Molecular Characterization of Coxsackievirus A24v from Feces and Conjunctiva Reveals Epidemiological Links
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. PCR Amplification and Partial Sequencing of VP1 and 3C Regions
2.3. Identity Analysis
2.4. Phylogenetic Analysis
2.4.1. Dataset and Multiple Alignment
2.4.2. Quality Control
2.5. Phylogenetic Analyses
3. Results
3.1. Analysis of Cuban CVA24v Sequences from Different Specimens
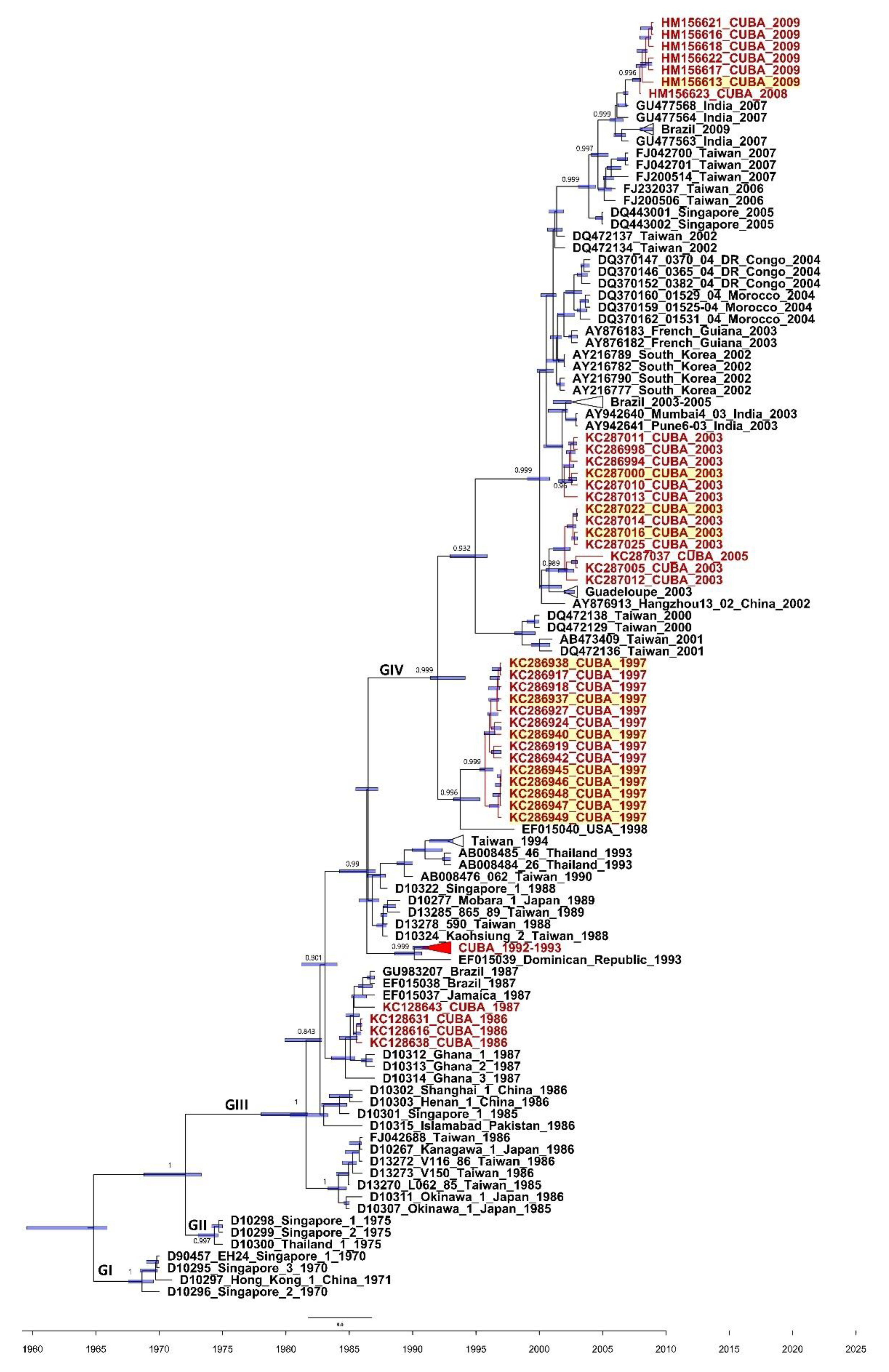
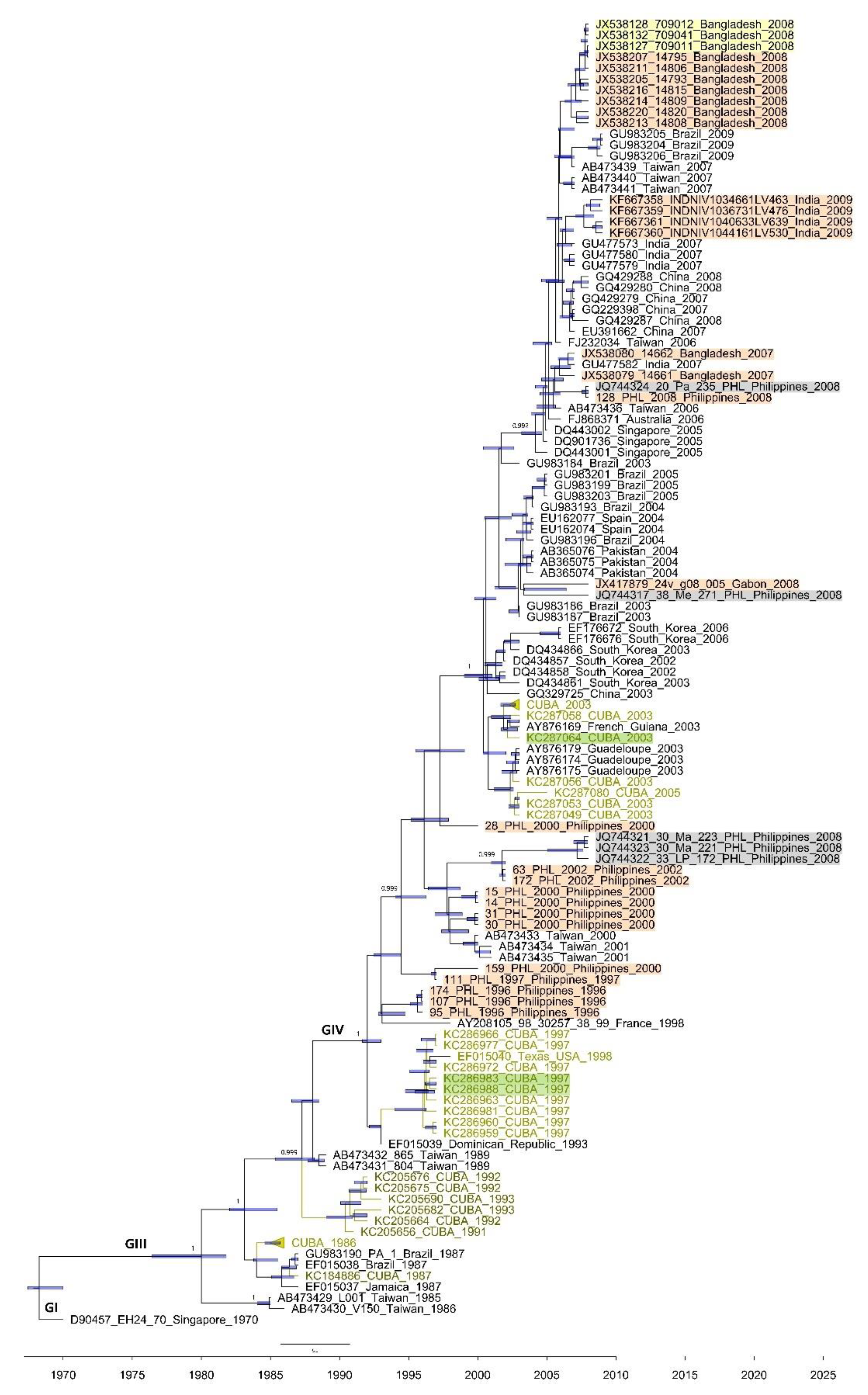
3.2. Phylogenetics of the 3C and VP1 Gene Sequences of CVA24v Strains Isolated from Feces
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yen, Y.C.; Chu, P.H.; Lu, P.L.; Lin, Y.C.; Shi, Y.Y.; Chou, L.C.; Wang, C.F.; Lin, Y.Y.; Su, H.J.; Lin, C.C.; et al. Phylodynamic Characterization of an Ocular-Tropism Coxsackievirus A24 Variant. PLoS ONE 2016, 11, e0160672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, N.; Huang, X.; Jin, X.; Geng, X.; Chan, T.C.; Liu, S. Molecular epidemiology of acute hemorrhagic conjunctivitis caused by coxsackie A type 24 variant in China, 2004–2014. Sci. Rep. 2017, 7, 45202. [Google Scholar] [CrossRef] [PubMed]
- Mirkovic, R.R.; Kono, R.; Yin-Murphy, M.; Sohier, R.; Schmidt, N.J.; Melnick, J.L. Enterovirus type 70: The etiologic agent of pandemic acute haemorrhagic conjunctivitis. Bull. World Health Organ. 1973, 49, 341–346. [Google Scholar] [PubMed]
- Yin-Murphy, M.; Lim, K.H.; Ho, Y.M. A coxsackievirus type A24 epidemic of acute conjunctivitis. Southeast Asian J. Trop. Med. Public Health 1976, 1, 1–5. [Google Scholar]
- Lim, K.H.; Yin-Murphy, M. An epidemic of conjunctivitis in Singapore in 1970. Singap. Med. J. 1971, 4, 119–127. [Google Scholar]
- Zocher, G.; Mistry, N.; Frank, M.; Hahnlein-Schick, I.; Ekstrom, J.O.; Arnberg, N.; Stehle, T. A Sialic Acid Binding Site in a Human Picornavirus. PLoS Pathog. 2014, 10, e1004401. [Google Scholar] [CrossRef] [PubMed]
- Baggen, J.; Hurdiss, D.L.; Zocher, G.; Mistry, N.; Roberts, R.W.; Slager, J.J.; Guo, H.; van Vliet, A.L.W.; Wahedi, M.; Benschop, K.; et al. Role of enhanced receptor engagement in the evolution of a pandemic acute hemorrhagic conjunctivitis virus. Proc. Natl. Acad. Sci. USA 2018, 115, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.C.; Jamshidi, F.; Johansson, S.M.; Oberste, M.S.; Arnberg, N. Sialic acid is a cellular receptor for coxsackievirus A24 variant, an emerging virus with pandemic potential. J. Virol. 2008, 82, 3061–3068. [Google Scholar] [CrossRef]
