The New Generation hDHODH Inhibitor MEDS433 Hinders the In Vitro Replication of SARS-CoV-2 and Other Human Coronaviruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Compounds
2.2. Cells and Viruses
2.3. Cytotoxicity Assays
2.4. Antiviral Assays
2.5. Kidney Organoid Generation and SARS-CoV-2 Infection
2.6. Immunofluorescence
2.7. Statistical Analysis
3. Results
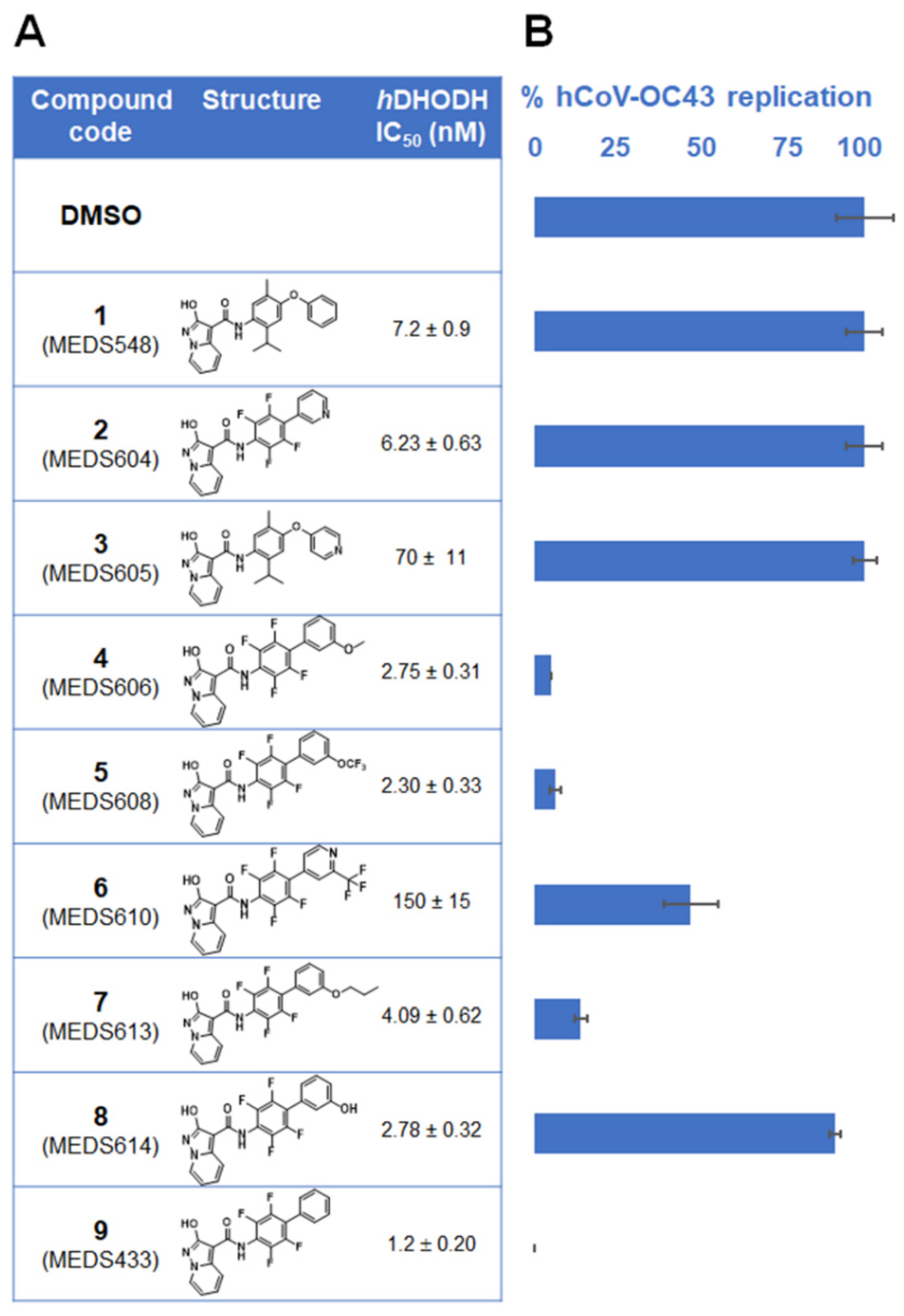
3.1. Identification of a New hDHODH Inhibitor Active against a Human Coronavirus
3.2. Inhibition of α- and β-Coronavirus Replication by the hDHODH Inhibitor MEDS433
3.3. MEDS433 Exerts an Antiviral Activity against SARS-CoV-2
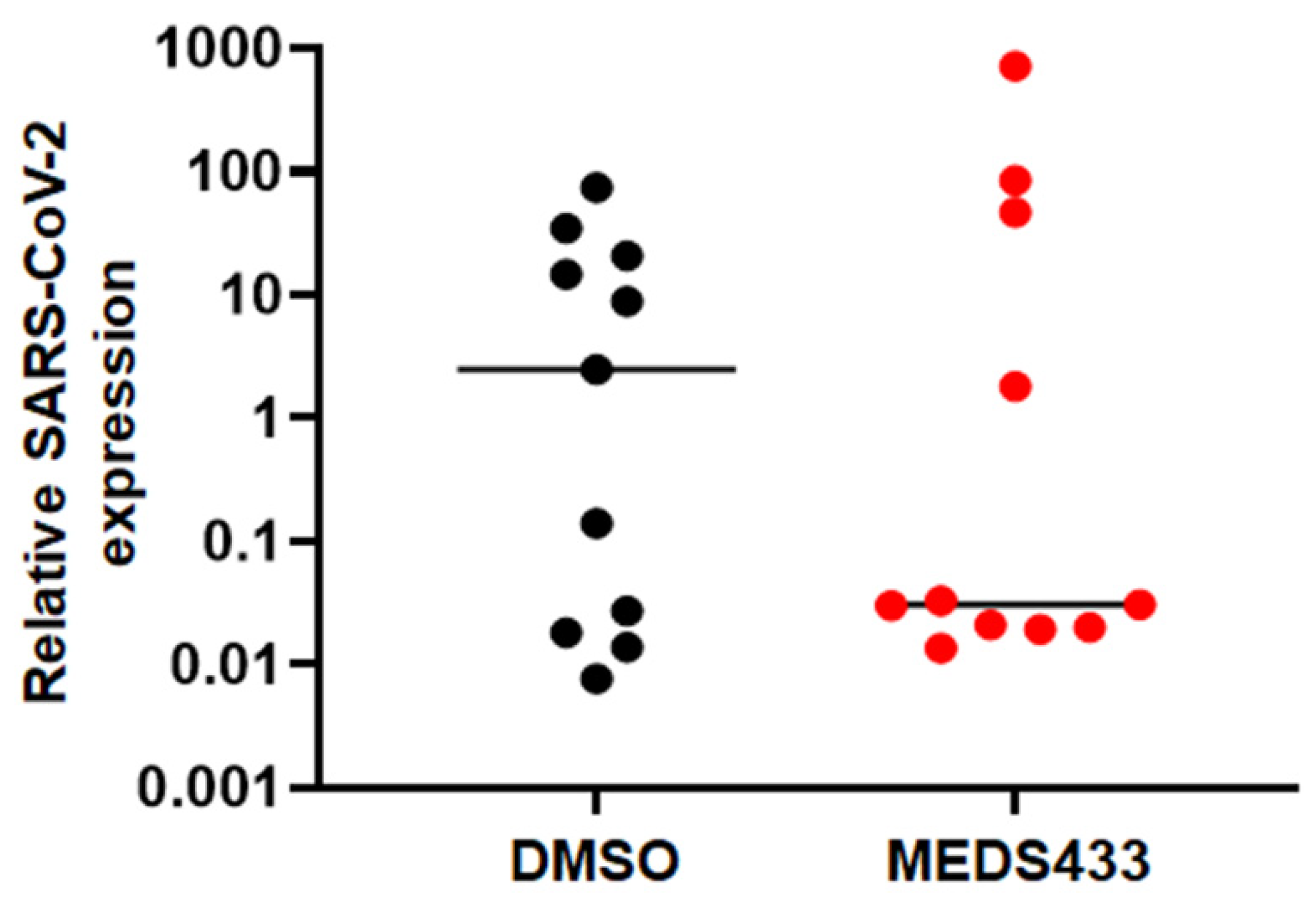
3.4. MEDS433 Affects SARS-CoV-2 Replication in Kidney Organoids
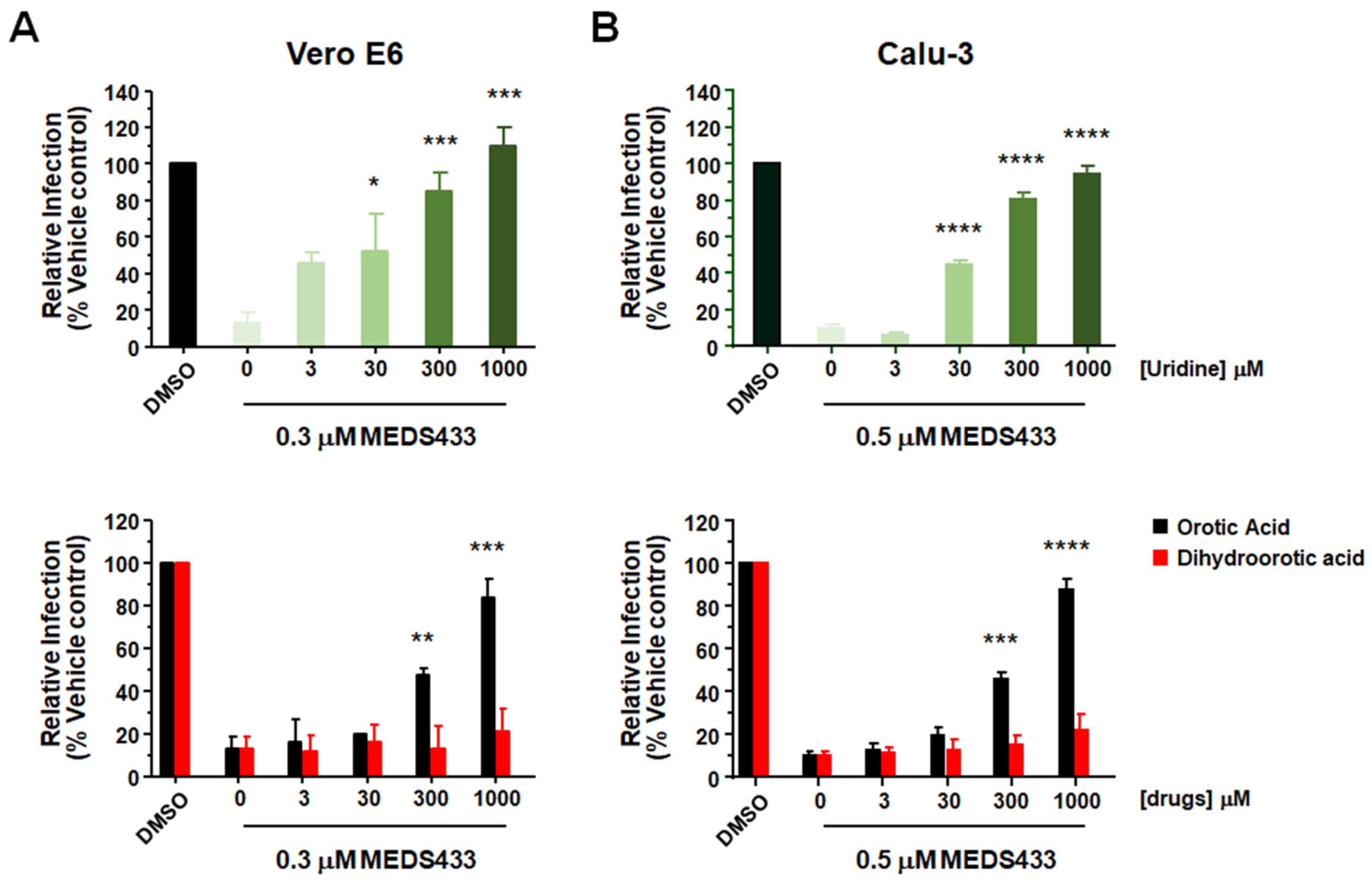
3.5. The Antiviral Activity of MEDS433 Is Reversed by Uridine and Orotic Acid
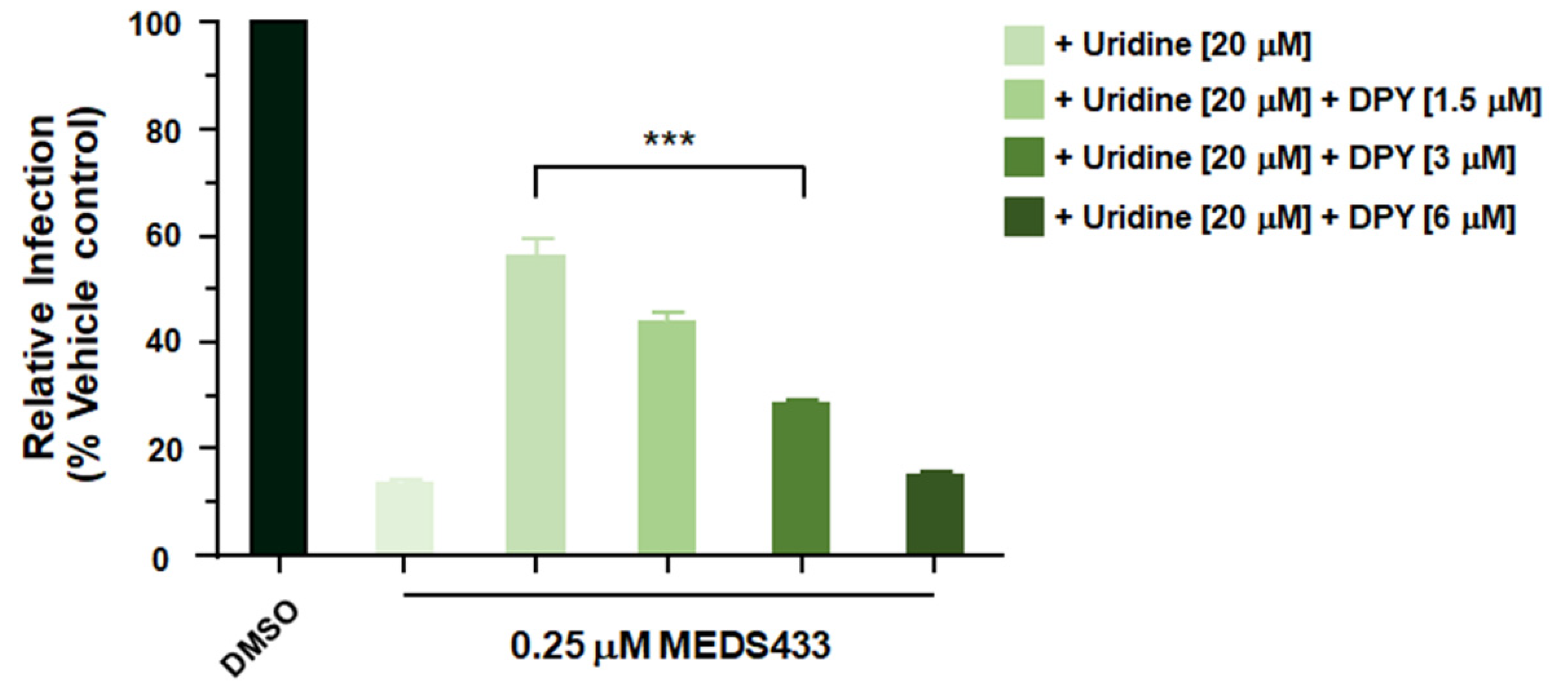
3.6. A Combination of MEDS433 with an Inhibitor of the Pyrimidine Salvage Pathway Is Effective against SARS-CoV-2 Replication Even in the Presence of Uridine
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Tripathi, T. One-year update on the COVID-19 pandemic: Where are we now? Acta Tropica 2021, 214, 105778. [Google Scholar] [CrossRef] [PubMed]
- Forni, G.; Mantovani, A.; on behalf of the COVID-19 Commission of Accademia Nazionale dei Lincei, Rome. COVID-19 vaccines: Where we stand and challenges ahead. Cell Death Differ. 2021, 28, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Richman, D.D. Antiviral drug discovery to address the COVID-19 pandemic. mBio 2020, 11, e02134-20. [Google Scholar] [CrossRef]
- Zumla, A.; Chan, J.F.W.; Azhar, E.I.; Hui, D.S.C.; Yuen, K.-Y. Coronaviruses. Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [Green Version]
