Enterovirus D: A Small but Versatile Species
Abstract
1. Introduction
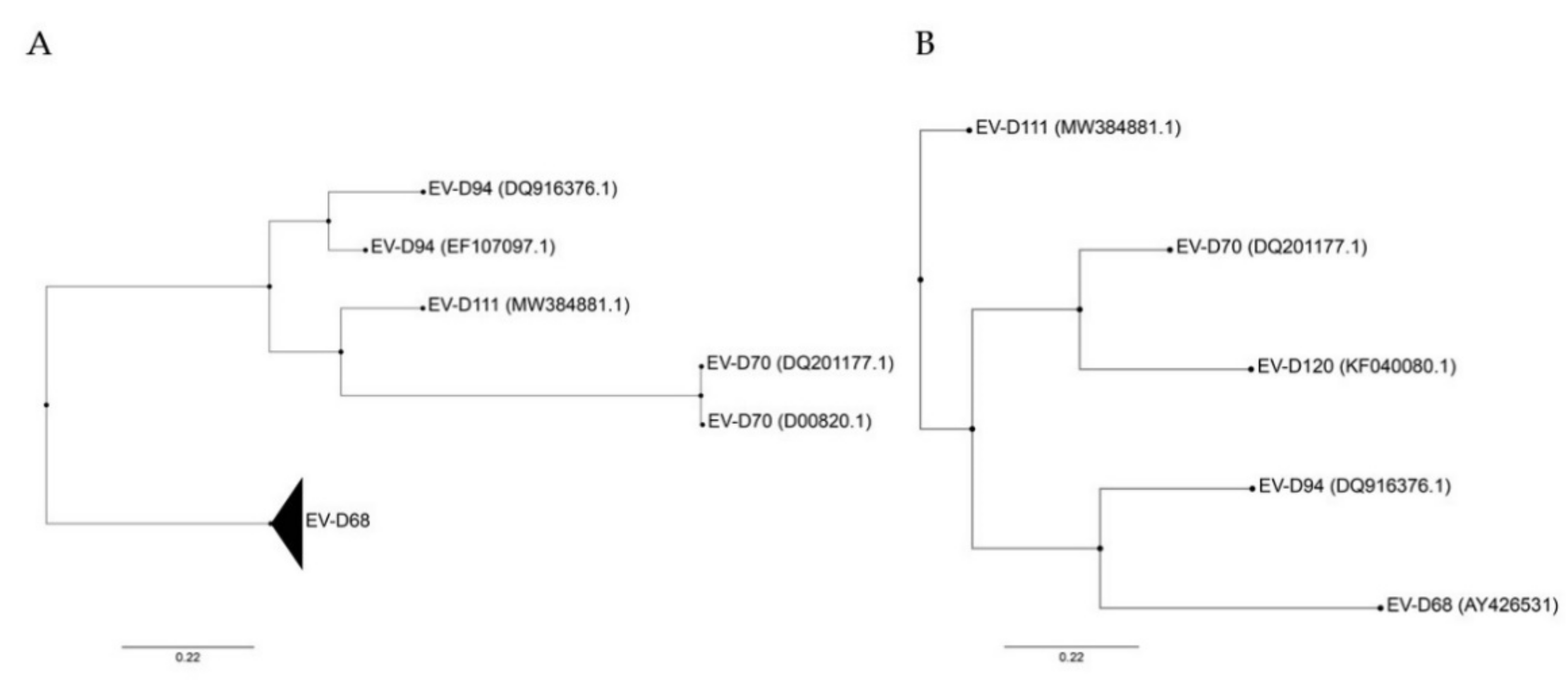
2. Enterovirus D Classification
3. Enterovirus D Genome Organization and Functional Elements
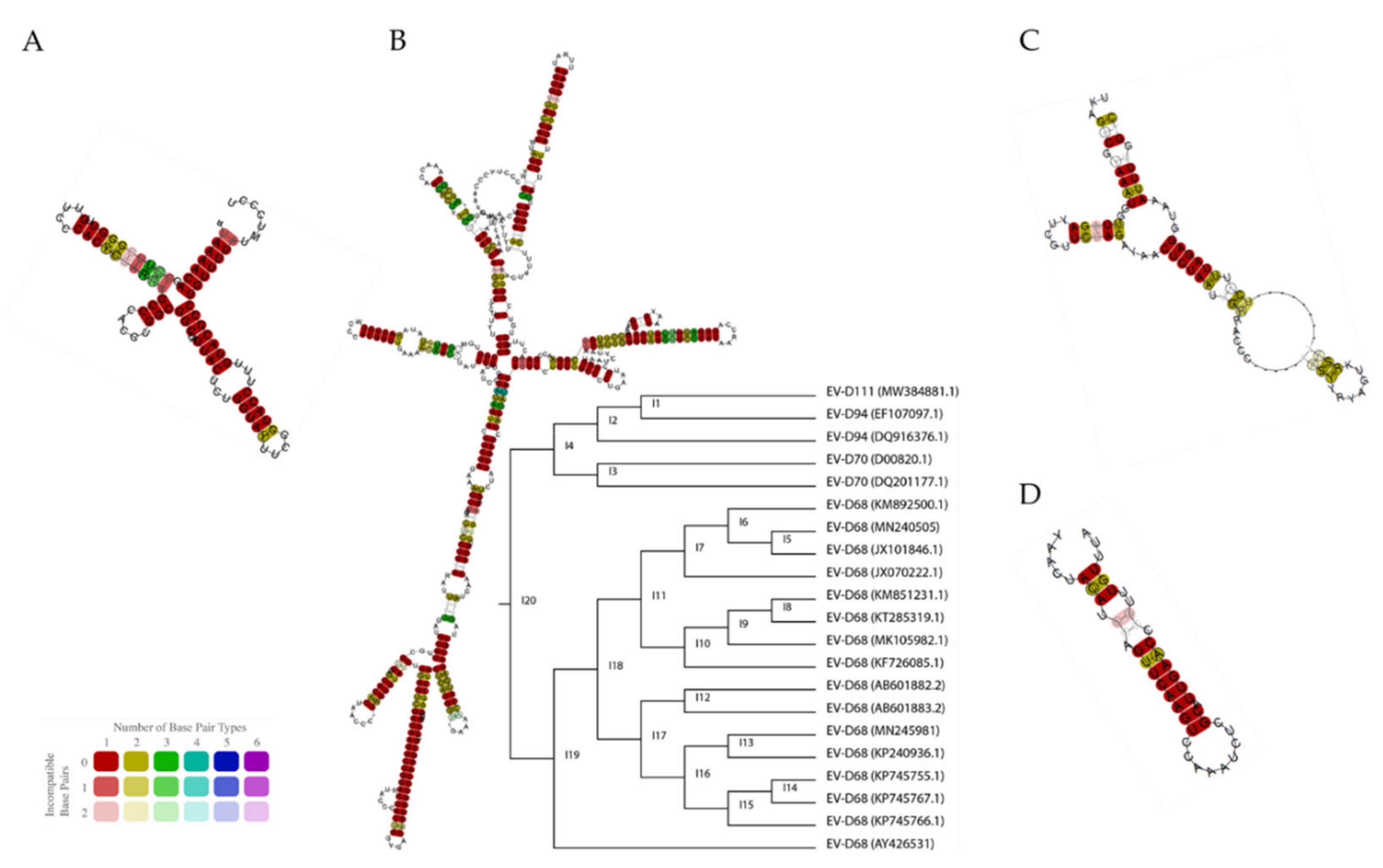
Cis-Acting RNA Structures
4. Structural and Non-Structural Proteins
4.1. Structural Proteins
4.2. Non-Structural Proteins
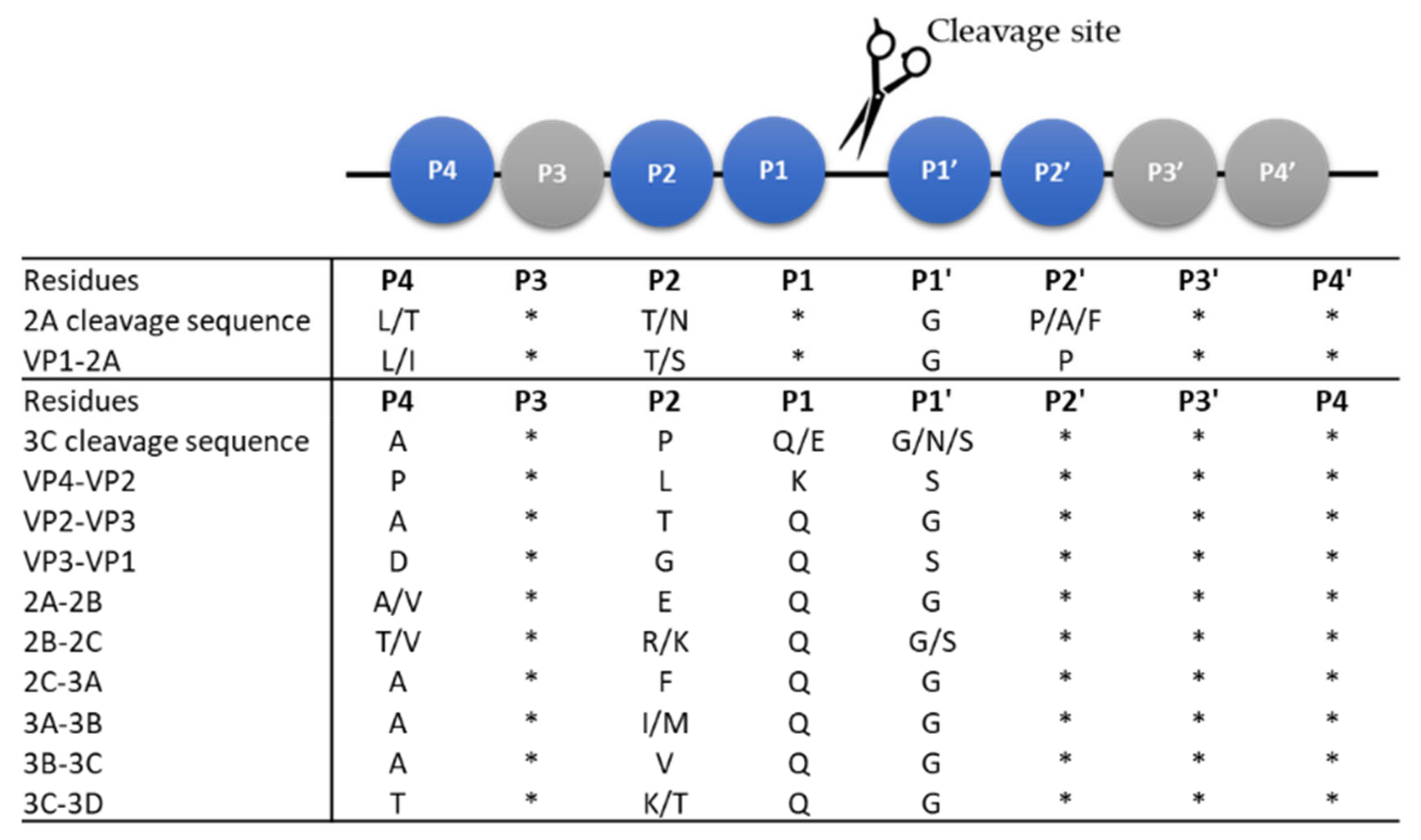
4.2.1. The Viral Proteases: 2A and 3C
4.2.2. The Non-Structural Proteins: 2B, 2C, and 2BC
4.2.3. The Non-Structural Proteins: 3A, 3B, and 3D and Their Precursors
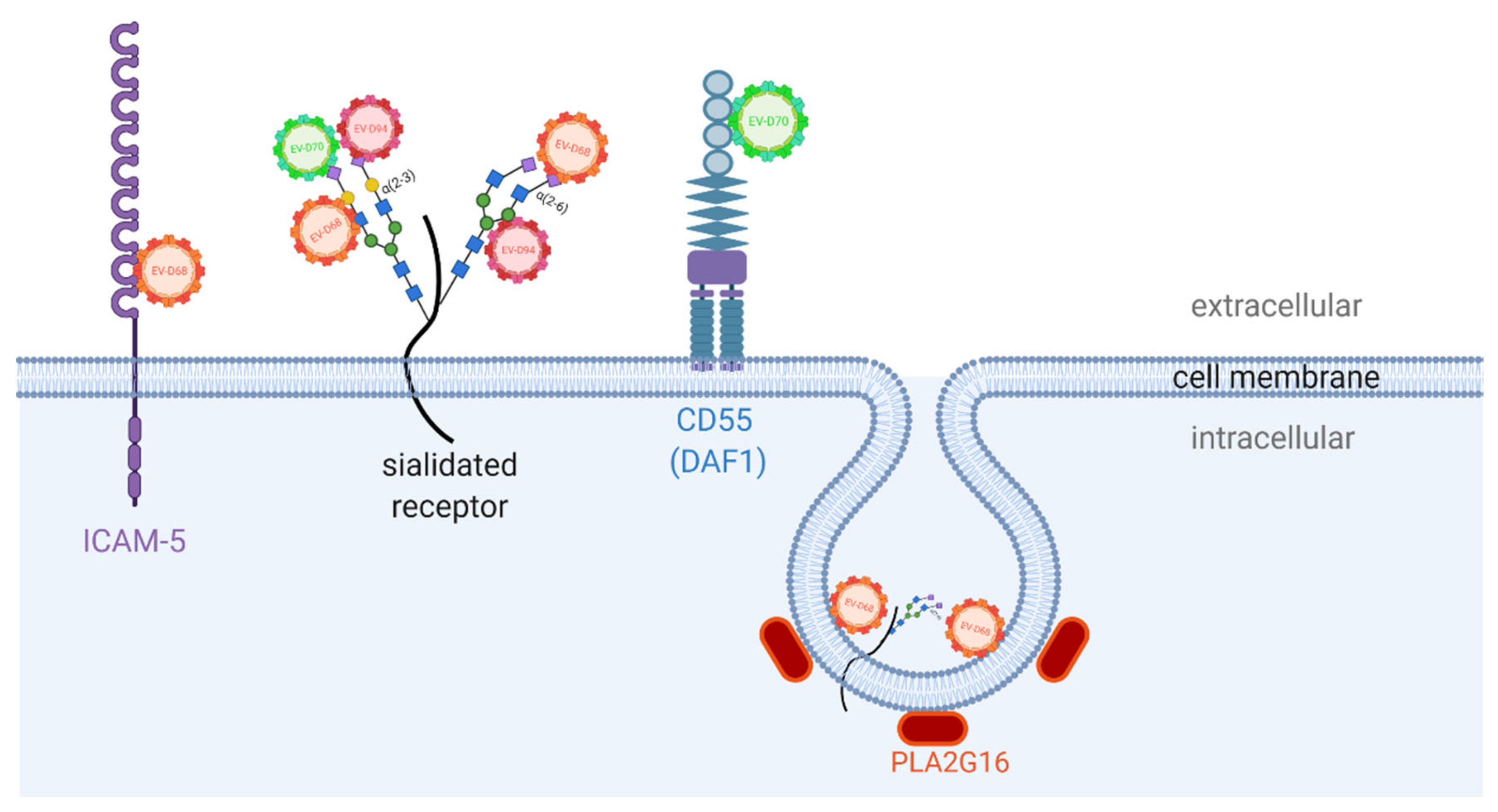
5. Enterovirus D Receptors
