Phytate and Microbial Suspension Amendments Increased Soybean Growth and Shifted Microbial Community Structure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design, Treatments and Sampling
2.2. Measurement of Total Phosphorus in Plant Shoot
2.3. DNA Extraction and Amplicon Sequencing
2.4. Bioinformatics Pipeline and Processing of Data
3. Results
3.1. Soybean Biomass, Nodulation and P Nutrition Response to Treatments
3.2. Bacterial and Fungal Community Structure in Different Biotopes
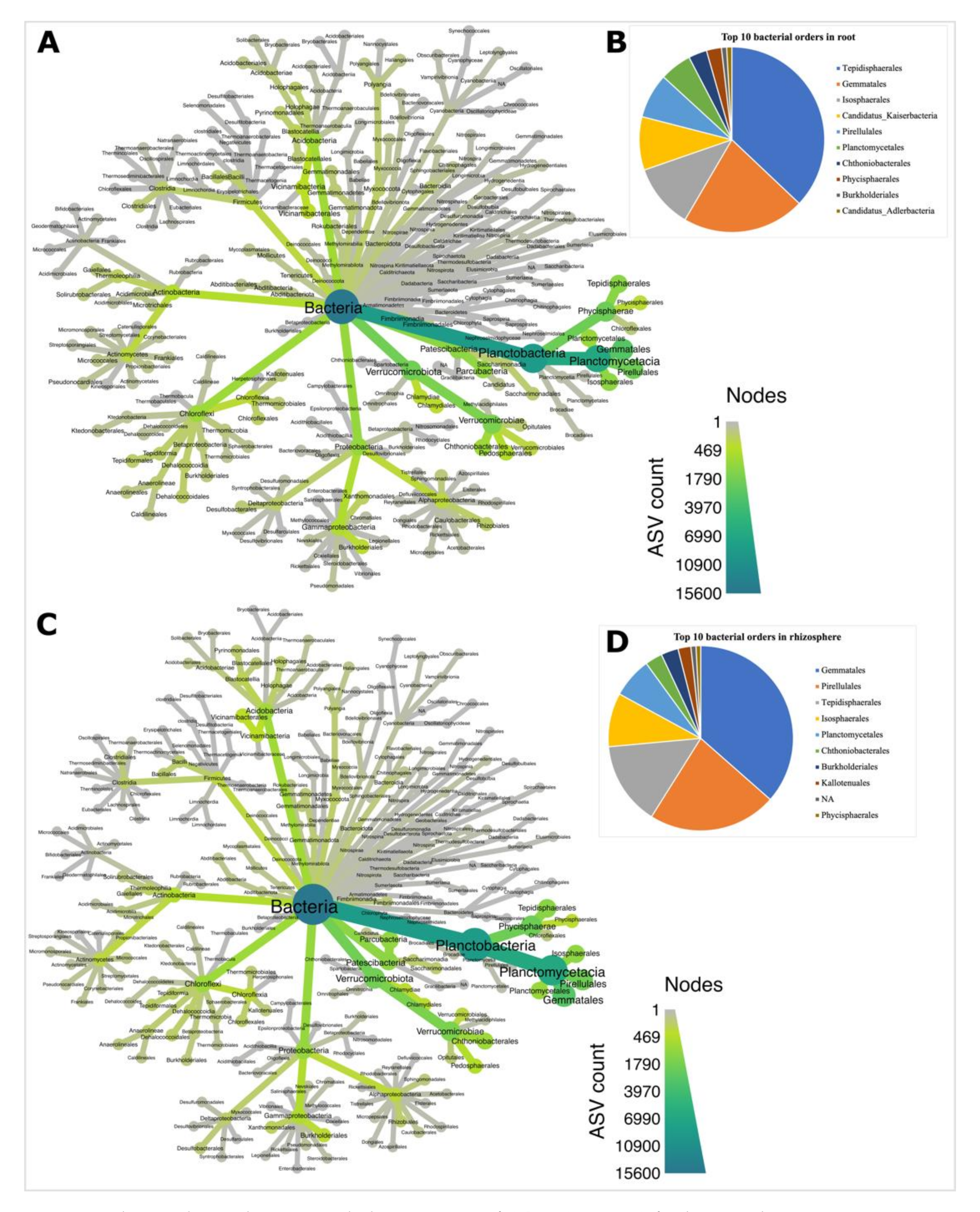
3.3. Planctobacteria and Ascomycota Dominated Soybean Microbiota
3.4. Determining Eco- and Core-Microbiota
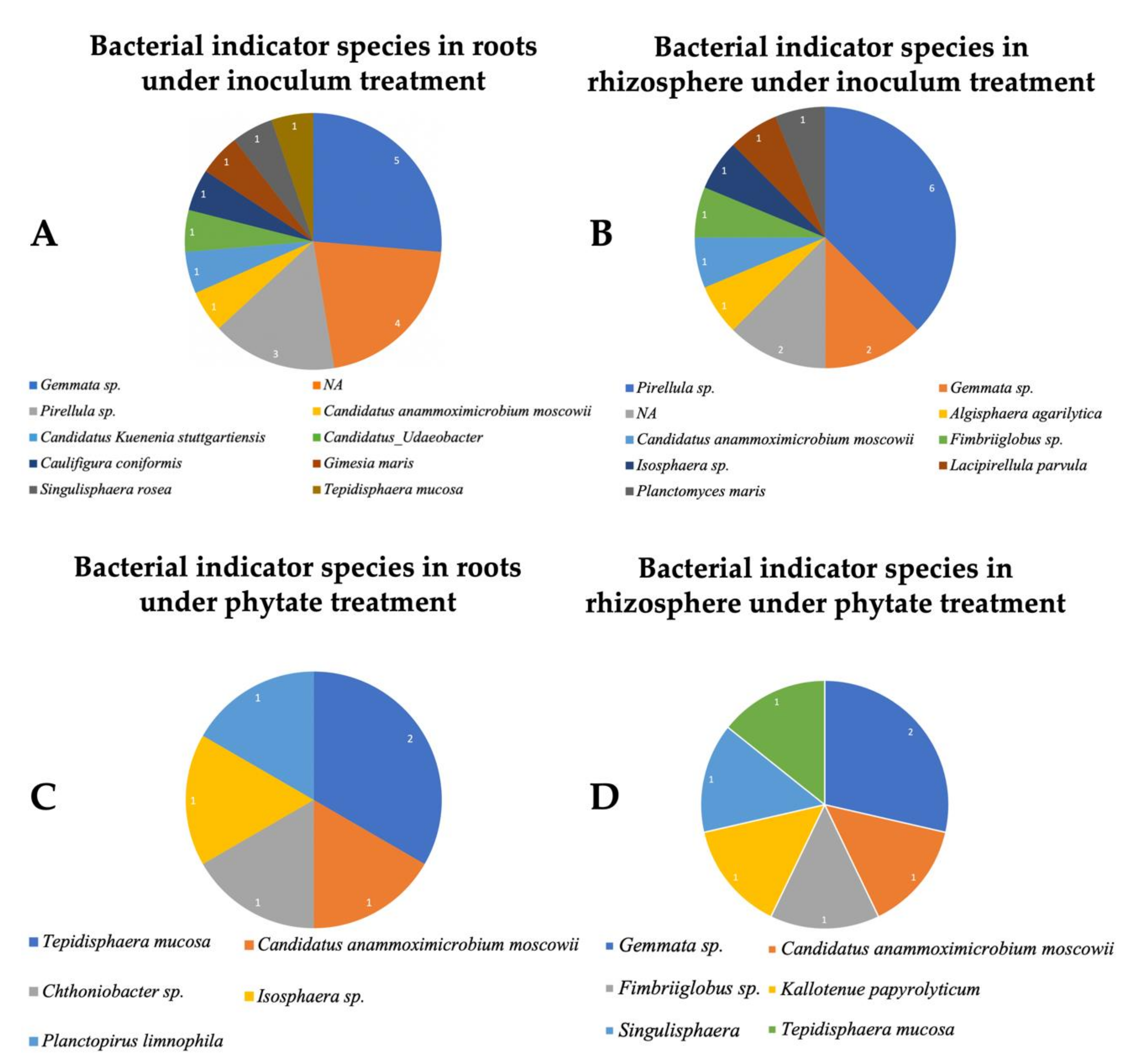
3.5. Bacteria Regulates the Connectivity of Soybean Microbiota
4. Discussion
4.1. Inoculation Tends to Influence Biomass, but Sample Types Sheltered Microbial Diversity
4.2. Planctobacteria and Ascomycota Predominate in the Soybean Microbiota
4.3. Microbial Amendment Influences the Community Composition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References and Notes
- Plaxton, W.C.; Tran, H.T. Metabolic adaptations of phosphate-starved plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef] [Green Version]
- FAO. World Fertilizer Trends and Outlook to 2020: Summary Report; FAO: Rome, Italy, 2017. [Google Scholar]
- Geissler, B.; Mew, M.C.; Steiner, G. Phosphate supply security for importing countries: Developments and the current situation. Sci. Total Environ. 2019, 677, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Obersteiner, M.; Peñuelas, J.; Ciais, P.; van der Velde, M.; Janssens, I.A. The phosphorus trilemma. Nat. Geosci. 2013, 6, 897–898. [Google Scholar] [CrossRef]
- Jarosch, K.A.; Kandeler, E.; Frossard, E.; Bünemann, E. Is the enzymatic hydrolysis of soil organic phosphorus compounds limited by enzyme or substrate availability? Soil Biol. Biochem. 2019, 139, 107628. [Google Scholar] [CrossRef] [Green Version]
- Castillo Villamizar, G.A.; Nacke, H.; Boehning, M.; Herz, K.; Daniel, R. Functional Metagenomics Reveals an Overlooked Diversity and Novel Features of Soil-Derived Bacterial Phosphatases and Phytases. mBio 2019, 10, e01966-18. [Google Scholar] [CrossRef] [Green Version]