- Totura, A.L.; Bavari, S. Broad-spectrum coronavirus antiviral drug discovery. Expert Opin. Drug Discov. 2019, 14, 397–412. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef] [Green Version]
- Tian, D.; Liu, Y.; Liang, C.; Xin, L.; Xie, X.; Zhang, D.; Wang, M.; Li, H.; Fu, X.; Liu, H.; et al. An update review of emerging small-molecule therapeutic options for COVID-19. Biomed. Pharmacother. 2021, 137, 111313. [Google Scholar] [CrossRef] [PubMed]
- Okesli, A.; Khosla, C.; Bassik, M.C. Human pyrimidine nucleotide biosynthesis as a target for antiviral chemotherapy. Curr. Opin. Biotech. 2017, 48, 127–134. [Google Scholar] [CrossRef]
- Reis, R.A.G.; Calil, F.A.; Feliciano, P.R.; Pinheiro, M.P.; Nonato, M.C. The dihydroorotate dehydrogenases: Past and present. Arch. Biochem. Biophys. 2017, 632, 175–191. [Google Scholar] [CrossRef]
- Loeffler, M.; Carrey, E.A.; Knecht, W. The pathway to pyrimidines: The essential focus on dihydroorotate dehydrogenase, the mitochondrial enzyme coupled to the respiratory chain. Nucleosides Nucleotides Nucleic Acids 2020, 39, 1281–1305. [Google Scholar] [CrossRef] [PubMed]
- Boschi, D.; Pippione, A.C.; Sainas, S.; Lolli, M.L. Dihydroorotate dehydrogenase inhibitors in anti-infective drug research. Eur. J. Med. Chem. 2019, 183, 111681. [Google Scholar] [CrossRef]
- Coehlo, A.R.; Oliveira, P.J. Dihydroorotate dehydrogenase inhibitors in SARS-CoV-2 infection. Eur. J. Clin. Investig. 2020, 50, e13366. [Google Scholar]
- Peters, G.J. Re-evaluation of Brequinar sodium, a dihydroorotate dehydrogenase inhibitor. Nucleosides Nucleotides Nucleic Acids 2018, 37, 666–678. [Google Scholar] [CrossRef]