- Yin-Murphy, M.; Baharuddin, I.; Phoon, M.C.; Chow, V.T. A recent epidemic of Coxsackie virus type A24 acute haemorrhagic conjunctivitis in Singapore. Br. J. Ophthalmol. 1986, 70, 869–873. [Google Scholar] [CrossRef]
- Kuo, P.C.; Lin, J.Y.; Chen, L.C.; Fang, Y.T.; Cheng, Y.C.; Wu, H.Y.; Tsai, C.Y.; Chen, Y.S.; Lin, S.Y.; Wu, C.L.; et al. Molecular and immunocytochemical identification of coxsackievirus A-24 variant from the acute haemorrhagic conjunctivitis outbreak in Taiwan in 2007. Eye 2010, 24, 131–136. [Google Scholar] [CrossRef]
- Redon, I.A.; Lago, P.J.; Perez, L.R.; Puentes, P.; Corredor, M.B. Outbreak of acute haemorrhagic conjunctivitis in Cuba. Mem. Inst. Oswaldo Cruz 1999, 94, 467–468. [Google Scholar] [CrossRef][Green Version]
- Fonseca, M.C.; Sarmiento, L.; Resik, S.; Pereda, N.; Rodríguez, H.; Kourí, V.; Martínez, P.A.; Piñón, A.; Limonta, D.; Más, P.; et al. Isolation of Coxsackievirus A24 variant from patients with hemorrhagic conjunctivitis in Cuba, 2008–2009. J. Clin. Virol. 2012, 53, 77–81. [Google Scholar] [CrossRef]
- Fonseca, M.C.; Pupo-Meriño, M.; García-González, L.A.; Resik, S.; Hung, L.H.; Muné, M.; Rodríguez, H.; Morier, L.; Norder, H.; Sarmiento, L. Molecular evolution of coxsackievirus A24v in Cuba over 23-years, 1986–2009. Sci. Rep. 2020, 10, 13761. [Google Scholar] [CrossRef]
- Nix, W.A.; Oberste, M.S.; Pallansch, M.A. Sensitive, Seminested PCR Amplification of VP1 Sequences for Direct Identification of All Enterovirus Serotypes from Original Clinical Specimens. J. Clin. Microbiol. 2006, 44, 2698–2704. [Google Scholar] [CrossRef]
- Lin, K.H.; Chern, C.L.; Chu, P.Y.; Chang, C.H.; Wang, H.L.; Sheu, M.M.; Huang, W.L.; Pongsuwanna, Y.; Yammoto, S.; Yoshino, S.; et al. Genetic analysis of recent Taiwanese isolates of a variant of coxsackievirus A24. J. Med. Virol. 2001, 64, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Norström, M.; Karlsson, A.; Salemi, M. Towards a new paradigm linking virus molecular evolution and pathogenesis: Experimental design and phylodynamic inference. New Microbiol. 2012, 35, 101–111. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z. DAMBE: Software Package for Data Analysis in Molecular Biology and Evolution. J. Hered. 2001, 92, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Strimmer, K.; von Haeseler, A. Likelihood-mapping: A simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 1997, 9, 6815–6819. [Google Scholar] [CrossRef]
