. Liver metabolomics reveals the effect of Lactobacillus reuteri on alcoholic liver disease. Front. Physiol. 2020, 11, 1494. [Google Scholar] [CrossRef]
- Kim, W. Diagnostic and therapeutic strategies for severe alcoholic hepatitis. Korean J. Gastroenterol. 2015, 65, 4–11. [Google Scholar] [CrossRef] [Green Version]
- Orywal, K.; Szmitkowski, M. Alcohol dehydrogenase and aldehyde dehydrogenase in malignant neoplasms. Clin. Exp. Pediatr. 2017, 17, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.E.; Koh, H.; Joo, D.J.; Nedumaran, B.; Jeon, H.J.; Park, C.S.; Harris, R.A.; Kim, Y.D. Induction of SIRT1 by melatonin improves alcohol-mediated oxidative liver injury by disrupting the CRBN-YY1-CYP2E1 signaling pathway. J. Pineal Res. 2020, 68, e12638. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.C.; Cross, C.E. Free radicals, antioxidants, and human disease: Where are we now? J. Lab. Clin. Med. 1992, 119, 598–620. [Google Scholar]
- Ducatelle, R.; Eeckhaut, V.; Haesebrouck, F.; Van Immerseel, F. A review on prebiotics and probiotics for the control of dysbiosis: Present status and future perspectives. Animal 2015, 9, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Soccol, C.R.; Vandenberghe, L.P.d.S.; Spier, M.R.; Medeiros, A.B.P.; Yamaguishi, C.T.; Lindner, J.D.D.; Pandey, A.; Thomaz-Soccol, V. The potential of probiotics: A review. Food Sci. Biotechnol. 2010, 48, 413–434. [Google Scholar]
- Li, F.; Duan, K.; Wang, C.; McClain, C.; Feng, W. Probiotics and alcoholic liver disease: Treatment and potential mechanisms. Gastroenterol. Res. Pract. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, Y.-T.; Kim, Y.-H.; Bae, J.-S.; Lim, K.-S.; Huh, C.-S.; Yang, W.-Y.; Kim, H.-S.; Baek, Y.-J. Effect of Lactobacillus brevis HY7401 intake on the serum ethanol concentration in rats. Korean J. Food Sci. Technol. 2004, 36, 604–608. [Google Scholar]
- Kim, J.-H.; Kim, H.-J.; Son, J.-H.; Chun, H.-N.; Yang, J.-O.; Choi, S.-J.; Paek, N.-S.; Choi, G.-H.; Kim, S.-K. Effect of Lactobacillus fermentum MG590 on alcohol metabolism and liver function in rats. J. Microbiol. Biotechnol. 2003, 13, 919–925. [Google Scholar]
- Segawa, S.; Wakita, Y.; Hirata, H.; Watari, J. Oral administration of heat-killed Lactobacillus brevis SBC8803 ameliorates alcoholic liver disease in ethanol-containing diet-fed C57BL/6N mice. Int. J. Food Microbiol. 2008, 128, 371–377. [Google Scholar] [CrossRef]
- Barone, R.; Rappa, F.; Macaluso, F.; Bavisotto, C.C.; Sangiorgi, C.; Di Paola, G.; Tomasello, G.; Di Felice, V.; Marcianò, V.; Farina, F. Alcoholic liver disease: A mouse model reveals protection by Lactobacillus Fermentum. Clin. Transl. Gastroenterol. 2016, 7, e138. [Google Scholar] [CrossRef]