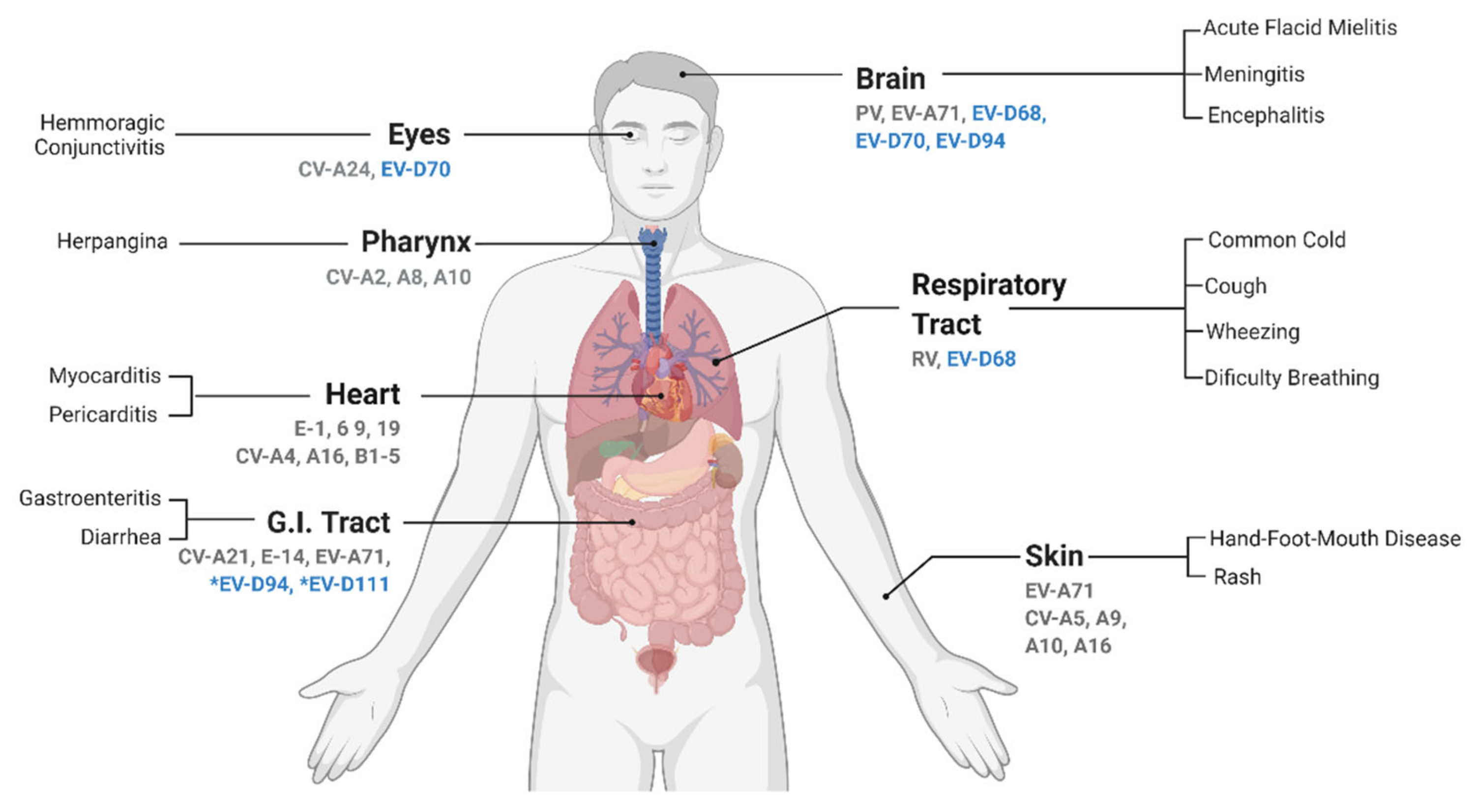
6. Enterovirus D Pathogenesis and Associated Symptoms
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Simmonds, P.; Gorbalenya, A.E.; Harvala, H.; Hovi, T.; Knowles, N.J.; Lindberg, A.M.; Oberste, M.S.; Palmenberg, A.C.; Reuter, G.; Skern, T.; et al. Recommendations for the nomenclature of enteroviruses and rhinoviruses. Arch. Virol. 2020, 165, 793–797. [Google Scholar] [CrossRef] [PubMed]
- ICTV. Picornaviridae, Genus Enterovirus. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/picornavirales/w/picornaviridae/681/genus-enterovirus (accessed on 15 July 2021).