- Sainas, S.; Pippione, A.C.; Lupino, E.; Giorgis, M.; Circosta, P.; Gaidano, V.; Goyal, P.; Bonanni, D.; Rolando, B.; Cignetti, A.; et al. Targeting myeloid differentiation using potent 2-Hydroxypyrazolo [1,5- a] pyridine scaffold-based human dihydroorotate dehydrogenase inhibitors. J. Med. Chem. 2018, 61, 6034–6055. [Google Scholar] [CrossRef] [PubMed]
- Sainas, S.; Giorgis, M.; Circosta, P.; Gaidano, V.; Bonanni, D.; Pippione, A.C.; Bagnati, R.; Passoni, A.; Qiu, Y.; Cojocaru, C.F.; et al. Targeting acute myelogenous leukemia using potent human dihydroorotate dehydrogenase inhibitors based on the 2-hydroxypyrazolo[1,5-a]pyridine scaffold: SAR of the biphenyl moiety. J. Med. Chem. 2021, 64, 5404–5428. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkruys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 2020, 181, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Gribaudo, G.; Riera, L.; Rudge, T.L.; Caposio, P.; Johnson, L.F.; Landolfo, S. Human cytomegalovirus infection induces cellular thymidylate synthase gene expression in quiescent fibroblasts. J. Gen. Virol. 2002, 83, 2983–2993. [Google Scholar] [CrossRef]
- Brown, A.J.; Wona, J.J.; Graham, R.L.; Dinnon III, K.H.; Sims, A.C.; Feng, J.Y.; Cihlarb, T.; Denison, M.R.; Baric, R.S.; Sheahan, T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNAdependent RNA polymerase. Antivir Res. 2019, 169, 104541. [Google Scholar] [CrossRef]
- Citarella, A.; Gentile, D.; Rescifina, A.; Piperno, A.; Mognetti, B.; Gribaudo, G.; Sciortino, M.T.; Holzer, W.; Pace, V.; Micale, N. Pseudo-dipeptide bearing α,α-difluoromethyl ketone moiety as electrophilic warhead with activity against coronaviruses. Int. J. Mol. Sci. 2021, 22, 1398. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020, 3, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Kenneth, J.; Livak, T.; Schmittgen, D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 4, 402–408. [Google Scholar]