- Martin, D.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Baele, G.; Lemey, P.; Bedford, T.; Rambaut, A.; Suchard, M.A.; Alekseyenko, A.V. Improving the Accuracy of Demographic and Molecular Clock Model Comparison While Accommodating Phylogenetic Uncertainty. Mol. Biol. Evol. 2012, 29, 2157–2167. [Google Scholar] [CrossRef]
- Minin, V.N.; Bloomquist, E.W.; Suchard, M.A. Smooth Skyride through a Rough Skyline: Bayesian Coalescent-Based Inference of Population Dynamics. Mol. Biol. Evol. 2008, 25, 1459–1471. [Google Scholar] [CrossRef]
- Oberste, M.S.; Feeroz, M.M.; Maher, K.; Nix, W.A.; Engel, G.A.; Hasan, K.M.; Begum, S.; Oh, G.; Chowdhury, A.H.; Pallansch, M.A.; et al. Characterizing the picornavirus landscape among synanthropic nonhuman primates in Bangladesh, 2007 to 2008. J. Virol. 2013, 87, 558–571. [Google Scholar] [CrossRef] [PubMed]
- Apostol, L.N.; Suzuki, A.; Bautista, A.; Galang, H.; Paladin, F.J.; Fuji, N.; Lupisan, S.; Olveda, R.; Oshitani, H. Detection of non-polio enteroviruses from 17 years of virological surveillance of acute flaccid paralysis in the philippines. J. Med. Virol. 2012, 84, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Sadeuh-Mba, S.A.; Bessaud, M.; Massenet, D.; Joffret, M.L.; Endegue, M.C.; Njouom, R.; Reynes, J.M.; Rousset, D.; Delpeyroux, F. High Frequency and Diversity of Species C Enteroviruses in Cameroon and Neighboring Countries. J. Clin. Microbiol. 2013, 51, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Laxmivandana, R.; Yergolkar, P.; Rajeshwari, M.; Chitambar, S.D. Genomic characterization of coxsackievirus type A24 strains associated with acute flaccid paralysis and rarely identified Hopkins syndrome. Arch. Virol. 2014, 159, 3125–3129. [Google Scholar] [CrossRef] [PubMed]
- Apostol, L.N.G.; Imagawa, T.; Suzuki, A.; Masago, Y.; Lupisan, S.; Olveda, R.; Saito, M.; Omura, T.; Oshitani, H. Genetic diversity and molecular characterization of enteroviruses from sewage-polluted urban and rural rivers in the Philippines. Virus Genes 2012, 45, 207–217. [Google Scholar] [CrossRef]