- Tapparel, C.; Siegrist, F.; Petty, T.J.; Kaiser, L. Picornavirus and Enterovirus Diversity with Associated Human Diseases. Infect. Genet. Evol. 2013, 14, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Pirbright Institute. Picornaviridae.com. Available online: https://www.picornaviridae.com/hosts.htm (accessed on 15 July 2021).
- Khan, F. Enterovirus D68: Acute Respiratory Illness and the 2014 Outbreak. Emerg. Med. Clin. 2015, 33, e19–e32. [Google Scholar] [CrossRef]
- Mirkovic, R.R.; Kono, R.; Yin-Murphy, M.; Sohier, R.; Schmidt, N.J.; Melnick, J.L. Enterovirus type 70: The etiologic agent of pandemic acute haemorrhagic conjunctivitis. Bull. World Health Organ. 1973, 49, 341–346. [Google Scholar]
- Pham, N.T.K.; Thongprachum, A.; Baba, T.; Okitsu, S.; Trinh, Q.D.; Komine-Aizawa, S.; Shimizu, H.; Hayakawa, S.; Ushijima, H. A 3-Month-Old Child with Acute Gastroenteritis with Enterovirus D68 Detected from Stool Specimen. Natl. Lab. Med. 2017, 63, 1269–1272. [Google Scholar] [CrossRef] [PubMed]
- Royston, L.; Essaidi-Laziosi, M.; Pérez-Rodríguez, F.J.; Piuz, I.; Geiser, J.; Krause, K.-H.; Huang, S.; Constant, S.; Kaiser, L.; Garcin, D.; et al. Viral chimeras decrypt the role of enterovirus capsid proteins in viral tropism, acid sensitivity and optimal growth temperature. PLoS Pathog. 2018, 14, e1006962. [Google Scholar] [CrossRef] [PubMed]
- Blomqvist, S.; Savolainen, C.; Råman, L.; Roivainen, M.; Hovi, T. Human Rhinovirus 87 and Enterovirus 68 Represent a Unique Serotype with Rhinovirus and Enterovirus Features. J. Clin. Microbiol. 2002, 40, 4218–4223. [Google Scholar] [CrossRef]
- Smura, T.; Junttila, N.; Blomqvist, S.; Norder, H.; Kaijalainen, S.; Paananen, A.; Magnius, L.O.; Hovi, T.; Roivainen, M. Enterovirus 94, a proposed new serotype in human enterovirus species D. J. Gen. Virol. 2007, 88, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Sadeuh-Mba, S.A.; Joffret, M.-L.; Mazitchi, A.; Endegue-Zanga, M.-C.; Njouom, R.; Delpeyroux, F.; Gouandjika-Vasilache, I.; Bessaud, M. Genetic and phenotypic characterization of recently discovered enterovirus D type 111. PLoS Neglected Trop. Dis. 2019, 13, e0007797. [Google Scholar] [CrossRef] [PubMed]
- Harvala, H.; Van Nguyen, D.; McIntyre, C.; Ahuka-Mundeke, S.; Ngole, E.M.; Delaporte, E.; Peeters, M.; Simmonds, P. Co-circulation of enteroviruses between apes and humans. J. Gen. Virol. 2014, 95, 403–407. [Google Scholar] [CrossRef]
- Kyriakopoulou, Z.; Pliaka, V.; Amoutzias, G.; Markoulatos, P. Recombination among human non-polio enteroviruses: Implications for epidemiology and evolution. Virus Genes 2014, 50, 177–188. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Lulla, V.; Dinan, A.M.; Hosmillo, M.; Chaudhry, Y.; Sherry, L.; Irigoyen, N.; Nayak, K.M.; Stonehouse, N.J.; Zilbauer, M.; Goodfellow, I.; et al. An upstream protein-coding region in enteroviruses modulates virus infection in gut epithelial cells. Nat. Microbiol. 2018, 4, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, B.; Horn, C.; Tapprich, W.E. Structure of the 5′ Untranslated Region of Enteroviral Genomic RNA. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Pascal, S.M.; Garimella, R.; Warden, M.S.; Ponniah, K. Structural Biology of the Enterovirus Replication-Linked 5′-Cloverleaf RNA and Associated Virus Proteins. Microbiol. Mol. Biol. Rev. 2020, 84. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, M.; Stachowiak, A.; Swiatkowska, A.; Ciesiołka, J. Structure and function of RNA elements present in enteroviral genomes. Acta Biochim. Pol. 2017, 63, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Barton, D.J.; O’Donnell, B.J.; Flanegan, J.B. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001, 20, 1439–1448. [Google Scholar] [CrossRef]
- Furuse, Y.; Chaimongkol, N.; Okamoto, M.; Oshitani, H. Evolutionary and Functional Diversity of the 5’ Untranslated Region of Enterovirus D68: Increased Activity of the Internal Ribosome Entry Site of Viral Strains during the 2010s. Viruses 2019, 11, 626. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Salas, E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol. 2008, 16, 230–237. [Google Scholar] [CrossRef]
- Melchers, W.J.; Bakkers, J.M.; Slot, H.J.B.; Galama, J.M.; Agol, V.I.; Pilipenko, E.V. Cross-talk between orientation-dependent recognition determinants of a complex control RNA element, the enterovirus oriR. RNA 2000, 6, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Pilipenko, E.V.; Poperechny, K.V.; Maslova, S.V.; Melchers, W.J.; Slot, H.J.; Agol, V.I. Cis-element, oriR, involved in the initiation of (-) strand poliovirus RNA: A quasi-globular multi-domain RNA structure maintained by tertiary (‘kissing’) interactions. EMBO J. 1996, 15, 5428–5436. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.V.; Wimmer, E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 2015, 206, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Dorsch-Häsler, K.; Yogo, Y.; Wimmer, E. Replication of picornaviruses. I. Evidence from in vitro RNA synthesis that poly (A) of the poliovirus genome is genetically coded. J. Virol. 1975, 16, 1512–1517. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Kerrigan, D.; Evans, D.J. Structure and function analysis of the poliovirus cis-acting replication element (CRE). RNA 2003, 9, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Gromeier, M.; Alexander, L.; Wimmer, E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 1996, 93, 2370–2375. [Google Scholar] [CrossRef]
- Guillot, S.; Otelea, D.; Delpeyroux, F.; Crainic, R. Point mutations involved in the attenuation/neurovirulence alternation in type 1 and 2 oral polio vaccine strains detected by site-specific polymerase chain reaction. Vaccine 1994, 12, 503–507. [Google Scholar] [CrossRef]
- Rezapkin, G.V.; Fan, L.; Asher, D.M.; Fibi, M.R.; Dragunsky, E.M.; Chumakov, K.M. Mutations in Sabin 2 Strain of Poliovirus and Stability of Attenuation Phenotype. Virology 1999, 258, 152–160. [Google Scholar] [CrossRef]
- DeJesus, N.; Franco, D.; Paul, A.; Wimmer, E.; Cello, J. Mutation of a Single Conserved Nucleotide between the Cloverleaf and Internal Ribosome Entry Site Attenuates Poliovirus Neurovirulence. J. Virol. 2005, 79, 14235–14243. [Google Scholar] [CrossRef]
- Cheng, J.; Gao, S.; Zhu, C.; Liu, S.; Li, J.; Kang, J.; Wang, Z.; Wang, T. Typical Stress Granule Proteins Interact with the 3′ Untranslated Region of Enterovirus D68 To Inhibit Viral Replication. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Dougherty, J.D.; Tsai, W.-C.; Lloyd, R.E. Multiple Poliovirus Proteins Repress Cytoplasmic RNA Granules. Viruses 2015, 7, 6127–6140. [Google Scholar] [CrossRef]
- Yang, X.; Hu, Z.; Fan, S.; Zhang, Q.; Zhong, Y.; Guo, D.; Qin, Y.; Chen, M. Picornavirus 2A protease regulates stress granule formation to facilitate viral translation. PLoS Pathog. 2018, 14, e1006901. [Google Scholar] [CrossRef]