- Park, J.-E.; Oh, S.-H.; Cha, Y.-S. Lactobacillus plantarum LG42 isolated from gajami sik-hae inhibits adipogenesis in 3T3-L1 adipocyte. Biomed Res. Int. 2013, 2013, 460927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blandino, A.; Caro, I.; Cantero, D. Comparative study of alcohol dehydrogenase activity in flor yeast extracts. Biotechnol. Lett. 1997, 19, 651–654. [Google Scholar] [CrossRef]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of cell viability by the MTT assay. Cold Spring Harb. Protoc. 2018, 2018, 95505. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Additives and Products; Substances Used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; Bastos, M.d.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [PubMed]
- Buxton, R. Blood agar plates and hemolysis protocols. Am. Soc. Microbiol. 2005, 30, 1–9. [Google Scholar]
- Jeong, Y.; Kim, H.; Lee, J.Y.; Won, G.; Choi, S.-I.; Kim, G.-H.; Kang, C.-H. The antioxidant, anti-diabetic, and anti-adipogenesis potential and probiotic properties of lactic acid bacteria isolated from human and fermented foods. Fermentation 2021, 7, 123. [Google Scholar] [CrossRef]
- Kim, S.; Choi, S.-I.; Jang, M.; Jeong, Y.; Kang, C.-H.; Kim, G.-H. Anti-adipogenic effect of Lactobacillus fermentum MG4231 and MG4244 through AMPK pathway in 3T3-L1 preadipocytes. Food Sci. Biotechnol. 2020, 29, 1541–1551. [Google Scholar] [CrossRef]
- Jang, J.Y.; Kim, D.J. Epidemiology of alcoholic liver disease in Korea. Clin. Mol. Hepatol. 2018, 24, 93–99. [Google Scholar] [CrossRef]
- Chávez-Tapia, N.C.; González-Rodríguez, L.; Jeong, M.; López-Ramírez, Y.; Barbero-Becerra, V.; Juárez-Hernández, E.; Romero-Flores, J.L.; Arrese, M.; Méndez-Sánchez, N.; Uribe, M. Current evidence on the use of probiotics in liver diseases. J. Funct. Foods 2015, 17, 137–151. [Google Scholar] [CrossRef]
- Cederbaum, A.I.; Lu, Y.; Wu, D. Role of oxidative stress in alcohol-induced liver injury. Arch. Toxicol. 2009, 83, 519–548. [Google Scholar] [CrossRef] [PubMed]
- Nagappan, A.; Jung, D.Y.; Kim, J.-H.; Lee, H.; Jung, M.H. Gomisin N alleviates ethanol-induced liver injury through ameliorating lipid metabolism and oxidative stress. Int. J. Mol. Sci. 2018, 19, 2601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Cederbaum, A.I. CYP2E1 and oxidative liver injury by alcohol. Free Radic. Biol. Med. 2008, 44, 723–738. [Google Scholar] [CrossRef] [Green Version]
- Patel, F.; Parwani, K.; Patel, D.; Mandal, P. Metformin and probiotics interplay in amelioration of ethanol-induced oxidative stress and inflammatory response in an in vitro and in vivo model of hepatic injury. Mediat. Inflamm. 2021, 2021, 6636152. [Google Scholar] [CrossRef]
- Kutlu, S.; Colakoglu, N.; Halifeoglu, I.; Sandal, S.; Seyran, A.D.; Aydin, M.; Yılmaz, B. Comparative evaluation of hepatotoxic and nephrotoxic effects of aroclors 1221 and 1254 in female rats. Cell Biochem. Funct. 2007, 25, 167–172. [Google Scholar] [CrossRef]
- Osna, N.A.; Donohue, T.M., Jr.; Kharbanda, K.K. Alcoholic liver disease: Pathogenesis and current management. Alcohol Res. Curr. Rev. 2017, 38, 147–161. [Google Scholar]