- Garreta, E.; Prado, P.; Tarantino, C.; Oria, R.; Fanlo, L.; Martì, E.; Zalvidea, D.; Trepat, X.; Roca-Cusachs, P.; Gavalda-Navarro, A.; et al. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat. Mater. 2019, 18, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K. In vitro and animal models for SARS-CoV-2 research. Trends Pharmacol. Sci. 2020, 41, 513–516. [Google Scholar] [CrossRef]
- Ahmadian, E.; Hosseiniyan Khatibi, S.M.; Razi Soofiyani, S.; Abediazar, S.; Shoja, M.M.; Ardalan, M. Covid-19 and kidney injury: Pathophysiology and molecular mechanisms. Rev. Med. Virol. 2021, 31, e2176. [Google Scholar] [CrossRef]
- Gabarre, P.; Dumas, G.; Dupont, T.; Darmon, M.; Azoulay, E.; Zafrani, L. Acute kidney injury in critically ill patients with COVID-19. Intensive Care Med. 2020, 46, 1339–1348. [Google Scholar] [CrossRef]
- Braun, B.; Lütgehetmann, M.; Pfefferle, S.; Wong, M.N.; Carsten, A.; Lindenmeyer, M.T.; Nörz, D.; Heinrich, F.; Meißner, K.; Wichmann, D.; et al. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet 2020, 396, 597–598. [Google Scholar] [CrossRef]
- Caceres, P.; Savickas, G.; Murray, S.; Umanath, K.; Uduman, J.; Yee, J.; Liao, T.D.; Bolin, S.; Levin, A.; Khan, M.; et al. High SARS-CoV-2 viral load in urine sediment correlates with acute kidney injury and poor COVID-19 outcome. J. Am. Soc. Nephrol. 2021, SN.2021010059, Online ahead of print. [Google Scholar] [CrossRef]
- Danilczyk, U.; Penninger, J.M. Angiotensin-converting enzyme II in the heart and the kidney. Circ. Res. 2006, 98, 463–471. [Google Scholar] [CrossRef]
- Fitzgerald, G.A. Dipyridamole. N. Engl. J. Med. 1987, 316, 1247–1257. [Google Scholar]