- Baggen, J.; Thibaut, H.J.; Strating, J.; Van Kuppeveld, F.J.M. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Genet. 2018, 16, 368–381. [Google Scholar] [CrossRef]
- Liu, Y.; Hill, M.G.; Klose, T.; Chen, Z.; Watters, K.; Bochkov, Y.; Jiang, W.; Palmenberg, A.C.; Rossmann, M.G. Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma. Proc. Natl. Acad. Sci. USA 2016, 113, 8997–9002. [Google Scholar] [CrossRef] [PubMed]
- Peischard, S.; Ho, H.T.; Theiss, C.; Strutz-Seebohm, N.; Seebohm, G. A Kidnapping Story: How Coxsackievirus B3 and Its Host Cell Interact. Cell. Physiol. Biochem. 2019, 53, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yu, B.; Lin, L.; Zhai, X.; Han, Y.; Qin, Y.; Guo, Z.; Wu, S.; Zhong, X.; Wang, Y.; et al. A functional nuclear localization sequence in the VP1 capsid protein of coxsackievirus B3. Virology 2012, 433, 513–521. [Google Scholar] [CrossRef]
- Laitinen, O.H.; Svedin, E.; Kapell, S.; Nurminen, A.; Hytönen, V.P.; Flodström-Tullberg, M. Enteroviral proteases: Structure, host interactions and pathogenicity. Rev. Med Virol. 2016, 26, 251–267. [Google Scholar] [CrossRef]
- Skern, T.; Hampölz, B.; Guarné, A.; Fita, I.; Bergmann, E.M.; Petersen, J.; James, M.N.G. Structure and Function of Picornavirus Proteinases. Mol. Biol. Picornavirus 2014, 199–212. [Google Scholar] [CrossRef]
- Seipelt, J.; Guarné, A.; Bergmann, E.; James, M.; Sommergruber, W.; Fita, I.; Skern, T. The structures of picornaviral proteinases. Virus Res. 1999, 62, 159–168. [Google Scholar] [CrossRef]
- Blom, N.S.; Hansen, J.; Brunak, S.; Blaas, D. Cleavage site analysis in picornaviral polyproteins: Discovering cellular targets by neural networks. Protein Sci. 1996, 5, 2203–2216. [Google Scholar] [CrossRef]
- Parsley, T.B.; Cornell, C.T.; Semler, B.L. Modulation of the RNA Binding and Protein Processing Activities of Poliovirus Polypeptide 3CD by the Viral RNA Polymerase Domain. J. Biol. Chem. 1999, 274, 12867–12876. [Google Scholar] [CrossRef] [PubMed]
- Lamphear, B.; Yan, R.; Yang, F.; Waters, D.; Liebig, H.; Klump, H.; Kuechler, E.; Skern, T.; Rhoads, R. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human Coxsackievirus and rhinovirus. J. Biol. Chem. 1993, 268, 19200–19203. [Google Scholar] [CrossRef]
- de Breyne, S.; Yu, Y.; Unbehaun, A.; Pestova, T.V.; Hellen, C.U.T. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. USA 2009, 106, 9197–9202. [Google Scholar] [CrossRef]
- Lamphear, B.J.; Kirchweger, R.; Skern, T.; Rhoads, R.E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases: Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995, 270, 21975–21983. [Google Scholar] [CrossRef]
- de Quinto, S.L.; Martínez-Salas, E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 2000, 6, 1380–1392. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.D.; Chase, A.J.; Cathcart, A.L.; Tran, G.P.; Semler, B.L. Viral Proteinase Requirements for the Nucleocytoplasmic Relocalization of Cellular Splicing Factor SRp20 during Picornavirus Infections. J. Virol. 2012, 87, 2390–2400. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, N.; Skern, T.; Gustin, K.E. Specific Cleavage of the Nuclear Pore Complex Protein Nup62 by a Viral Protease. J. Biol. Chem. 2010, 285, 28796–28805. [Google Scholar] [CrossRef] [PubMed]
- Yalamanchili, P.; Harris, K.; Wimmer, E.; Dasgupta, A. Inhibition of basal transcription by poliovirus: A virus- encoded protease (3Cpro) inhibits formation of TBP-TATA box complex in vitro. J. Virol. 1996, 70, 2922–2929. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.E.; Lieberman, P.M.; Berk, A.J.; Dasgupta, A. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol. 1993, 13, 1232. [Google Scholar] [CrossRef] [PubMed]
- Weidman, M.K.; Yalamanchili, P.; Ng, B.; Tsai, W.; Dasgupta, A. Poliovirus 3C Protease-Mediated Degradation of Transcriptional Activator p53 Requires a Cellular Activity. Virology 2001, 291, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Yalamanchili, P.; Weidman, K.; Dasgupta, A. Cleavage of Transcriptional Activator Oct-1 by Poliovirus Encoded Protease 3Cpro. Virology 1997, 239, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Cui, Z.; Zhang, Z.; Wei, H.; Zhang, X. Poliovirus 2Apro induces the nucleic translocation of poliovirus 3CD and 3C’ proteins. Acta Biochim. et Biophys. Sin. 2010, 43, 38–44. [Google Scholar] [CrossRef]
- Mukherjee, A.; Morosky, S.A.; Delorme-Axford, E.; Dybdahl-Sissoko, N.; Oberste, M.S.; Wang, T.; Coyne, C. B The coxsackievirus B 3C pro protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 2011, 7, e1001311. [Google Scholar] [CrossRef] [PubMed]
- Barral, P.M.; Morrison, J.M.; Drahos, J.; Gupta, P.; Sarkar, D.; Fisher, P.B.; Racaniello, V.R. MDA-5 Is Cleaved in Poliovirus-Infected Cells. J. Virol. 2007, 81, 3677–3684. [Google Scholar] [CrossRef]
- Feng, Q.; Langereis, M.A.; Lork, M.; Nguyen, M.; Hato, S.V.; Lanke, K.; Emdad, L.; Bhoopathi, P.; Fisher, P.B.; Lloyd, R.E. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J. Virol. 2014, 88, 3369. [Google Scholar] [CrossRef]
- Xiang, Z.; Li, L.; Lei, X.; Zhou, H.; Zhou, Z.; He, B.; Wang, J. Enterovirus 68 3C Protease Cleaves TRIF To Attenuate Antiviral Responses Mediated by Toll-Like Receptor 3. J. Virol. 2014, 88, 6650–6659. [Google Scholar] [CrossRef]
- Rui, Y.; Su, J.; Wang, H.; Chang, J.; Wang, S.; Zheng, W.; Cai, Y.; Wei, W.; Gordy, J.; Markham, R.; et al. Disruption of MDA5-Mediated Innate Immune Responses by the 3C Proteins of Coxsackievirus A16, Coxsackievirus A6, and Enterovirus D68. J. Virol. 2017, 91, e00546-17. [Google Scholar] [CrossRef]
- Xiang, Z.; Liu, L.; Lei, X.; Zhou, Z.; He, B.; Wang, J. 3C Protease of Enterovirus D68 Inhibits Cellular Defense Mediated by Interferon Regulatory Factor 7. J. Virol. 2016, 90, 1613–1621. [Google Scholar] [CrossRef]
- Kang, J.; Pang, Z.; Zhou, Z.; Li, X.; Liu, S.; Cheng, J.; Liu, P.; Tan, W.; Wang, Z.; Wang, T. Enterovirus D68 Protease 2Apro Targets TRAF3 to Subvert Host Innate Immune Responses. J. Virol. 2020, 95, e01856-20. [Google Scholar] [CrossRef] [PubMed]
- Visser, L.J.; Langereis, M.A.; Rabouw, H.H.; Wahedi, M.; Muntjewerff, E.M.; de Groot, R.J.; van Kuppeveld, F.J.M. Essential Role of Enterovirus 2A Protease in Counteracting Stress Granule Formation and the Induction of Type I Interferon. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Bienz, K.; Egger, D.; Pasamontes, L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 1987, 160, 220–226. [Google Scholar] [CrossRef]
- De Jong, A.S.; Wessels, E.; Dijkman, H.B.P.M.; Galama, J.M.D.; Melchers, W.; Willems, P.H.; van Kuppeveld, F.J.M. Determinants for Membrane Association and Permeabilization of the Coxsackievirus 2B Protein and the Identification of the Golgi Complex as the Target Organelle. J. Biol. Chem. 2003, 278, 1012–1021. [Google Scholar] [CrossRef]
- Sandoval, I.V.; Carrasco, L. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J. Virol. 1997, 71, 4679–4693. [Google Scholar] [CrossRef]