- Tarafdar, J.C.; Chhonkar, P.K. Phosphatase production by microorganisms isolated from diverse types of soils. Zent. Bakteriol. Nat. 1979, 134, 119–124. [Google Scholar] [CrossRef]
- Giles, C.D.; Hsu, P.-C.; Richardson, A.E.; Hurst, M.R.H.; Hill, J. Plant assimilation of phosphorus from an insoluble organic form is improved by addition of an organic anion producing Pseudomonas sp. Soil Biol. Biochem. 2014, 68, 263–269. [Google Scholar] [CrossRef]
- Unno, Y.; Okubo, K.; Wasaki, J.; Shinano, T.; Osaki, M. Plant growth promotion abilities and microscale bacterial dynamics in the rhizosphere of Lupin analysed by phytate utilization ability. Environ. Microbiol. 2005, 7, 396–404. [Google Scholar] [CrossRef]
- Wang, X.X.; Hoffland, E.; Feng, G.; Kuyper, T.W. Phosphate Uptake from Phytate Due to Hyphae-Mediated Phytase Activity by Arbuscular Mycorrhizal Maize. Front. Plant Sci. 2017, 8, 684. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Feng, G.; Declerck, S. Signal beyond nutrient, fructose, exuded by an arbuscular mycorrhizal fungus triggers phytate mineralization by a phosphate solubilizing bacterium. ISME J. 2018, 12, 2339–2351. [Google Scholar] [CrossRef] [Green Version]
- Middleton, H.; Yergeau, E.; Monard, C.; Combier, J.P.; El Amrani, A. Rhizospheric Plant-Microbe Interactions: miRNAs as a Key Mediator. Trends Plant Sci. 2021, 26, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, Z.D.; Muller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, M.K.; Wong, C.K.; Chu, K.H.; Kwan, H. Community Structure, Dynamics and Interactions of Bacteria, Archaea and Fungi in Subtropical Coastal Wetland Sediments. Sci. Rep. 2018, 8, 14397. [Google Scholar] [CrossRef] [Green Version]
- Hartman, K.; van der Heijden, M.G.A.; Wittwer, R.A.; Banerjee, S.; Walser, J.C.; Schlaeppi, K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2018, 6, 14. [Google Scholar] [CrossRef]
- Bononi, L.; Chiaramonte, J.B.; Pansa, C.C.; Moitinho, M.A.; Melo, I. Phosphorus-solubilizing Trichoderma spp. from Amazon soils improve soybean plant growth. Sci. Rep. 2020, 10, 2858. [Google Scholar] [CrossRef] [Green Version]