- Pizzorno, G.; Cao, D.; Leffert, J.J.; Russell, R.L.; Zhang, D.; Handschumacher, R.E. Homeostatic control of uridine and the role of uridine phosphorylase: A biological and clinical update. Biochim. Biophys. Acta 2002, 1587, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Deans, R.M.; Morgens, D.W.; Okesli, A.; Pillay, S.; Horlbeck, M.A.; Kampmann, M.; Gilbert, L.A.; Li, A.; Mateo, R.; Smith, M.; et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat. Chem. Biol. 2016, 12, 361–366. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Cao, L.; Gao, H.; Wu, Y.; Wang, Y.; Fang, F.; Lan, T.; Lou, Z.; Rao, Y. Discovery, optimization, and target identification of novel potent broad-spectrum antiviral inhibitors. J. Med. Chem. 2019, 62, 4056–4073. [Google Scholar] [CrossRef]
- Adalja, A.; Inglesby, T. Broad-spectrum antiviral agents: A crucial pandemic tool. Expert Rev. Anti Infec. Ther. 2019, 17, 467–470. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Jiang, H. Potential treatment of COVID-19 by inhibitors of human dihydroorotate dehydrogenase. Protein Cell 2020, 11, 699–702. [Google Scholar] [CrossRef]
- Luban, J.; Sattler, R.; Mühlberger, E.; Graci, J.D.; Cao, L.; Weetall, M.; Trotta, C.; Colacino, J.M.; Bavari, S.; Strambio-De-Castillia, C.; et al. The DHODH inhibitor PTC299 arrests SARS-CoV-2 replication and suppresses induction of inflammatory cytokines. Virus Res. 2020, 292, 198246. [Google Scholar] [CrossRef]
- Hahn, F.; Wangen, C.; Häge, S.; Peter, A.S.; Dobler, G.; Hurst, B.; Julander, J.; Fuchs, J.; Ruzsics, Z.; Überla, K.; et al. IMU-838, a developmental DHODH inhibitor in phase II for autoimmune disease, shows anti-SARS-CoV-2 and broad-spectrum antiviral efficacy in vitro. Viruses 2020, 12, 1394. [Google Scholar] [CrossRef]
- Immunic Therapeutics USA. Main Phase 2 Analysis CALVID-1 Trial of IMU-838 in Moderate COVID-19. Available online: https://www.immunic-therapeutics.com (accessed on 15 July 2021).