- De Jong, A.S.; de Mattia, F.; Van Dommelen, M.M.; Lanke, K.; Melchers, W.J.G.; Willems, P.H.; van Kuppeveld, F.J.M. Functional Analysis of Picornavirus 2B Proteins: Effects on Calcium Homeostasis and Intracellular Protein Trafficking. J. Virol. 2008, 82, 3782–3790. [Google Scholar] [CrossRef]
- Lama, J.; Carrasco, L. Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J. Biol. Chem. 1992, 267, 15932–15937. [Google Scholar] [CrossRef]
- Doedens, J.; Kirkegaard, K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995, 14, 894–907. [Google Scholar] [CrossRef]
- Aldabe, R.; Barco, A.; Carrasco, L. Membrane Permeabilization by Poliovirus Proteins 2B and 2BC. J. Biol. Chem. 1996, 271, 23134–23137. [Google Scholar] [CrossRef]
- Van Kuppeveld, F.J.; Hoenderop, J.G.; Smeets, R.L.; Willems, P.H.; Dijkman, H.B.; Galama, J.M.; Melchers, W.J. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 1997, 16, 3519–3532. [Google Scholar] [CrossRef]
- Van Kuppeveld, F.J.; de Jong, A.S.; Melchers, W.J.; Willems, P.H. Enterovirus protein 2B po (u) res out the calcium: A viral strategy to survive? Trends Microbiol. 2005, 13, 41–44. [Google Scholar] [CrossRef]
- Agirre, A.; Barco, A.; Carrasco, L.; Nieva, J.L. Viroporin-mediated membrane permeabilization: Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 2002, 277, 40434–40441. [Google Scholar] [CrossRef] [PubMed]
- Cuconati, A.; Xiang, W.; Lahser, F.; Pfister, T.; Wimmer, E. A Protein Linkage Map of the P2 Nonstructural Proteins of Poliovirus. J. Virol. 1998, 72, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- De Jong, A.S.; Schrama, I.W.J.; Willems, P.H.G.M.; Galama, J.M.D.; Melchers, W.J.G.; Van Kuppeveld, F.J.M. Multimerization reactions of coxsackievirus proteins 2B, 2C and 2BC: A mammalian two-hybrid analysis. J. Gen. Virol. 2002, 83, 783–793. [Google Scholar] [CrossRef] [PubMed]
- van Kuppeveld, F.J.M.; Melchers, W.J.G.; Willems, P.H.; Gadella, J.T.W.J. Homomultimerization of the Coxsackievirus 2B Protein in Living Cells Visualized by Fluorescence Resonance Energy Transfer Microscopy. J. Virol. 2002, 76, 9446–9456. [Google Scholar] [CrossRef][Green Version]
- De Jong, A.S.; Melchers, W.J.; Glaudemans, D.H.; Willems, P.H.; van Kuppeveld, F.J. Mutational analysis of different regions in the coxsackievirus 2B protein: Requirements for homo-multimerization, membrane permeabilization, subcellular localization, and virus replication. J. Biol. Chem. 2004, 279, 19924–19935. [Google Scholar] [CrossRef] [PubMed]
- Bienz, K.; Egger, D.; Pfister, T. Characteristics of the poliovirus replication complex. In Positive-Strand RNA Viruses; Springer: Berlin/Heidleberg, Germany, 1994; pp. 147–157. [Google Scholar]
- Nieva, J.L.; Agirre, A.; Nir, S.; Carrasco, L. Mechanisms of membrane permeabilization by picornavirus 2B viroporin. FEBS Lett. 2003, 552, 68–73. [Google Scholar] [CrossRef]
- Campanella, M.; de Jong, A.S.; Lanke, K.W.H.; Melchers, W.; Willems, P.H.; Pinton, P.; Rizzuto, R.; van Kuppeveld, F.J.M. The Coxsackievirus 2B Protein Suppresses Apoptotic Host Cell Responses by Manipulating Intracellular Ca2+ Homeostasis. J. Biol. Chem. 2004, 279, 18440–18450. [Google Scholar] [CrossRef]
- Carthy, C.M.; Yanagawa, B.; Luo, H.; Granville, D.J.; Yang, D.; Cheung, P.; Cheung, C.; Esfandiarei, M.; Rudin, C.M.; Thompson, C.B. Bcl-2 and Bcl-xL overexpression inhibits cytochrome c release, activation of multiple caspases, and virus release following coxsackievirus B3 infection. Virology 2003, 313, 147–157. [Google Scholar] [CrossRef]
- Wu, H.; Zhai, X.; Chen, Y.; Wang, R.; Lin, L.; Chen, S.; Wang, T.; Zhong, X.; Wu, X.; Wang, Y.; et al. Protein 2B of Coxsackievirus B3 Induces Autophagy Relying on Its Transmembrane Hydrophobic Sequences. Viruses 2016, 8, 131. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-C.; Chang, C.-L.; Wang, P.-S.; Tsai, Y.; Liu, H.-S. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J. Med Virol. 2009, 81, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Zhang, J.; Si, X.; Gao, G.; Mao, I.; McManus, B.M.; Luo, H. Autophagosome Supports Coxsackievirus B3 Replication in Host Cells. J. Virol. 2008, 82, 9143–9153. [Google Scholar] [CrossRef]
- Jackson, W.T.; Jr, T.H.G.; Taylor, M.; Mulinyawe, S.; Rabinovitch, M.; Kopito, R.R.; Kirkegaard, K. Subversion of Cellular Autophagosomal Machinery by RNA Viruses. PLoS Biol. 2005, 3, e156. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.K.F.; Sam, I.-C.; Verlhac, P.; Baguet, J.; Eskelinen, E.-L.; Faure, M.; Chan, Y.F. 2BC Non-Structural Protein of Enterovirus A71 Interacts with SNARE Proteins to Trigger Autolysosome Formation. Viruses 2017, 9, 169. [Google Scholar] [CrossRef]
- Norder, H.; De Palma, A.M.; Selisko, B.; Costenaro, L.; Papageorgiou, N.; Arnan, C.; Coutard, B.; Lantez, V.; De Lamballerie, X.; Baronti, C.; et al. Picornavirus non-structural proteins as targets for new anti-virals with broad activity. Antivir. Res. 2011, 89, 204–218. [Google Scholar] [CrossRef]
- Xia, H.; Wang, P.; Wang, G.; Yang, J.; Sun, X.; Wu, W.; Qiu, Y.; Shu, T.; Zhao, X.; Yin, L.; et al. Human Enterovirus Nonstructural Protein 2CATPase Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone. PLoS Pathog. 2015, 11, e1005067. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.L.; Carrasco, L. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 1993, 268, 8105–8110. [Google Scholar] [CrossRef]
- Mirzayan, C.; Wimmer, E. Biochemical Studies on Poliovirus Polypeptide 2C: Evidence for ATPase Activity. Virology 1994, 199, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-H.; Wang, K.; Zhao, K.; Hua, S.-C.; Du, J. The Structure, Function, and Mechanisms of Action of Enterovirus Non-structural Protein 2C. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Wang, C.; Jiang, P.; Sand, C.; Paul, A.V.; Wimmer, E. Alanine Scanning of Poliovirus 2CATPaseReveals New Genetic Evidence that Capsid Protein/2CATPaseInteractions Are Essential for Morphogenesis. J. Virol. 2012, 86, 9964–9975. [Google Scholar] [CrossRef]
- Asare, E.; Mugavero, J.; Jiang, P.; Wimmer, E.; Paul, A.V. A single amino acid substitution in poliovirus nonstructural protein 2CATPase causes conditional defects in encapsidation and uncoating. J. Virol. 2016, 90, 6174. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, C.; Mueller, S.; Paul, A.V.; Wimmer, E.; Jiang, P. Direct interaction between two viral proteins, the nonstructural protein 2C ATPase and the capsid protein VP3, is required for enterovirus morphogenesis. PLoS Pathog. 2010, 6, e1001066. [Google Scholar] [CrossRef] [PubMed]
- Pfister, T.; Jones, K.W.; Wimmer, E. A cysteine-rich motif in poliovirus protein 2CATPase is involved in RNA replication and binds zinc in vitro. J. Virol. 2000, 74, 334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Du, H.; Yin, P.; Yang, X.; Zhang, L.; Jin, Q.; Zhu, G. Enterovirus 71 2C protein inhibits NF-κB activation by binding to RelA (p65). Sci. Rep. 2015, 5, 14302. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zheng, Z.; Liu, Y.; Zhang, Z.; Liu, Q.; Meng, J.; Ke, X.; Hu, Q.; Wang, H. 2C Proteins of Enteroviruses Suppress IKKβ Phosphorylation by Recruiting Protein Phosphatase 1. J. Virol. 2016, 90, 5141–5151. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.S.; Dodd, D.A.; Kirkegaard, K. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 2005, 337, 18–29. [Google Scholar] [CrossRef]