- Pallansch, M.; Oberste, M.S.; Whitton, L. Enteroviruses: Polioviruses, Coxsackieviruses, Echoviruses, and Newer Enteroviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wikins: Philadelphia, PA, USA; New York, NY, USA, 2013; pp. 490–529. [Google Scholar]
- Mirkovic, R.R.; Schmidt, N.J.; Yin-Murphy, M.; Melnick, J.L. Enterovirus etiology of the 1970 Singapore epidemic of acute conjunctivitis. Intervirology 1974, 4, 119–127. [Google Scholar] [CrossRef]
- Kosrirukvongs, P.; Kanyok, R.; Sitritantikorn, S.; Wasi, C. Acute hemorrhagic conjunctivitis outbreak in Thailand, 1992. Southeast Asian J. Trop. Med. Public Health 1996, 27, 244–249. [Google Scholar] [PubMed]
- Bahri, O.; Rezig, D.; Nejma-Oueslati, B.B.; Yahia, A.B.; Sassi, J.B.; Hogga, N.; Sadraoui, A.; Triki, H. Enteroviruses in Tunisia: Virological surveillance over 12 years (1992–2003). J. Med. Microbiol. 2005, 54, 63–69. [Google Scholar] [CrossRef]
- Junttila, N.; Lévêque, N.; Kabue, J.P.; Cartet, G.; Mushiya, F.; Muyembe-Tamfum, J.J.; Trompette, A.; Lina, B.; Magnius, L.O.; Chomel, J.J.; et al. New enteroviruses, EV-93 and EV-94, associated with acute flaccid paralysis in the Democratic Republic of the Congo. J. Med. Virol. 2007, 79, 393–400. [Google Scholar] [CrossRef]
- Bessaud, M.; Pillet, S.; Ibrahim, W.; Joffret, M.L.; Pozzetto, B.; Delpeyroux, F.; Gouandjika-Vasilache, I. Molecular characterization of human enteroviruses in the Central African Republic: Uncovering wide diversity and identification of a new human enterovirus A71 genogroup. J. Clin. Microbiol. 2012, 50, 1650–1658. [Google Scholar] [CrossRef] [PubMed]
- Kelly, H.; Brussen, K.A.; Lawrence, A.; Elliot, E.; Pearn, J.; Thorley, B. Polioviruses and other enteroviruses isolated from faecal samples of patients with acute flaccid paralysis in Australia, 1996–2004. J. Paediatr. Child Health 2006, 42, 370–376. [Google Scholar] [CrossRef]
- Bingjun, T.; Yoshida, H.; Yan, W.; Lin, L.; Tsuji, T.; Shimizu, H.; Miyamura, T. Molecular typing and epidemiology of non-polio enteroviruses isolated from Yunnan Province, the People’s Republic of China. J. Med. Virol. 2008, 80, 670–679. [Google Scholar] [CrossRef]
- Nix, W.A.; Khetsuriani, N.; Peñaranda, S.; Maher, K.; Venczel, L.; Cselkó, Z.; Freire, M.C.; Cisterna, D.; Lema, C.L.; Rosales, P.; et al. Diversity of picornaviruses in rural Bolivia. J. Gen. Virol. 2013, 94, 2017–2028. [Google Scholar] [CrossRef] [PubMed]