- Schaper, W. Dipyridamole, an underestimated vascular protective drug. Cardiovasc. Drugs Ther. 2005, 19, 357–363. [Google Scholar] [CrossRef]
- Ward, J.L.; Sherali, A.; Mo, Z.P.; Tse, C.M. Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter- deficient PK15 cells: ENT2 exhibits a low affinity for guanosine and cytidine but a high affinity for inosine. J. Biol. Chem. 2000, 275, 8375–8381. [Google Scholar] [PubMed] [Green Version]
- Wang, Q.Y.; Bushell, S.; Qing, M.; Xu, H.Y.; Bonavia, A.; Nunes, S.; Zhou, J.; Poh, M.K.; Florez de Sessions, P.; Niyomrattanakit, P.; et al. Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J. Virol. 2011, 85, 6548–6556. [Google Scholar] [CrossRef] [Green Version]
- Smee, D.F.; Hurst, B.L.; Day, C.W. D282, a non-nucleoside inhibitor of influenza virus infection that interferes with de novo pyrimidine biosynthesis. Antivir. Chem. Chemother. 2012, 22, 263–272. [Google Scholar] [CrossRef]
- Grandin, C.; Lucas-Hourani, M.; Janin, Y.L.; Dauzonne, D.; Munier-Lehmann, H.; Paturet, A.; Taborik, F.; Vabret, A.; Contamin, H.; Tangy, F.; et al. Respiratory syncytial virus infection in macaques is not suppressed by intranasal sprays of pyrimidine biosynthesis inhibitors. Antiviral Res. 2016, 125, 58–62. [Google Scholar] [CrossRef]
- Gaidano, V.; Houshmand, M.; Vitale, N.; Carrà, G.; Morotti, A.; Tenace, V.; Rapelli, S.; Sainas, S.; Pippione, A.C.; Giorgis, M.; et al. The synergism between DHODH inhibitors and dipyridamole leads to metabolic lethality in acute myeloid leukemia. Cancers 2021, 13, 1003. [Google Scholar] [CrossRef]