- Wessels, E.; Notebaart, R.A.; Duijsings, D.; Lanke, K.; Vergeer, B.; Melchers, W.J.; van Kuppeveld, F.J.M. Structure-function analysis of the coxsackievirus protein 3A: Identification of residues important for dimerization, viral RNA replication, and transport inhibition. J. Biol. Chem. 2006, 281, 28232–28243. [Google Scholar] [CrossRef]
- Towner, J.S.; Ho, T.V.; Semler, B.L. Determinants of Membrane Association for Poliovirus Protein 3AB. J. Biol. Chem. 1996, 271, 26810–26818. [Google Scholar] [CrossRef]
- Jackson, T.; Belsham, G. Picornaviruses: A View from 3A. Viruses 2021, 13, 456. [Google Scholar] [CrossRef]
- Wessels, E.; Duijsings, D.; Notebaart, R.A.; Melchers, W.J.G.; van Kuppeveld, F.J.M. A Proline-Rich Region in the Coxsackievirus 3A Protein Is Required for the Protein To Inhibit Endoplasmic Reticulum-to-Golgi Transport. J. Virol. 2005, 79, 5163–5173. [Google Scholar] [CrossRef] [PubMed]
- Wessels, E.; Duijsings, D.; Lanke, K.H.W.; van Dooren, S.H.J.; Jackson, C.L.; Melchers, W.J.G.; van Kuppeveld, F.J.M. Effects of Picornavirus 3A Proteins on Protein Transport and GBF1-Dependent COP-I Recruitment. J. Virol. 2006, 80, 11852–11860. [Google Scholar] [CrossRef][Green Version]
- Wessels, E.; Duijsings, D.; Niu, T.-K.; Neumann, S.; Oorschot, V.M.; de Lange, F.; Lanke, K.H.; Klumperman, J.; Henke, A.; Jackson, C.; et al. A Viral Protein that Blocks Arf1-Mediated COP-I Assembly by Inhibiting the Guanine Nucleotide Exchange Factor GBF1. Dev. Cell 2006, 11, 191–201. [Google Scholar] [CrossRef]
- Wessels, E.; Duijsings, D.; Lanke, K.H.W.; Melchers, W.J.G.; Jackson, C.; van Kuppeveld, F.J.M. Molecular Determinants of the Interaction between Coxsackievirus Protein 3A and Guanine Nucleotide Exchange Factor GBF1. J. Virol. 2007, 81, 5238–5245. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, F.; Xia, H.; Wang, P.; Yang, J.; Zhao, T.; Zhang, Q.; Hu, Y.; Zhou, X. The identification and characterization of nucleic acid chaperone activity of human enterovirus 71 nonstructural protein 3AB. Virology 2014, 464-465, 353–364. [Google Scholar] [CrossRef]
- DeStefano, J.J.; Titilope, O. Poliovirus Protein 3AB Displays Nucleic Acid Chaperone and Helix-Destabilizing Activities. J. Virol. 2006, 80, 1662–1671. [Google Scholar] [CrossRef] [PubMed]
- Xiang, W.; Harris, K.S.; Alexander, L.; Wimmer, E. Interaction between the 5’-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 1995, 69, 3658–3667. [Google Scholar] [CrossRef] [PubMed]
- Spear, A.; Ogram, S.A.; Morasco, B.J.; Smerage, L.E.; Flanegan, J.B. Viral precursor protein P3 and its processed products perform discrete and essential functions in the poliovirus RNA replication complex. Virology 2015, 485, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Xiang, W.; Cuconati, A.; Paul, A.V.; Cao, X.; Wimmer, E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1995, 1, 892–904. [Google Scholar]
- Paul, A.V.; Cao, X.; Harris, K.S.; Lama, J.; Wimmer, E. Studies with poliovirus polymerase 3Dpol. Stimulation of poly (U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 1994, 269, 29173–29181. [Google Scholar] [CrossRef]
- Lama, J.; Paul, A.; Harris, K.; Wimmer, E. Properties of purified recombinant poliovirus protein 3aB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J. Biol. Chem. 1994, 269, 66–70. [Google Scholar] [CrossRef]
- Richards, O.C.; Ehrenfeld, E. Effects of Poliovirus 3AB Protein on 3D Polymerase-catalyzed Reaction. J. Biol. Chem. 1998, 273, 12832–12840. [Google Scholar] [CrossRef]
- Rodriguez-Wells, V.; Plotch, S.J.; DeStefano, J.J. Primer-dependent synthesis by poliovirus RNA-dependent RNA polymerase (3Dpol). Nucleic Acids Res. 2001, 29, 2715–2724. [Google Scholar] [CrossRef]
- Lyle, J.M.; Clewell, A.; Richmond, K.; Richards, O.C.; Hope, D.A.; Schultz, S.C.; Kirkegaard, K. Similar Structural Basis for Membrane Localization and Protein Priming by an RNA-dependent RNA Polymerase. J. Biol. Chem. 2002, 277, 16324–16331. [Google Scholar] [CrossRef]
- Paul, A.V.; Van Boom, J.H.; Filippov, D.V.; Wimmer, E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nat. Cell Biol. 1998, 393, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Franco, D.; Paul, A.V.; Wimmer, E. Tyrosine 3 of Poliovirus Terminal Peptide VPg(3B) Has an Essential Function in RNA Replication in the Context of Its Precursor Protein, 3AB. J. Virol. 2007, 81, 5669–5684. [Google Scholar] [CrossRef] [PubMed]
- Pathak, H.B.; Oh, H.S.; Goodfellow, I.G.; Arnold, J.J.; Cameron, C.E. Picornavirus genome replication: Roles of precursor proteins and rate-limiting steps in oriI-dependent VPg uridylylation. J. Biol. Chem. 2008, 283, 30677–30688. [Google Scholar] [CrossRef] [PubMed]
- Cameron, C.E.; Oh, H.S.; Moustafa, I.M. Expanding knowledge of P3 proteins in the poliovirus lifecycle. Future Microbiol. 2010, 5, 867–881. [Google Scholar] [CrossRef]
- Sadeuh-Mba, S.A.; Bessaud, M.; Massenet, D.; Joffret, M.-L.; Endegue, M.-C.; Njouom, R.; Reynes, J.-M.; Rousset, D.; Delpeyroux, F. High Frequency and Diversity of Species C Enteroviruses in Cameroon and Neighboring Countries. J. Clin. Microbiol. 2012, 51, 759–770. [Google Scholar] [CrossRef]
- Alexander, D.A.; Dimock, K. Sialic Acid Functions in Enterovirus 70 Binding and Infection. J. Virol. 2002, 76, 11265–11272. [Google Scholar] [CrossRef]
- Baggen, J.; Thibaut, H.J.; Staring, J.; Jae, L.; Liu, Y.; Guo, H.; Slager, J.J.; de Bruin, J.W.; van Vliet, A.L.W.; Blomen, V.A.; et al. Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc. Natl. Acad. Sci. USA 2016, 113, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T.; Okamoto, M.; Nakakita, S.-I.; Suzuki, A.; Saito, M.; Tamaki, R.; Lupisan, S.; Roy, C.N.; Hiramatsu, H.; Sugawara, K.-E.; et al. Antigenic and Receptor Binding Properties of Enterovirus 68. J. Virol. 2014, 88, 2374–2384. [Google Scholar] [CrossRef]
- Liu, Y.; Sheng, J.; Baggen, J.; Meng, G.; Xiao, C.; Thibaut, H.J.; Van Kuppeveld, F.J.M.; Rossmann, M.G. Sialic acid-dependent cell entry of human enterovirus D68. Nat. Commun. 2015, 6, 8865. [Google Scholar] [CrossRef]
- Nokhbeh, M.R.; Hazra, S.; Alexander, D.A.; Khan, A.; McAllister, M.; Suuronen, E.J.; Griffith, M.; Dimock, K. Enterovirus 70 Binds to Different Glycoconjugates Containing α2,3-Linked Sialic Acid on Different Cell Lines. J. Virol. 2005, 79, 7087–7094. [Google Scholar] [CrossRef] [PubMed]
- Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respir. Viruses 2008, 2, 147–154. [CrossRef] [PubMed]
- Kuchipudi, S.; Nelli, R.; Gontu, A.; Satyakumar, R.; Nair, M.S.; Subbiah, M. Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover. Viruses 2021, 13, 262. [Google Scholar] [CrossRef]