- Ali, A.T.; Hochfeld, W.E.; Myburgh, R.; Pepper, M.S. Adipocyte and adipogenesis. Eur. J. Cell Biol. 2013, 92, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Ghazali, R. Mechanisms into the development of fatty liver disease: Role of free fatty acids and alcohol. Ph.D. Thesis, University of Westminster, London, UK, 2017. [Google Scholar]
- Chu, J.; Joung, H.; Kim, B.-K.; Choi, I.-S.; Park, T.-S. Inhibitory effects of Lactobacillus plantarum Q180 on lipid accumulation in HepG2 cells. Korean J. Food Nutr. 2019, 32, 738–744. [Google Scholar]
- Hong, S.-M.; Chung, E.-C.; Kim, C.-H. Anti-obesity effect of fermented whey beverage using lactic acid bacteria in diet-induced obese rats. Korean J. Food Sci. Anim. 2015, 35, 653–659. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.; Lee, Y.; Kang, H.J.; Ju, J.; Ji, Y.; Park, H.; Park, H.; Lee, H.; Holzapfel, W.H. Two putative probiotic strains improve diet-induced hypercholesterolemia through modulating intestinal cholesterol uptake and hepatic cholesterol efflux. J. Appl. Microbiol. 2021, 16, 1–9. [Google Scholar]
- Bang, J.-H.; Shin, H.-J.; Choi, H.-J.; Kim, D.-W.; Ahn, C.-S.; Jeong, Y.-K.; Joo, W.-H. Probiotic potential of Lactobacillus isolates. J. Life Sci. 2012, 22, 251–258. [Google Scholar] [CrossRef]
- Delgado, S.; O’sullivan, E.; Fitzgerald, G.; Mayo, B. Subtractive screening for probiotic properties of Lactobacillus species from the human gastrointestinal tract in the search for new probiotics. J. Food Sci. Technol. 2007, 72, M310–M315. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kanwar, S.S. Adherence potential of indigenous lactic acid bacterial isolates obtained from fermented foods of Western Himalayas to intestinal epithelial Caco-2 and HT-29 cell lines. J. Food Sci. Technol. 2017, 54, 3504–3511. [Google Scholar] [CrossRef]
- Dudík, B.; Kiňová Sepová, H.; Bilka, F.; Pašková, Ľ.; Bilková, A. Mucin pre-cultivated Lactobacillus reuteri E shows enhanced adhesion and increases mucin expression in HT-29 cells. Antonie Leeuwenhoek 2020, 113, 1191–1200. [Google Scholar] [CrossRef]





| Lactic Acid Bacteria (μg/mL) | Cell Viability (%) | |||
|---|---|---|---|---|
| Control | + 3% Ethanol | |||
| Untreated | 100.00 ± 5.68 | 55.40 ± 2.07 | ||
| Levilactobacillus brevis | MG5280 | 50 | 99.13 ± 7.37 | 79.99 ± 5.68 *** |
| 100 | 97.26 ± 5.94 | 73.27 ± 4.13 *** | ||
| MG5306 | 50 | 96.91 ± 4.38 | 76.36 ± 3.47 *** | |
| 100 | 94.43 ± 2.04 | 75.04 ± 4.24 *** | ||
| MG5311 | 50 | 98.17 ± 5.43 | 83.47 ± 1.53 *** | |
| 100 | 99.63 ± 3.52 | 86.66 ± 2.17 *** | ||
| Limosilactobacillus reuteri | MG4224 | 50 | 96.26 ± 5.03 | 85.02 ± 1.84 *** |
| 100 | 99.49 ± 3.76 | 87.85 ± 0.89 *** | ||
| MG505 | 50 | 97.44 ± 5.55 | 85.02 ± 0.70 *** | |
| 100 | 96.58 ± 0.53 | 87.85 ± 1.91 *** | ||
| MG5149 | 50 | 92.51 ± 6.83 | 83.47 ± 8.61 *** | |
| 100 | 94.72 ± 11.25 | 86.66 ± 7.77 *** | ||
| MG5458 | 50 | 91.26 ± 1.18 | 82.60 ± 1.31 *** | |