- Sadeuh-Mba, S.A.; Bessaud, M.; Joffret, M.L.; Endegue Zanga, M.C.; Balanant, J.; Mpoudi Ngole, E.; Njouom, R.; Reynes, J.M.; Delpeyroux, F.; Rousset, D. Characterization of Enteroviruses from Nonhuman Primates in Cameroon Revealed Virus Types Widespread in Humans along with Candidate New Types and Species. PLoS Negl. Trop. Dis. 2014, 8, e3052. [Google Scholar] [CrossRef] [PubMed]
- Aw, T.G.; Gin, K.Y. Environmental surveillance and molecular characterization of human enteric viruses in tropical urban wastewaters. J. Appl. Microbiol. 2010, 109, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, M.; Martin, J. Detection by Direct Next Generation Sequencing Analysis of Emerging Enterovirus D68 and C109 Strains in an Environmental Sample From Scotland. Front. Microbiol. 2018, 9, 1956. [Google Scholar] [CrossRef] [PubMed]
| Years | No. of Sequences from Feces | No. of Sequences from Conjunctival Swabs | Nucleotide Identity (%) | |||
|---|---|---|---|---|---|---|
| 3C | VP1 | 3C | VP1 | 3C | VP1 | |
| 1997 | 16 | 14 | 25 | 22 | 99.0–100 | 99.1–100 |
| 2003 | 16 | 16 | 23 | 23 | 97.7–100 | 97.3–100 |
| 2008–2009 | 2 | 2 | 14 | - | 99.4–100 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca, M.C.; Pupo-Meriño, M.; García-González, L.A.; Muné, M.; Resik, S.; Norder, H.; Sarmiento, L. Molecular Characterization of Coxsackievirus A24v from Feces and Conjunctiva Reveals Epidemiological Links. Microorganisms 2021, 9, 531. https://doi.org/10.3390/microorganisms9030531
Fonseca MC, Pupo-Meriño M, García-González LA, Muné M, Resik S, Norder H, Sarmiento L. Molecular Characterization of Coxsackievirus A24v from Feces and Conjunctiva Reveals Epidemiological Links. Microorganisms. 2021; 9(3):531. https://doi.org/10.3390/microorganisms9030531
Chicago/Turabian StyleFonseca, Magilé C., Mario Pupo-Meriño, Luis A. García-González, Mayra Muné, Sonia Resik, Heléne Norder, and Luis Sarmiento. 2021. "Molecular Characterization of Coxsackievirus A24v from Feces and Conjunctiva Reveals Epidemiological Links" Microorganisms 9, no. 3: 531. https://doi.org/10.3390/microorganisms9030531
APA StyleFonseca, M. C., Pupo-Meriño, M., García-González, L. A., Muné, M., Resik, S., Norder, H., & Sarmiento, L. (2021). Molecular Characterization of Coxsackievirus A24v from Feces and Conjunctiva Reveals Epidemiological Links. Microorganisms, 9(3), 531. https://doi.org/10.3390/microorganisms9030531