- Bertram, S.; Heurich, A.; Lavender, H.; Gierer, S.; Danisch, S.; Perin, P.; Lucas, J.M.; Nelson, P.S.; Pöhlmann, S.; Soilleux, E.J. Influenza and SARS-Coronavirus Activating Proteases TMPRSS2 and HAT Are Expressed at Multiple Sites in Human Respiratory and Gastrointestinal Tracts. PLoS ONE 2012, 7, e35876. [Google Scholar] [CrossRef]
- Sata, T.; Roth, J.; Zuber, C.; Stamm, B.; Heitz, P.U. Expression of alpha 2,6-linked sialic acid residues in neoplastic but not in normal human colonic mucosa. A lectin-gold cytochemical study with Sambucus nigra and Maackia amurensis lectins. Am. J. Pathol. 1991, 139, 1435–1448. [Google Scholar]
- Karnauchow, T.M.; Tolson, D.L.; A Harrison, B.; Altman, E.; Lublin, D.M.; Dimock, K. The HeLa cell receptor for enterovirus 70 is decay-accelerating factor (CD55). J. Virol. 1996, 70, 5143–5152. [Google Scholar] [CrossRef] [PubMed]
- Karnauchow, T.M.; Dawe, S.; Lublin, D.M.; Dimock, K. Short Consensus Repeat Domain 1 of Decay-Accelerating Factor Is Required for Enterovirus 70 Binding. J. Virol. 1998, 72, 9380–9383. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lin, F.; Chipman, P.R.; Bator, C.M.; Baker, T.S.; Shoham, M.; Kuhn, R.J.; Medof, M.E.; Rossmann, M.G. Structure of decay-accelerating factor bound to echovirus 7: A virus-receptor complex. Proc. Natl. Acad. Sci. USA 2002, 99, 10325–10329. [Google Scholar] [CrossRef]
- Bergelson, J.M.; Mohanty, J.G.; Crowell, R.L.; John, N.F.S.; Lublin, D.M.; Finberg, R.W. Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55). J. Virol. 1995, 69, 1903–1906. [Google Scholar] [CrossRef]
- Shafren, D.R.; Bates, R.C.; Agrez, M.V.; Herd, R.L.; Burns, G.F.; Barry, R.D. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J. Virol. 1995, 69, 3873–3877. [Google Scholar] [CrossRef]
- Shafren, D.R.; Dorahy, D.J.; A Ingham, R.; Burns, G.F.; Barry, R.D. Coxsackievirus A21 binds to decay-accelerating factor but requires intercellular adhesion molecule 1 for cell entry. J. Virol. 1997, 71, 4736–4743. [Google Scholar] [CrossRef] [PubMed]
- Haddad, A.; Nokhbeh, M.R.; Alexander, D.A.; Dawe, S.J.; Grisé, C.; Gulzar, N.; Dimock, K. Binding to Decay-Accelerating Factor Is Not Required for Infection of Human Leukocyte Cell Lines by Enterovirus 70. J. Virol. 2004, 78, 2674–2681. [Google Scholar] [CrossRef]
- Martino, T.A.; Petric, M.; Weingartl, H.; Bergelson, J.M.; Opavsky, M.A.; Richardson, C.D.; Modlin, J.F.; Finberg, R.W.; Kain, K.C.; Willis, N.; et al. The Coxsackie-Adenovirus Receptor (CAR) Is Used by Reference Strains and Clinical Isolates Representing All Six Serotypes of Coxsackievirus Group B and by Swine Vesicular Disease Virus. Virology 2000, 271, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Guo, H.; Chang, J.; Yu, Y.; Liu, G.; Zhang, N.; Willard, S.H.; Zheng, S.; Yu, X.-F. ICAM-5/Telencephalin Is a Functional Entry Receptor for Enterovirus D68. Cell Host Microbe 2016, 20, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Hixon, A.M.; Clarke, P.; Tyler, K.L. Contemporary Circulating Enterovirus D68 Strains Infect and Undergo Retrograde Axonal Transport in Spinal Motor Neurons Independent of Sialic Acid. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, A.B.; Warren, A.L.; Racaniello, V.R. Neurotropism of Enterovirus D68 Isolates Is Independent of Sialic Acid and Is Not a Recently Acquired Phenotype. mBio 2019, 10, e02370-19. [Google Scholar] [CrossRef] [PubMed]
- Baggen, J.; Liu, Y.; Lyoo, H.; Van Vliet, A.L.W.; Wahedi, M.; De Bruin, J.W.; Roberts, R.W.; Overduin, P.; Meijer, A.; Rossmann, M.G.; et al. Bypassing pan-enterovirus host factor PLA2G16. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Staring, J.; von Castelmur, E.; Blomen, V.A.; Hengel, L.G.V.D.; Brockmann, M.; Baggen, J.; Thibaut, H.J.; Nieuwenhuis, J.; Janssen, H.; Van Kuppeveld, F.J.M.; et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nat. Cell Biol. 2017, 541, 412–416. [Google Scholar] [CrossRef]
- Na Ayudhya, S.S.; Meijer, A.; Bauer, L.; Munnink, B.O.; Embregts, C.; Leijten, L.; Siegers, J.Y.; Laksono, B.M.; van Kuppeveld, F.; Kuiken, T.; et al. Enhanced Enterovirus D68 Replication in Neuroblastoma Cells Is Associated with a Cell Culture-Adaptive Amino Acid Substitution in VP1. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Liu, Y.; Sheng, J.; Fokine, A.; Meng, G.; Shin, W.-H.; Long, F.; Kuhn, R.J.; Kihara, D.; Rossmann, M.G. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children. Science 2015, 347, 71–74. [Google Scholar] [CrossRef]
- Liu, Y.; Sheng, J.; van Vliet, A.L.W.; Buda, G.; van Kuppeveld, F.J.M.; Rossmann, M.G. Molecular basis for the acid-initiated uncoating of human enterovirus D68. Proc. Natl. Acad. Sci. USA 2018, 115, E12209–E12217. [Google Scholar] [CrossRef]
- Holm-Hansen, C.C.; Midgley, S.E.; Fischer, T.K. Global emergence of enterovirus D68: A systematic review. Lancet Infect. Dis. 2016, 16, e64–e75. [Google Scholar] [CrossRef]
- Moss, R.B. Enterovirus 68 Infection–Association with Asthma. J. Allergy Clin. Immunol. Pr. 2016, 4, 226–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-S.; Lee, H.-C.; Lee, K.-M.; Gong, Y.-N.; Shih, S.-R. Enterovirus and Encephalitis. Front. Microbiol. 2020, 11, 261. [Google Scholar] [CrossRef] [PubMed]
- Smura, T.; Ylipaasto, P.; Klemola, P.; Kaijalainen, S.; Kyllönen, L.; Sordi, V.; Piemonti, L.; Roivainen, M. Cellular tropism of human enterovirus D species serotypes EV-94, EV-70, and EV-68 in vitro: Implications for pathogenesis. J. Med Virol. 2010, 82, 1940–1949. [Google Scholar] [CrossRef]
- Palacios, G.; Oberste, M.S. Enteroviruses as agents of emerging infectious diseases. J. NeuroVirology 2005, 11, 424–433. [Google Scholar] [CrossRef]
- Hixon, A.M.; Frost, J.; Rudy, M.J.; Messacar, K.; Clarke, P.; Tyler, K.L. Understanding Enterovirus D68-Induced Neurologic Disease: A Basic Science Review. Viruses 2019, 11, 821. [Google Scholar] [CrossRef]
- Chatterjee, S.; O Quarcoopome, C.; Apenteng, A. Unusual type of epidemic conjunctivitis in Ghana. Br. J. Ophthalmol. 1970, 54, 628–630. [Google Scholar] [CrossRef]
- Higgins, P.G. Enteroviral conjunctivitis and its neurological complications. Arch. Virol. 1982, 73, 91–101. [Google Scholar] [CrossRef]
- Chen, D.; Texada, D.E.; Duggan, C.; Deng, Y.; Redens, T.B.; Langford, M.P. Caspase-3 and -7 mediate apoptosis of human Chang’s conjunctival cells induced by enterovirus 70. Virology 2006, 347, 307–322. [Google Scholar] [CrossRef]
- Bharucha, E.; Mondkar, V.; Wadia, N.; Irani, P.; Katrak, S. Neurological complications of a new conjunctivitis. Lancet 1972, 300, 970–971. [Google Scholar] [CrossRef]
- Junttila, N.; Leveque, N.; Kabue, J.P.; Cartet, G.; Mushiya, F.; Muyembe-Tamfum, J.-J.; Trompette, A.; Lina, B.; Magnius, L.O.; Chomel, J.-J.; et al. New enteroviruses, EV-93 and EV-94, associated with acute flaccid paralysis in the Democratic Republic of the Congo. J. Med Virol. 2007, 79, 393–400. [Google Scholar] [CrossRef]