- Luganini, A.; Sibille, G.; Mognetti, B.; Sainas, S.; Pippione, A.C.; Giorgis, M.; Boschi, D.; Lolli, M.L.; Gribaudo, G. Effective deploying of a novel DHODH inhibitor against herpes simplex type 1 and type 2 replication. Antiviral Res. 2021, 189, 105057. [Google Scholar] [CrossRef]
- Gregov, D.; Jenkins, A.; Duncan, E.; Sieber, D.; Rodgers, S.; Duncan, B.; Bochner, F.; Lloyd, J. Dipyridamole: Pharmacokinetics and effects on aspects of platelet function in man. Br. J. Clin. Pharmac. 1987, 24, 425–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, R.; Zhang, L.; Li, S.; Sun, Y.; Ding, M.; Wang, Y.; Zhao, Y.; Wu, Y.; Shang, W.; Jiang, X.; et al. Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly emerged coronavirus SARS-CoV-2. Protein Cell 2020, 11, 723–739. [Google Scholar] [CrossRef] [PubMed]
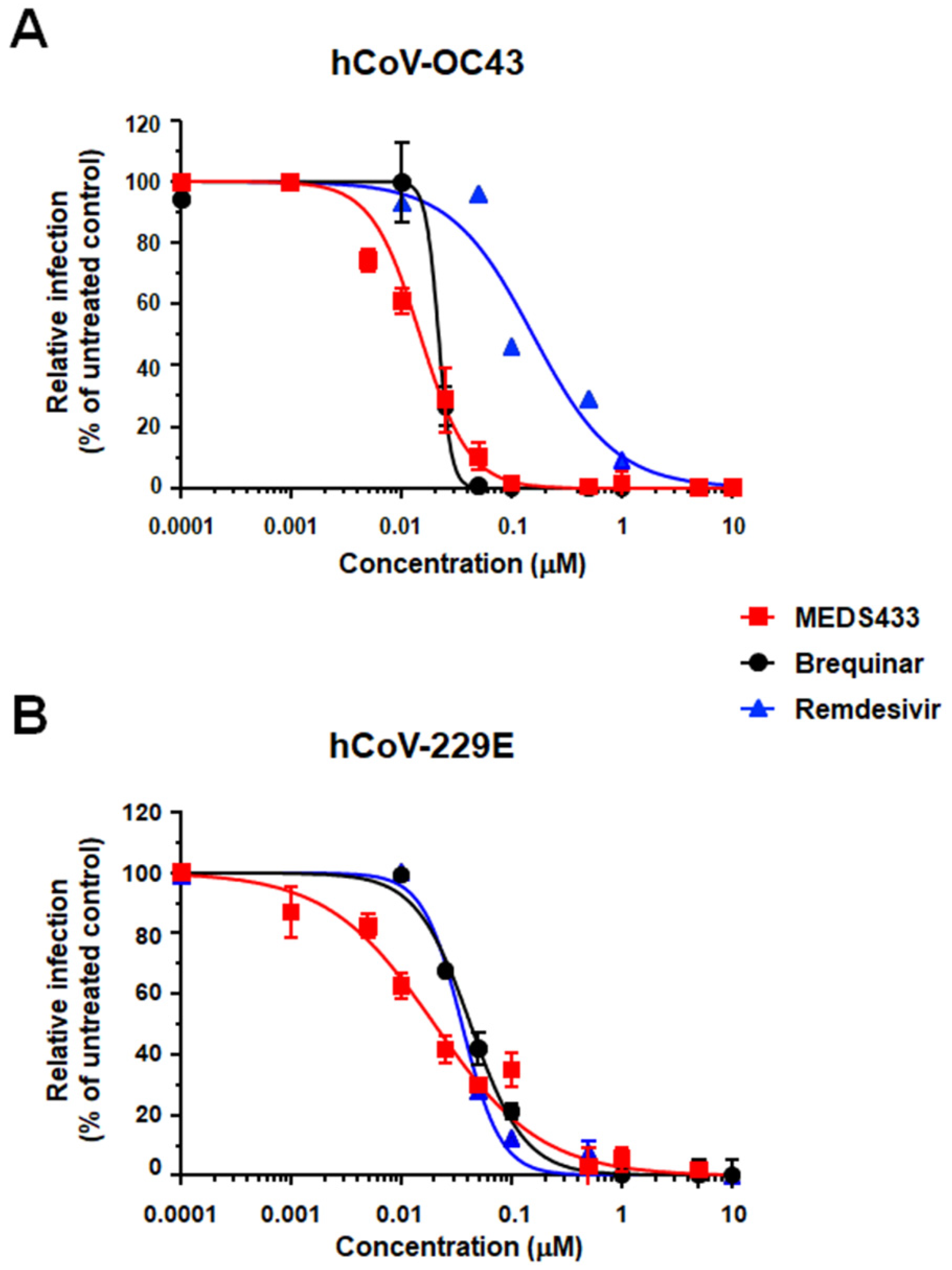
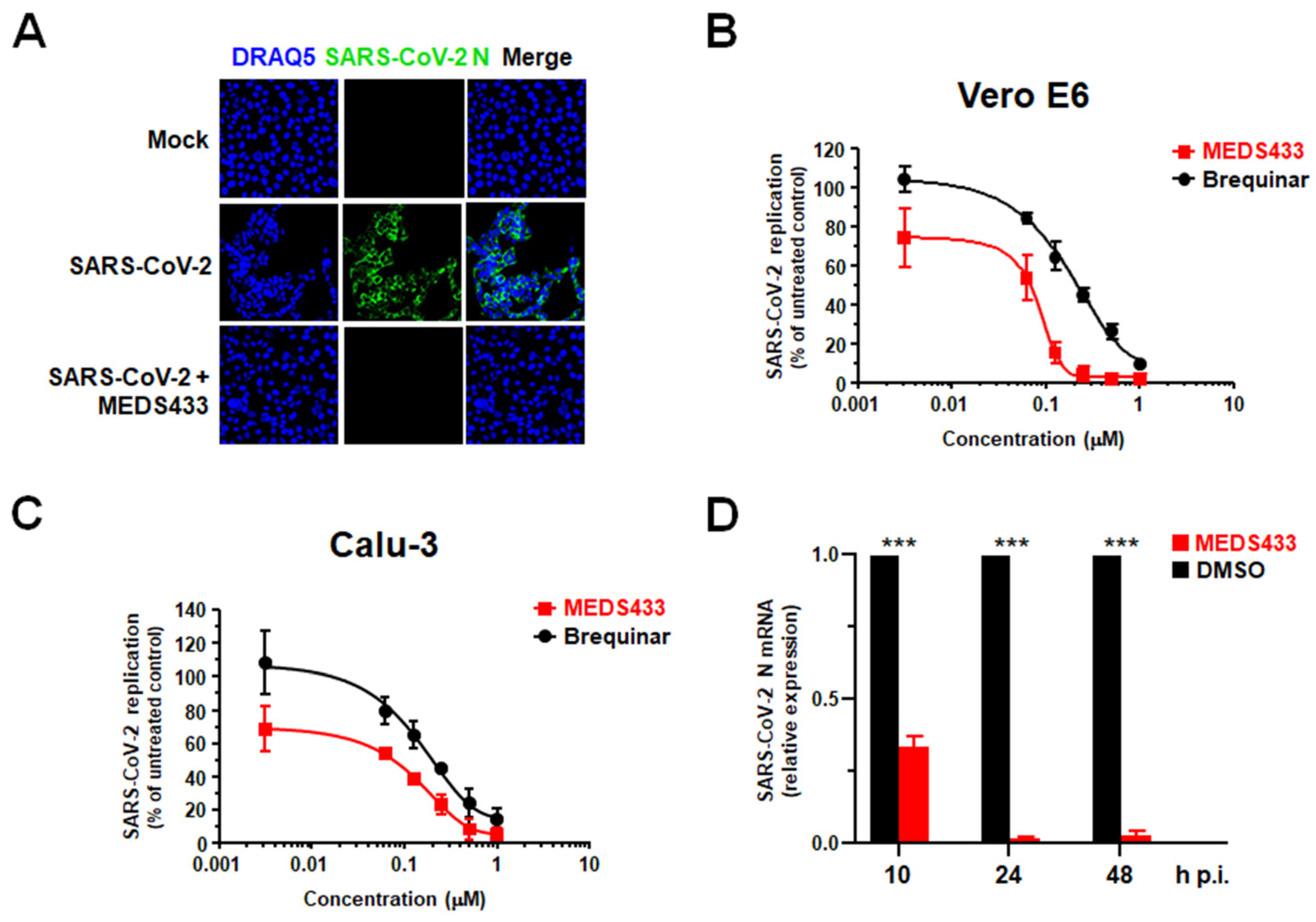
| hCoV | Cell Line | EC50 (μM) a | EC90 (μM) b | CC50 (μM) c | SI d |
|---|---|---|---|---|---|
| hCoV-OC43 | HCT-8 | 0.012 ± 0.003 | 0.044 ± 0.021 | 78.48 ± 4.60 | 6329 |
| hCoV-229E | MRC5 | 0.022 ± 0.003 | 0.288 ± 0.040 | 104.80 ± 19.75 | 4763 |
| SARS-CoV-2 | Vero E6 | 0.063 ± 0.004 | 0.136 ± 0.007 | >500 | >7900 |