- Helgason, T.; Feng, H.; Sherlock, D.J.; Young, J.P.W.; Fitter, A.H. Arbuscular mycorrhizal communities associated with maples (Acer spp.) in a common garden are influenced by season and host plant. Botany 2014, 92, 321–326. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 2nd ed.; Academic Press: San Diego, CA, USA, 1997. [Google Scholar]
- Brundrett, M.C.; Kendrick, B. The mycorrhizal status, root anatomy, and phenology of plants in a sugar maple forest. Can. J. Bot. 1988, 66, 1153–1173. [Google Scholar] [CrossRef]
- Lang, A.K.; Jevon, F.V.; Vietorisz, C.R.; Ayres, M.P.; Hatala Matthes, J. Fine roots and mycorrhizal fungi accelerate leaf litter decomposition in a northern hardwood forest regardless of dominant tree mycorrhizal associations. New Phytol. 2021, 230, 316–326. [Google Scholar] [CrossRef]
- St Clair, S.B.; Lynch, J.P. Base cation stimulation of mycorrhization and photosynthesis of sugar maple on acid soils are coupled by foliar nutrient dynamics. New Phytol. 2005, 165, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Goron, T.L.; Watts, S.; Shearer, C.; Raizada, M.N. Growth in Turface® clay permits root hair phenotyping along the entire crown root in cereal crops and demonstrates that root hair growth can extend well beyond the root hair zone. BMC Res. Notes 2015, 8, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaut, S.; Masse, J.; Norrie, J.P.; Blal, B.; Hijri, M. A commercial seaweed extract structured microbial communities associated with tomato and pepper roots and significantly increased crop yield. Microb. Biotechnol. 2019, 12, 1346–1358. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, B.; Hijri, M. Potential impacts of soil microbiota manipulation on secondary metabolites production in cannabis. J. Cannabis Res. 2021, 3, 25. [Google Scholar] [CrossRef]
- Davies, J.F.T.; Olalde-Portugal, V.; Alvarado, M.; Escamilla, H.; Ferrera-Cerrato, R.; Espinosa, J. Alleviating phosphorus stress of chile ancho pepper (Capsicum annuum L. ‘San Luis’) by arbuscular mycorrhizal inoculation. J. Hortic. Sci. Biotechnol. 2000, 75, 655–661. [Google Scholar] [CrossRef]
- Gujar, P.D.; Bhavsar, K.P.; Khire, J.M. Effect of phytase from Aspergillusniger on plant growth and mineral assimilation in wheat (Triticum aestivum Linn.) and its potential for use as a soil amendment. J. Sci. Food Agric. 2013, 93, 2242–2247. [Google Scholar] [PubMed]