| 100 | 91.14 ± 6.32 | 89.59 ± 0.96 *** | ||
| Limosilactobacillus fermentum | MG4237 | 50 | 95.37 ± 2.16 | 80.33 ± 2.04 *** |
| 100 | 91.71 ± 0.46 | 91.47 ± 1.23 *** | ||
| MG4294 | 50 | 93.89 ± 2.90 | 80.53 ± 1.92 *** | |
| 100 | 93.89 ± 3.59 | 80.53 ± 0.84 *** | ||
| MG590 | 50 | 100.89 ± 2.31 | 68.82 ± 1.46 ** | |
| 100 | 100.63 ± 2.29 | 75.00 ± 2.73 *** | ||
| Antimicrobiotics 1 | L. brevis | L. reuteri | L. fermentum | ||
|---|---|---|---|---|---|
| MG5280 | MG5311 | MG5458 | MG4237 | MG4294 | |
| Ampicillin | S (0.75) | S (0.5) | S (0.75) | S (0.094) | S (0.094) |
| Gentamicin | S (0.094) | S (0.047) | S (2) | S (0.19) | S (0.19) |
| Kanamycin | S (3) | S (3) | S (6) | S (4) | S (4) |
| Streptomycin | S (4) | S (6) | S (24) | S (3) | S (6) |
| Tetracycline | S (6) | R (>256) | S (2) | S (3) | S (1.5) |
| Chloramphenicol | S (2) | S (4) | S (3) | S (3) | S (3) |
| Erythromycin | S (0.047) | S (0.047) | S (0.25) | R (3) | S (0.25) |
| Clindamycin | S (1.5) | S (2) | S (0.016) | S (0.023) | S (0.016) |
| Experiment | L. brevis | L. reuteri | L. fermentum | |||
|---|---|---|---|---|---|---|
| MG5280 | MG5311 | MG5458 | MG4237 | MG4294 | ||
| Stimulated gastrointestinal fluid (Log CFU/mL) | Initial | 7.66 ± 0.02 | 7.50 ± 0.04 | 7.57 ± 0.04 | 7.75 ± 0.03 | 7.63 ± 0.01 |
| pH 3 | 7.65 ± 0.05 | 7.70 ± 0.02 | 7.58 ± 0.07 | 7.85 ± 0.03 | 7.71 ± 0.07 | |
| pH 4 | 7.64 ± 0.01 | 7.72 ± 0.06 | 7.58 ± 0.06 | 7.82 ± 0.08 | 7.72 ± 0.07 | |
| pH 7 | 7.65 ± 0.06 | 7.70 ± 0.04 | 7.51 ± 0.00 | 7.83 ± 0.01 | 7.56 ± 0.07 | |
| pH 8 | 7.63 ± 0.11 | 7.66 ± 0.10 | 7.44 ± 0.06 | 7.73 ± 0.01 | 7.55 ± 0.10 | |
| Adhesion ability | Initial | 8.52 ± 0.04 | 7.23 ± 0.04 | 7.23 ± 0.04 | 7.23 ± 0.04 | 7.23 ± 0.04 |
| (Log CFU/mL) | Adherent | 8.77 ± 0.02 | 6.96 ± 0.03 | 6.96 ± 0.03 | 6.96 ± 0.03 | 6.96 ± 0.03 |
| Cell adhesion (%) | 84.21 ± 0.26 | 84.77 ± 0.45 | 70.76 ± 0.865 | 79.29 ± 0.32 | 55.36 ± 1.00 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.Y.; Kim, H.; Jeong, Y.; Kang, C.-H. Lactic Acid Bacteria Exert a Hepatoprotective Effect against Ethanol-Induced Liver Injury in HepG2 Cells. Microorganisms 2021, 9, 1844. https://doi.org/10.3390/microorganisms9091844
Lee JY, Kim H, Jeong Y, Kang C-H. Lactic Acid Bacteria Exert a Hepatoprotective Effect against Ethanol-Induced Liver Injury in HepG2 Cells. Microorganisms. 2021; 9(9):1844. https://doi.org/10.3390/microorganisms9091844
Chicago/Turabian StyleLee, Ji Yeon, Hyemin Kim, Yulah Jeong, and Chang-Ho Kang. 2021. "Lactic Acid Bacteria Exert a Hepatoprotective Effect against Ethanol-Induced Liver Injury in HepG2 Cells" Microorganisms 9, no. 9: 1844. https://doi.org/10.3390/microorganisms9091844
APA StyleLee, J. Y., Kim, H., Jeong, Y., & Kang, C.-H. (2021). Lactic Acid Bacteria Exert a Hepatoprotective Effect against Ethanol-Induced Liver Injury in HepG2 Cells. Microorganisms, 9(9), 1844. https://doi.org/10.3390/microorganisms9091844