- Schieble, J.H.; Fox, V.L.; Lennette, E.H. A probable new human picornavirus associated with resoiratory disease. Am. J. Epidemiol. 1967, 85, 297–310. [Google Scholar] [CrossRef]
- Royston, L.; Tapparel, C. Rhinoviruses and Respiratory Enteroviruses: Not as Simple as ABC. Viruses 2016, 8, 16. [Google Scholar] [CrossRef]
- Kidd, S.; Lopez, A.S.; Konopka-Anstadt, J.L.; Nix, W.A.; Routh, J.A.; Oberste, M.S. Enterovirus D68-associated acute flaccid myelitis, United States, 2020. Emerg. Infect. Dis. 2020, 26, e201630. [Google Scholar] [CrossRef]
- Smith, B.D.; Pekosz, A. Contemporary enterovirus D68 strains show enhanced replication and translation at 37 °C. bioRxiv 2020. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, S.; Shen, S.; Guo, H.; Huang, H.; Wei, W. Methyl-β-cyclodextrin inhibits EV-D68 virus entry by perturbing the accumulation of virus particles and ICAM-5 in lipid rafts. Antivir. Res. 2020, 176, 104752. [Google Scholar] [CrossRef]
- Brown, B.A.; Nix, W.A.; Sheth, M.; Frace, M.; Oberste, M.S. Seven Strains of Enterovirus D68 Detected in the United States during the 2014 Severe Respiratory Disease Outbreak. Genome Announc. 2014, 2, e01201-14. [Google Scholar] [CrossRef]
- Hixon, A.M.; Yu, G.; Leser, J.S.; Yagi, S.; Clarke, P.; Chiu, C.Y.; Tyler, K.L. A mouse model of paralytic myelitis caused by enterovirus D68. PLoS Pathog. 2017, 13, e1006199. [Google Scholar] [CrossRef]
- Hu, Y.; Musharrafieh, R.; Zheng, M.; Wang, J. Enterovirus D68 Antivirals: Past, Present, and Future. ACS Infect. Dis. 2020, 6, 1572–1586. [Google Scholar] [CrossRef]
- Elrick, M.J.; Pekosz, A.; Duggal, P. Enterovirus D68 molecular and cellular biology and pathogenesis. J. Biol. Chem. 2021, 296, 100317. [Google Scholar] [CrossRef]
- Sun, L.; Meijer, A.; Froeyen, M.; Zhang, L.; Thibaut, H.J.; Baggen, J.; George, S.; Vernachio, J.; Van Kuppeveld, F.J.M.; Leyssen, P.; et al. Antiviral Activity of Broad-Spectrum and Enterovirus-Specific Inhibitors against Clinical Isolates of Enterovirus D68. Antimicrob. Agents Chemother. 2015, 59, 7782–7785. [Google Scholar] [CrossRef]
- Rhoden, E.; Zhang, M.; Nix, W.A.; Oberste, M.S. InVitroEfficacy of Antiviral Compounds against Enterovirus D68. Antimicrob. Agents Chemother. 2015, 59, 7779–7781. [Google Scholar] [CrossRef] [PubMed]
- Smee, D.F.; Evans, W.J.; Nicolaou, K.; Tarbet, E.B.; Day, C.W. Susceptibilities of enterovirus D68, enterovirus 71, and rhinovirus 87 strains to various antiviral compounds. Antivir. Res. 2016, 131, 61–65. [Google Scholar] [CrossRef]
- Ma, C.; Hu, Y.; Zhang, J.; Musharrafieh, R.; Wang, J. A Novel Capsid Binding Inhibitor Displays Potent Antiviral Activity against Enterovirus D68. ACS Infect. Dis. 2019, 5, 1952–1962. [Google Scholar] [CrossRef]
- Arita, M.; Fuchino, H.; Kawakami, H.; Ezaki, M.; Kawahara, N. Characterization of a New Antienterovirus D68 Compound Purified from Avocado. ACS Infect. Dis. 2020, 6, 2291–2300. [Google Scholar] [CrossRef]
- Musharrafieh, R.; Zhang, J.; Tuohy, P.; Kitamura, N.; Bellampalli, S.S.; Hu, Y.; Khanna, R.; Wang, J. Discovery of Quinoline Analogues as Potent Antivirals against Enterovirus D68 (EV-D68). J. Med. Chem. 2018, 62, 4074–4090. [Google Scholar] [CrossRef]
- Kim, Y.; Kankanamalage, A.C.G.; Damalanka, V.C.; Weerawarna, P.; Groutas, W.C.; Chang, K.-O. Potent inhibition of enterovirus D68 and human rhinoviruses by dipeptidyl aldehydes and α-ketoamides. Antivir. Res. 2015, 125, 84–91. [Google Scholar] [CrossRef]
- Musharrafieh, R.; Ma, C.; Zhang, J.; Hu, Y.; Diesing, J.M.; Marty, M.T.; Wang, J. Validating enterovirus D68-2Apro as an antiviral drug target and the discovery of telaprevir as a potent D68-2Apro inhibitor. J. Virol. 2019, 93, e02221-18. [Google Scholar] [CrossRef]
- Hurst, B.L.; Evans, W.J.; Smee, D.F.; Van Wettere, A.J.; Tarbet, E.B. Evaluation of antiviral therapies in respiratory and neurological disease models of Enterovirus D68 infection in mice. Virology 2018, 526, 146–154. [Google Scholar] [CrossRef]
- Ulferts, R.; Van Der Linden, L.; Thibaut, H.J.; Lanke, K.H.W.; Leyssen, P.; Coutard, B.; De Palma, A.M.; Canard, B.; Neyts, J.; Van Kuppeveld, F.J.M. Selective Serotonin Reuptake Inhibitor Fluoxetine Inhibits Replication of Human Enteroviruses B and D by Targeting Viral Protein 2C. Antimicrob. Agents Chemother. 2013, 57, 1952–1956. [Google Scholar] [CrossRef]
- Gao, Q.; Yuan, S.; Zhang, C.; Wang, Y.; Wang, Y.; He, G.; Zhang, S.; Altmeyer, R.; Zou, G. Discovery of Itraconazole with Broad-SpectrumIn VitroAntienterovirus Activity That Targets Nonstructural Protein 3A. Antimicrob. Agents Chemother. 2015, 59, 2654–2665. [Google Scholar] [CrossRef]
- Xu, N.; Yang, J.; Zheng, B.; Zhang, Y.; Cao, Y.; Huan, C.; Wang, S.; Chang, J.; Zhang, W. The Pyrimidine Analog FNC Potently Inhibits the Replication of Multiple Enteroviruses. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Van Der Linden, L.; Adrián, L.V.; Selisko, B.; Ferrer-Orta, C.; Liu, X.; Lanke, K.; Ulferts, R.; De Palma, A.M.; Tanchis, F.; Goris, N.; et al. The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family. PLoS Pathog. 2015, 11, e1004733. [Google Scholar] [CrossRef]
- Tang, Q.; Li, S.; Du, L.; Chen, S.; Gao, J.; Cai, Y.; Xu, Z.; Zhao, Z.; Lan, K.; Wu, S. Emetine protects mice from enterovirus infection by inhibiting viral translation. Antivir. Res. 2019, 173, 104650. [Google Scholar] [CrossRef]
- Hixon, A.M.; Clarke, P.; Tyler, K.L. Evaluating Treatment Efficacy in a Mouse Model of Enterovirus D68–Associated Paralytic Myelitis. J. Infect. Dis. 2017, 216, 1245–1253. [Google Scholar] [CrossRef]
- Takeda, N.; Tanimura, M.; Miyamura, K. Molecular evolution of the major capsid protein VP1 of enterovirus 70. J. Virol. 1994, 68, 854–862. [Google Scholar] [CrossRef]
- Smura, T.; Savolainen-Kopra, C.; Roivainen, M. Evolution of newly described enteroviruses. Future Virol. 2011, 6, 109–131. [Google Scholar] [CrossRef]
- Mombo, I.M.; Lukashev, A.N.; Bleicker, T.; Brünink, S.; Berthet, N.; Maganga, G.D.; Durand, P.; Arnathau, C.; Boundenga, L.; Ngoubangoye, B.; et al. African Non-Human Primates Host Diverse Enteroviruses. PLoS ONE 2017, 12, e0169067. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filipe, I.C.; Guedes, M.S.; Zdobnov, E.M.; Tapparel, C. Enterovirus D: A Small but Versatile Species. Microorganisms 2021, 9, 1758. https://doi.org/10.3390/microorganisms9081758
Filipe IC, Guedes MS, Zdobnov EM, Tapparel C. Enterovirus D: A Small but Versatile Species. Microorganisms. 2021; 9(8):1758. https://doi.org/10.3390/microorganisms9081758
Chicago/Turabian StyleFilipe, Ines Cordeiro, Mariana Soares Guedes, Evgeny M. Zdobnov, and Caroline Tapparel. 2021. "Enterovirus D: A Small but Versatile Species" Microorganisms 9, no. 8: 1758. https://doi.org/10.3390/microorganisms9081758
APA StyleFilipe, I. C., Guedes, M. S., Zdobnov, E. M., & Tapparel, C. (2021). Enterovirus D: A Small but Versatile Species. Microorganisms, 9(8), 1758. https://doi.org/10.3390/microorganisms9081758