- Singh, B.; Satyanarayana, T. Plant growth promotion by an extracellular HAP-phytase of a thermophilic mold Sporotrichum thermophile. Appl. Biochem. Biotechnol. 2010, 160, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Pequerul, A.; Perez, C.; Madero, P.; Val, J.; Monge, E. A Rapid Wet Digestion Method for Plant Analysis. In Optimization of Plant Nutrition; Springer: Dordrecht, The Netherlands, 1993; pp. 3–6. [Google Scholar]
- Holman, W. A new technique for the determination of phosphorus by the molybdenum blue method. Biochem. J. 1943, 37, 256–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farnelid, H.; Turk-Kubo, K.; Ploug, H.; Ossolinski, J.E.; Collins, J.R.; Van Mooy, B.A.S.; Zehr, J.P. Diverse diazotrophs are present on sinking particles in the North Pacific Subtropical Gyre. ISME J. 2019, 13, 170–182. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [Green Version]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Project: Vienna, Austria, 2020. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P. Vegan: Community Ecology Package. R Package Version 2019; pp. 2–5.
- Wickham, H.; Wickham, M.H. Package ‘plyr’. Available online: https://cran.r-project.org/web/packages/plyr/index.html (accessed on 14 August 2021).
- Peșteanu, A.; Bostan, M. Perfecţionarea unor elemente tehnologice la producerea materialului săditor pentru fondarea livezilor moderne de măr. Stiinta Agric. 2020, 52–59. [Google Scholar]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Stevens, M. Package—Vegan: Community ecology package. R package version 2.5-6. 2020. [Google Scholar]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Poisot, T.; Foster, Z.S.L.; Sharpton, T.J.; Grünwald, N.J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017, 13, e1005404. [Google Scholar]
- De Cáceres, M.; Jansen, F. Package ‘Indicspecies’. Studying the Statistical Relationship between Species and Groups of Sites. Available online: https://rdrr.io/cran/indicspecies/man/indicspecies-package.html (accessed on 14 August 2021).
- Hervé, M.; Hervé, M.M. Package ‘RVAideMemoire’. Available online: https://cran.r-project.org/web/packages/RVAideMemoire/index.html (accessed on 14 August 2021).