| SARS-CoV-2 | Calu-3 | 0.076 ± 0.005 | 0.513 ± 0.016 | >125 | >1600 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calistri, A.; Luganini, A.; Mognetti, B.; Elder, E.; Sibille, G.; Conciatori, V.; Del Vecchio, C.; Sainas, S.; Boschi, D.; Montserrat, N.; et al. The New Generation hDHODH Inhibitor MEDS433 Hinders the In Vitro Replication of SARS-CoV-2 and Other Human Coronaviruses. Microorganisms 2021, 9, 1731. https://doi.org/10.3390/microorganisms9081731
Calistri A, Luganini A, Mognetti B, Elder E, Sibille G, Conciatori V, Del Vecchio C, Sainas S, Boschi D, Montserrat N, et al. The New Generation hDHODH Inhibitor MEDS433 Hinders the In Vitro Replication of SARS-CoV-2 and Other Human Coronaviruses. Microorganisms. 2021; 9(8):1731. https://doi.org/10.3390/microorganisms9081731
Chicago/Turabian StyleCalistri, Arianna, Anna Luganini, Barbara Mognetti, Elizabeth Elder, Giulia Sibille, Valeria Conciatori, Claudia Del Vecchio, Stefano Sainas, Donatella Boschi, Nuria Montserrat, and et al. 2021. "The New Generation hDHODH Inhibitor MEDS433 Hinders the In Vitro Replication of SARS-CoV-2 and Other Human Coronaviruses" Microorganisms 9, no. 8: 1731. https://doi.org/10.3390/microorganisms9081731
APA StyleCalistri, A., Luganini, A., Mognetti, B., Elder, E., Sibille, G., Conciatori, V., Del Vecchio, C., Sainas, S., Boschi, D., Montserrat, N., Mirazimi, A., Lolli, M. L., Gribaudo, G., & Parolin, C. (2021). The New Generation hDHODH Inhibitor MEDS433 Hinders the In Vitro Replication of SARS-CoV-2 and Other Human Coronaviruses. Microorganisms, 9(8), 1731. https://doi.org/10.3390/microorganisms9081731