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Balaban, N.P.; Suleimanova, A.D.; Valeeva, L.R.; Chastukhina, I.B.; Rudakova, N.L.; Sharipova, M.R.; Shakirov, E.V. Microbial Phytases and Phytate: Exploring Opportunities for Sustainable Phosphorus Management in Agriculture. Am. J. Mol. Biol. 2017, 7, 11–29. [Google Scholar] [CrossRef] [Green Version]
- Patel, K.J.; Singh, A.K.; Nareshkumar, G.; Archana, G. Organic-acid-producing, phytate-mineralizing rhizobacteria and their effect on growth of pigeon pea (Cajanus cajan). Appl. Soil Ecol. 2010, 44, 252–261. [Google Scholar] [CrossRef]
- Srinivasan, R.; Alagawadi, A.R.; Yandigeri, M.S.; Meena, K.K.; Saxena, A.K. Characterization of phosphate-solubilizing microorganisms from salt-affected soils of India and their effect on growth of sorghum plants [Sorghum bicolor (L.) Moench]. Ann. Microbiol. 2011, 62, 93–105. [Google Scholar] [CrossRef]
- Chabot, R.; Antoun, H.; Cescas, M.P. Stimulation de la croissance du maïs et de la laitue romaine par des microorganismes dissolvant le phosphore inorganique. Can. J. Microbiol. 1993, 39, 941–947. [Google Scholar] [CrossRef]
- Ge, C. Effect of Biofertilizers and Plant Growth Promoting Bacteria on the Growth Characteristics of the Herb Asparagus Officinalis. Appl. Ecol. Environ. Res. 2016, 14, 547–558. [Google Scholar] [CrossRef]
- Wang, J.; Li, R.; Zhang, H.; Wei, G.; Li, Z. Beneficial bacteria activate nutrients and promote wheat growth under conditions of reduced fertilizer application. BMC Microbiol. 2020, 20, 38. [Google Scholar] [CrossRef] [Green Version]
- Korir, H.; Mungai, N.W.; Thuita, M.; Hamba, Y.; Masso, C. Co-inoculation Effect of Rhizobia and Plant Growth Promoting Rhizobacteria on Common Bean Growth in a Low Phosphorus Soil. Front. Plant Sci. 2017, 8, 141. [Google Scholar] [CrossRef] [Green Version]
- Zayed, M.S. Improvement of growth and nutritional quality of Moringa oleifera using different biofertilizers. Ann. Agric. Sci. 2012, 57, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Q.; Ma, Q.; Chen, Y.; Tian, B.; Xu, L.; Bai, Y.; Chen, W.; Li, X. Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J. 2020, 14, 1915–1928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, S.; Zhao, H.; Pan, S.; Zhang, N.; Qiao, Y. Elevated CO2 weakens the shift in bacterial community structure in response to 8-year soybean straw return in the same experiment. Int. J. Phytoremediat. 2020, 23, 505–510. [Google Scholar] [CrossRef]
- Zachow, C.; Muller, H.; Tilcher, R.; Berg, G. Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima-ancestor of all beet crops-and modern sugar beets. Front. Microbiol. 2014, 5, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barraza, A.; Vizuet-de-Rueda, J.C.; Alvarez-Venegas, R. Highly diverse root endophyte bacterial community is driven by growth substrate and is plant genotype-independent in common bean (Phaseolus vulgaris L.). PeerJ 2020, 8, e9423. [Google Scholar] [CrossRef] [PubMed]
- French, E.; Tran, T.; Iyer-Pascuzzi, A.S. Tomato Genotype Modulates Selection and Responses to Root Microbiota. Phytobiomes J. 2020, 4, 314–326. [Google Scholar] [CrossRef]
- Pierson, B.K.; Giovannoni, S.J.; Stahl, D.A.; Castenholz, R.W. Heliothrix oregonensis, gen. nov., sp. nov., a phototrophic filamentous gliding bacterium containing bacteriochlorophyll a. Arch. Microbiol. 1985, 142, 164–167. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Kulichevskaya, I.S.; Beletsky, A.V.; Ivanova, A.A.; Rijpstra, W.I.C.; Damsté, J.S.S.; Mardanov, A.V.; Ravin, N.V. Lacipirellula parvula gen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulales ord. nov., Gemmatales ord. nov. and Isosphaerales ord. nov. Syst. Appl. Microbiol. 2020, 43, 126050. [Google Scholar]
- Wang, J.; Jenkins, C.; Webb, R.I.; Fuerst, J.A. Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl. Environ. Microbiol. 2002, 68, 417–422. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Zhang, Q.; Ai, C.; He, P.; Zhou, W. Microbial utilization of rice root residue-derived carbon explored by DNA stable-isotope probing. Eur. J. Soil Sci. 2021, 72, 460–473. [Google Scholar] [CrossRef]
- Wironen, M.; Moore, T.R. Exotic earthworm invasion increases soil carbon and nitrogen in an old-growth forest in southern Quebec. Can. J. For. Res. 2006, 36, 845–854. [Google Scholar] [CrossRef]
- Guo, Q.; Yan, L.; Korpelainen, H.; Niinemets, Ü.; Li, C. Plant-plant interactions and N fertilization shape soil bacterial and fungal communities. Soil Biol. Biochem. 2019, 128, 127–138. [Google Scholar] [CrossRef]
- Leroy, C.; Jauneau, A.; Martinez, Y.; Cabin-Flaman, A.; Gibouin, D.; Orivel, J.; Sejalon-Delmas, N. Exploring fungus-plant N transfer in a tripartite ant-plant-fungus mutualism. Ann. Bot. 2017, 120, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Joner, E.J.; Johansen, A. Phosphatase activity of external hyphae of two arbuscular mycorrhizal fungi. Mycol. Res. 2000, 104, 81–86. [Google Scholar] [CrossRef]
- Feng, G.; Song, Y.; Li, X.; Christie, P. Contribution of arbuscular mycorrhizal fungi to utilization of organic sources of phosphorus by red clover in a calcareous soil. Appl. Soil Ecol. 2003, 22, 139–148. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Ivanova, A.A. Planctomycetes in boreal and subarctic wetlands: Diversity patterns and potential ecological functions. FEMS Microbiol. Ecol. 2019, 95, fiy227. [Google Scholar] [CrossRef] [Green Version]
- Kabore, O.D.; Godreuil, S.; Drancourt, M. Planctomycetes as Host-Associated Bacteria: A Perspective That Holds Promise for Their Future Isolations, by Mimicking Their Native Environmental Niches in Clinical Microbiology Laboratories. Front. Cell Infect. Microbiol. 2020, 10, 519301. [Google Scholar] [CrossRef]
- Hu, Y.; Duan, C.; Fu, D.; Wu, X.; Yan, K.; Fernando, E.; Karunarathna, S.C.; Promputtha, I.; Mortimer, P.E.; Xu, J. Structure of Bacterial Communities in Phosphorus-Enriched Rhizosphere Soils. Appl. Sci. 2020, 10, 6387. [Google Scholar] [CrossRef]
- Nassal, D.; Spohn, M.; Eltlbany, N.; Jacquiod, S.; Smalla, K.; Marhan, S.; Kandeler, E. Effects of phosphorus-mobilizing bacteria on tomato growth and soil microbial activity. Plant Soil 2017, 427, 17–37. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Koide, R.T.; Adams, T.S.; DeForest, J.L.; Cheng, L.; Eissenstat, D.M. Root morphology and mycorrhizal symbioses together shape nutrient foraging strategies of temperate trees. Proc. Natl. Acad. Sci. 2016, 113, 8741–8746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moutoglis, P.; Klironomos, J.; Widden, P.; Kendrick, B. Direct observation of spores of vesicular-arbuscular mycorrhizal fungi growing on sugar maple roots in the field, using sodium hexametaphosphate as a soil dispersant. Mycologia 1995, 87, 419–423. [Google Scholar] [CrossRef]
- Wallace, J.; Laforest-Lapointe, I.; Kembel, S.W. Variation in the leaf and root microbiome of sugar maple (Acer saccharum) at an elevational range limit. PeerJ 2018, 6, e5293. [Google Scholar] [CrossRef] [Green Version]
- Buckley, D.H.; Huangyutitham, V.; Nelson, T.A.; Rumberger, A.; Thies, J.E. Diversity of Planctomycetes in Soil in Relation to Soil History and Environmental Heterogeneity. Appl. Environ. Microbiol. 2006, 72, 4522–4531. [Google Scholar] [CrossRef] [Green Version]
- Pollet, T.; Humbert, J.F.; Tadonleke, R.D. Planctomycetes in lakes: Poor or strong competitors for phosphorus? Appl. Environ. Microbiol. 2014, 80, 819–828. [Google Scholar] [CrossRef] [Green Version]
- Arii, K.; Hamel, B.R.; Lechowicz, M.J. Environmental correlates of canopy composition at Mont St. Hilaire, Quebec, Canada1. J. Torrey Bot. Soc. 2005, 132, 90–102. [Google Scholar] [CrossRef]
- Marie-Victorin, F. Aperçus sur la flore de la Montagne de Sainte-Hilaire. Bull. Soc. Géol. Que. 1913, 7, 163–168. [Google Scholar]
- Maycock, P.F. Botanical Studies on Mont St. Hilaire, Rouvillecounty, Quebec: General Description of the Area and a Floristic Survey. Can. J. Bot. 1961, 39, 1293–1325. [Google Scholar] [CrossRef]




| Source of Variation | df | Shoot Dry Weight | Root Dry Weight | Number of Nodules | Total P | ||||
|---|---|---|---|---|---|---|---|---|---|
| F | Pr(>F) | F | Pr(>F) | F | Pr(>F) | F | Pr(>F) | ||
| Inoculum | 1 | 3.060 | 0.088 | 3.899 | 0.058 | 0.171 | 0.683 | 3.391 | 0.098 |
| Phytate | 1 | 8.018 | 0.007 | 0.037 | 0.847 | 8.525 | 0.017 | 2.901 | 0.122 |
| Inoculum: Phytate | 1 | 0.983 | 0.327 | 0.009 | 0.921 | 0.773 | 0.390 | 1.465 | 0.256 |
| (A) Bacteria. | ||||||
| Variable | Source | DF | SumOfSqs | R2 | F | Pr(>F) |
| Roots | Inoculation | 1 | 0.2347 | 0.04366 | 1.7477 | 0.001 *** |
| Phytate | 1 | 0.1686 | 0.03136 | 1.2555 | 0.024 * | |
| Inoculation: Phytate | 1 | 0.1384 | 0.02574 | 1.0306 | 0.354 | |
| Residual | 36 | 4.8353 | 0.89924 | |||
| Total | 39 | 5.3771 | 1.00000 | |||
| Rhizosphere | Inoculation | 1 | 0.2676 | 0.05702 | 2.2687 | 0.004 ** |
| Phytate | 1 | 0.0878 | 0.01871 | 0.7446 | 0.929 | |
| Inoculation: Phytate | 1 | 0.0908 | 0.01934 | 0.7696 | 0.849 | |
| Residual | 36 | 4.2474 | 0.90490 | |||
| Total | 39 | 4.6937 | 1.00000 | |||
| (B) Fungi. | ||||||
| Variable | Source | DF | SumOfSqs | R2 | F | Pr(>F) |
| Roots | Inoculation | 1 | 0.1592 | 0.01993 | 0.7662 | 0.751 |
| Phytate | 1 | 0.1368 | 0.01713 | 0.6551 | 0.878 | |
| Inoculation: Phytate | 1 | 0.1730 | 0.02166 | 0.8283 | 0.653 | |
| Residual | 36 | 7.5181 | 0.94128 | |||
| Total | 39 | 7.9870 | 1.00000 | |||
| Rhizosphere | Inoculation | 1 | 0.1502 | 0.04113 | 1.6240 | 0.007 ** |
| Phytate | 1 | 0.0789 | 0.02161 | 0.8532 | 0.791 | |
| Inoculation: Phytate | 1 | 0.0928 | 0.02542 | 1.0035 | 0.428 | |
| Residual | 36 | 3.3296 | 0.91184 | |||
| Total | 39 | 3.6515 | 1.00000 | |||
| Bacteria | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Microbial Inoculation | Phytate Addition | ||||||||||
| Roots | Rhizosphere | Roots | Rhizosphere | ||||||||
| Number of ASVs = 19 | Number of ASVs = 16 | Number of ASVs = 7 | Number of ASVs = 6 | ||||||||
| BASV5 | 0.0001 | *** | BASV590 | 0.0007 | *** | BASV1248 | 0.0073 | ** | BASV374 | 0.003 | ** |
| BASV8 | 0.0001 | *** | BASV861 | 0.0021 | ** | BASV107 | 0.0045 | ** | BASV761 | 0.0083 | ** |
| BASV114 | 0.0088 | ** | BASV569 | 0.0056 | ** | BASV1377 | 0.0418 | * | BASV1028 | 0.0203 | * |
| BASV713 | 0.004 | ** | BASV1092 | 0.0049 | ** | BASV738 | 0.0396 | * | BASV798 | 0.0232 | * |
| BASV36 | 0.0077 | ** | BASV571 | 0.0084 | ** | BASV185 | 0.0359 | * | BASV599 | 0.0252 | * |
| BASV1149 | 0.0163 | * | BASV585 | 0.0086 | ** | BASV1514 | 0.0465 | * | BASV619 | 0.0497 | * |
| BASV264 | 0.018 | * | BASV421 | 0.0183 | * | BASV27 | 0.0426 | * | |||
| BASV590 | 0.0206 | * | BASV537 | 0.0176 | * | ||||||
| BASV993 | 0.0205 | * | BASV20 | 0.0153 | * | ||||||
| BASV354 | 0.025 | * | BASV559 | 0.0265 | * | ||||||
| BASV669 | 0.0131 | * | BASV375 | 0.0333 | * | ||||||
| BASV605 | 0.0188 | * | BASV621 | 0.0357 | * | ||||||
| BASV250 | 0.0337 | * | BASV587 | 0.03 | * | ||||||
| BASV381 | 0.0285 | * | BASV882 | 0.0419 | * | ||||||
| BASV576 | 0.0168 | * | BASV21 | 0.0302 | * | ||||||
| BASV489 | 0.0231 | * | BASV1256 | 0.0359 | * | ||||||
| BASV1439 | 0.0495 | * | |||||||||
| BASV904 | 0.0499 | * | |||||||||
| BASV967 | 0.0247 | * | |||||||||
| Fungi | |||||||||||
| Microbial inoculation | Phytate addition | ||||||||||
| Roots | Rhizosphere | Roots | Rhizosphere | ||||||||
| Number of species = 0 | Number of species = 48 | Number of species = 1 | Number of species = 9 | ||||||||
| FASV410 | 0.002 | ** | FASV113 | 0.045 | * | FASV1196 | 0.023 | * | |||
| FASV131 | 0.004 | ** | FASV338 | 0.020 | * | ||||||
| FASV479 | 0.001 | *** | FASV413 | 0.021 | * | ||||||
| FASV508 | 0.003 | ** | FASV798 | 0.034 | * | ||||||
| FASV400 | 0.009 | ** | FASV151 | 0.018 | * | ||||||
| FASV839 | 0.014 | * | FASV1227 | 0.020 | * | ||||||
| FASV513 | 0.004 | ** | FASV749 | 0.050 | * | ||||||
| FASV373 | 0.015 | * | FASV209 | 0.042 | * | ||||||
| FASV493 | 0.019 | * | FASV366 | 0.029 | * | ||||||
| FASV503 | 0.007 | ** | |||||||||
| FASV706 | 0.018 | * | |||||||||
| FASV439 | 0.007 | ** | |||||||||
| FASV1286 | 0.015 | * | |||||||||
| FASV944 | 0.011 | * | |||||||||
| FASV502 | 0.023 | * | |||||||||
| FASV512 | 0.012 | * | |||||||||
| FASV551 | 0.027 | * | |||||||||
| FASV530 | 0.022 | * | |||||||||
| FASV193 | 0.003 | ** | |||||||||
| FASV471 | 0.021 | * | |||||||||
| FASV885 | 0.016 | * | |||||||||
| FASV248 | 0.034 | * | |||||||||
| FASV207 | 0.045 | * | |||||||||
| FASV606 | 0.046 | * | |||||||||
| FASV328 | 0.038 | * | |||||||||
| FASV1119 | 0.014 | * | |||||||||
| FASV370 | 0.027 | * | |||||||||
| FASV261 | 0.035 | * | |||||||||
| FASV628 | 0.042 | * | |||||||||
| FASV98 | 0.003 | ** | |||||||||
| FASV703 | 0.022 | * | |||||||||
| FASV790 | 0.006 | ** | |||||||||
| FASV1169 | 0.050 | * | |||||||||
| FASV662 | 0.040 | * | |||||||||
| FASV1078 | 0.042 | * | |||||||||
| FASV1855 | 0.040 | * | |||||||||
| FASV702 | 0.045 | * | |||||||||
| FASV945 | 0.037 | * | |||||||||
| FASV324 | 0.024 | * | |||||||||
| FASV552 | 0.024 | * | |||||||||
| FASV465 | 0.041 | * | |||||||||
| FASV183 | 0.008 | ** | |||||||||
| FASV1306 | 0.048 | * | |||||||||
| FASV299 | 0.040 | * | |||||||||
| FASV1097 | 0.017 | * | |||||||||
| FASV569 | 0.004 | ** | |||||||||
| FASV108 | 0.029 | * | |||||||||
| FASV87 | 0.046 | * | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, B.; Floc’h, J.-B.; Lahrach, Z.; Hijri, M. Phytate and Microbial Suspension Amendments Increased Soybean Growth and Shifted Microbial Community Structure. Microorganisms 2021, 9, 1803. https://doi.org/10.3390/microorganisms9091803
Ahmed B, Floc’h J-B, Lahrach Z, Hijri M. Phytate and Microbial Suspension Amendments Increased Soybean Growth and Shifted Microbial Community Structure. Microorganisms. 2021; 9(9):1803. https://doi.org/10.3390/microorganisms9091803
Chicago/Turabian StyleAhmed, Bulbul, Jean-Baptiste Floc’h, Zakaria Lahrach, and Mohamed Hijri. 2021. "Phytate and Microbial Suspension Amendments Increased Soybean Growth and Shifted Microbial Community Structure" Microorganisms 9, no. 9: 1803. https://doi.org/10.3390/microorganisms9091803
APA StyleAhmed, B., Floc’h, J.-B., Lahrach, Z., & Hijri, M. (2021). Phytate and Microbial Suspension Amendments Increased Soybean Growth and Shifted Microbial Community Structure. Microorganisms, 9(9), 1803. https://doi.org/10.3390/microorganisms9091803









