Cultivation of Spore-Forming Gut Microbes Using a Combination of Bile Acids and Amino Acids
Abstract
1. Introduction
2. Materials and Methods
2.1. Fecal Sample Collection
2.2. Spore Purification
2.3. Germination-Inducing Substances
2.4. Germination and Culturing
2.5. 16S rRNA Gene Amplicon Sequencing and Analysis
2.6. Statistical Analyses
2.7. Accession Number of 16S rRNA Gene Sequences
2.8. Ethics Approval
3. Results
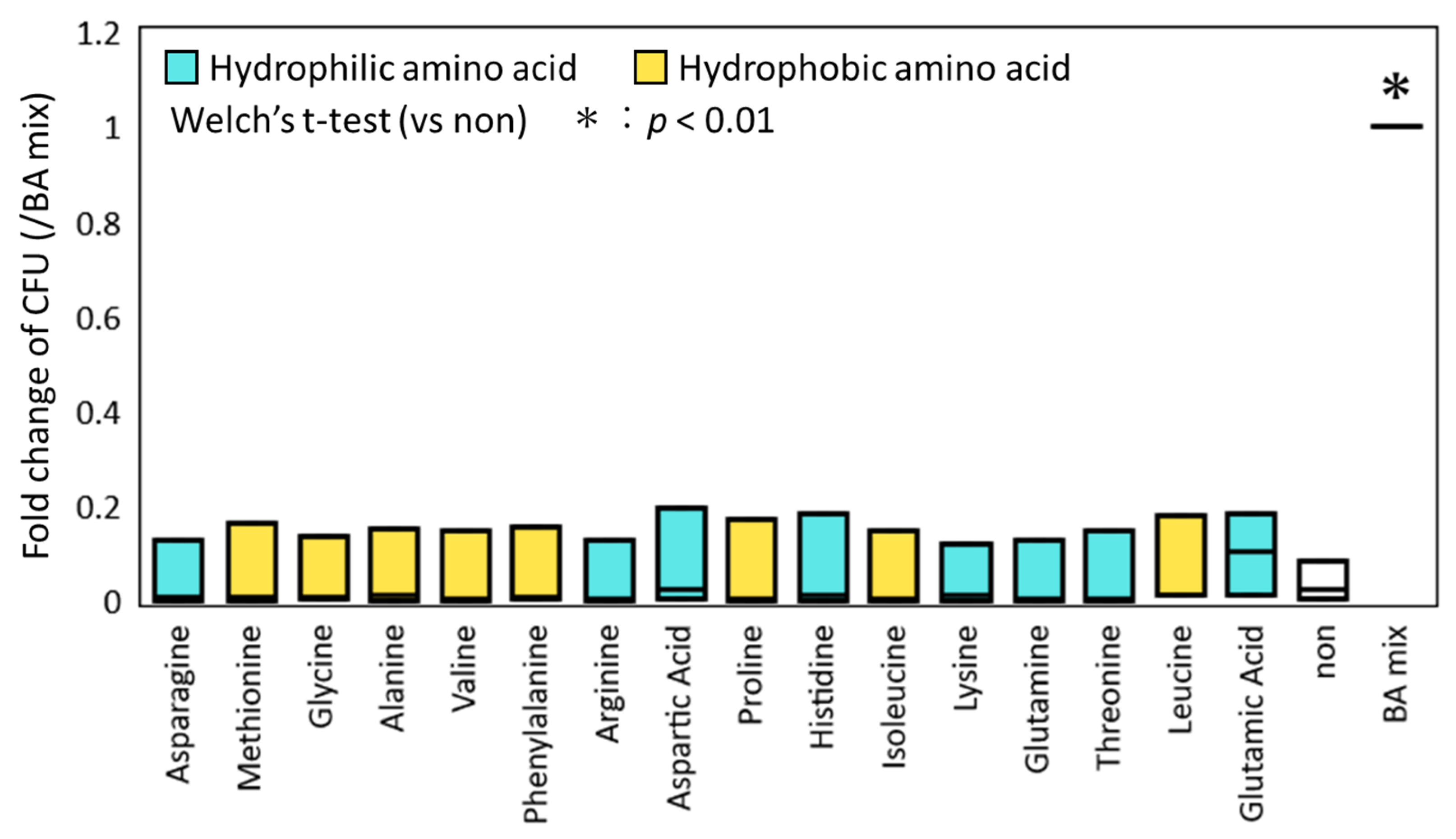
3.1. Effect of Inidividual AAs on the Germination and Colony-Inducing Activities for Spore-Forming Bacteria in Feces
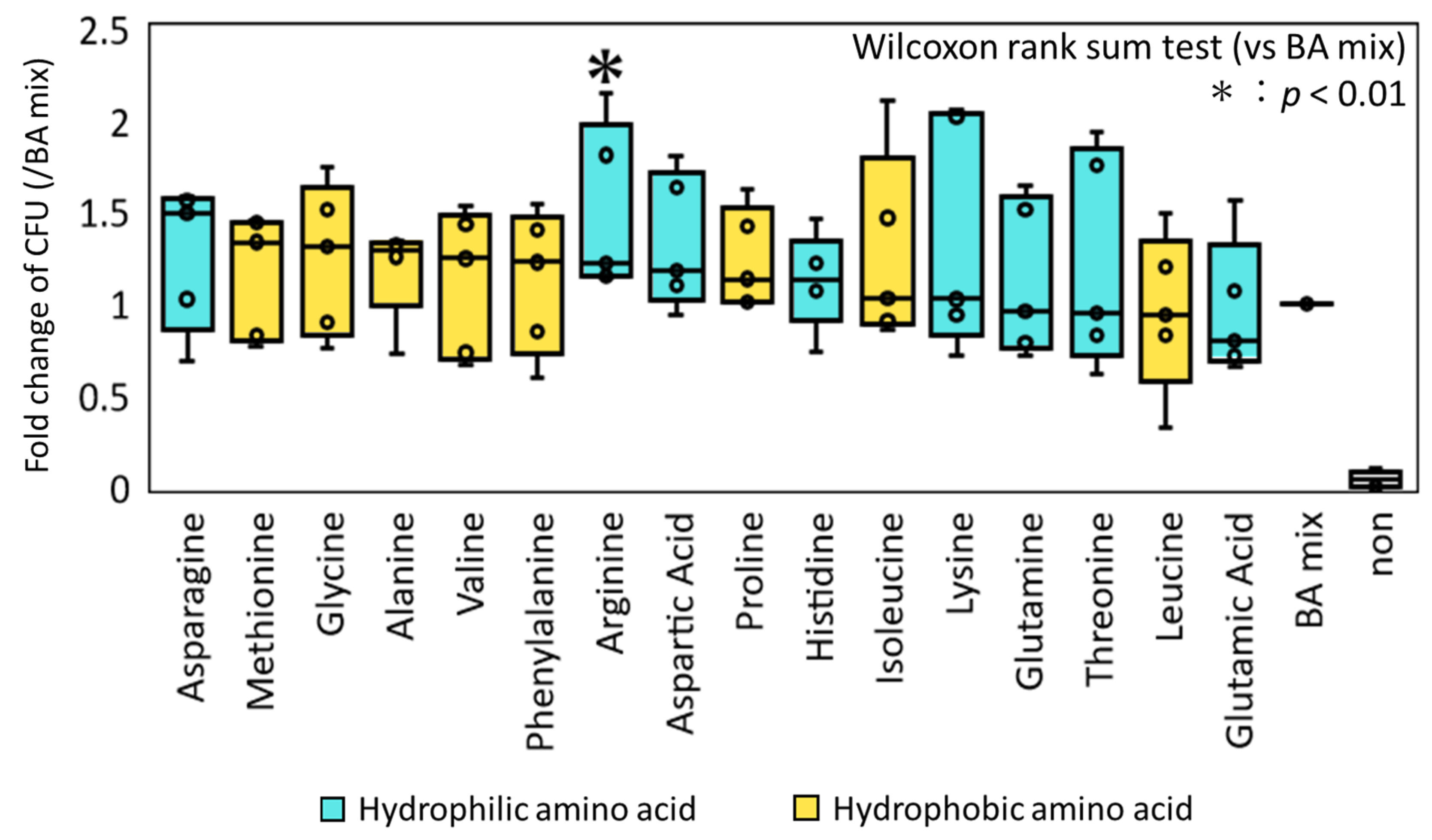
3.2. Culturing of Spore-Forming Bacteria in Feces Using a Combination of Various AAs with BAs
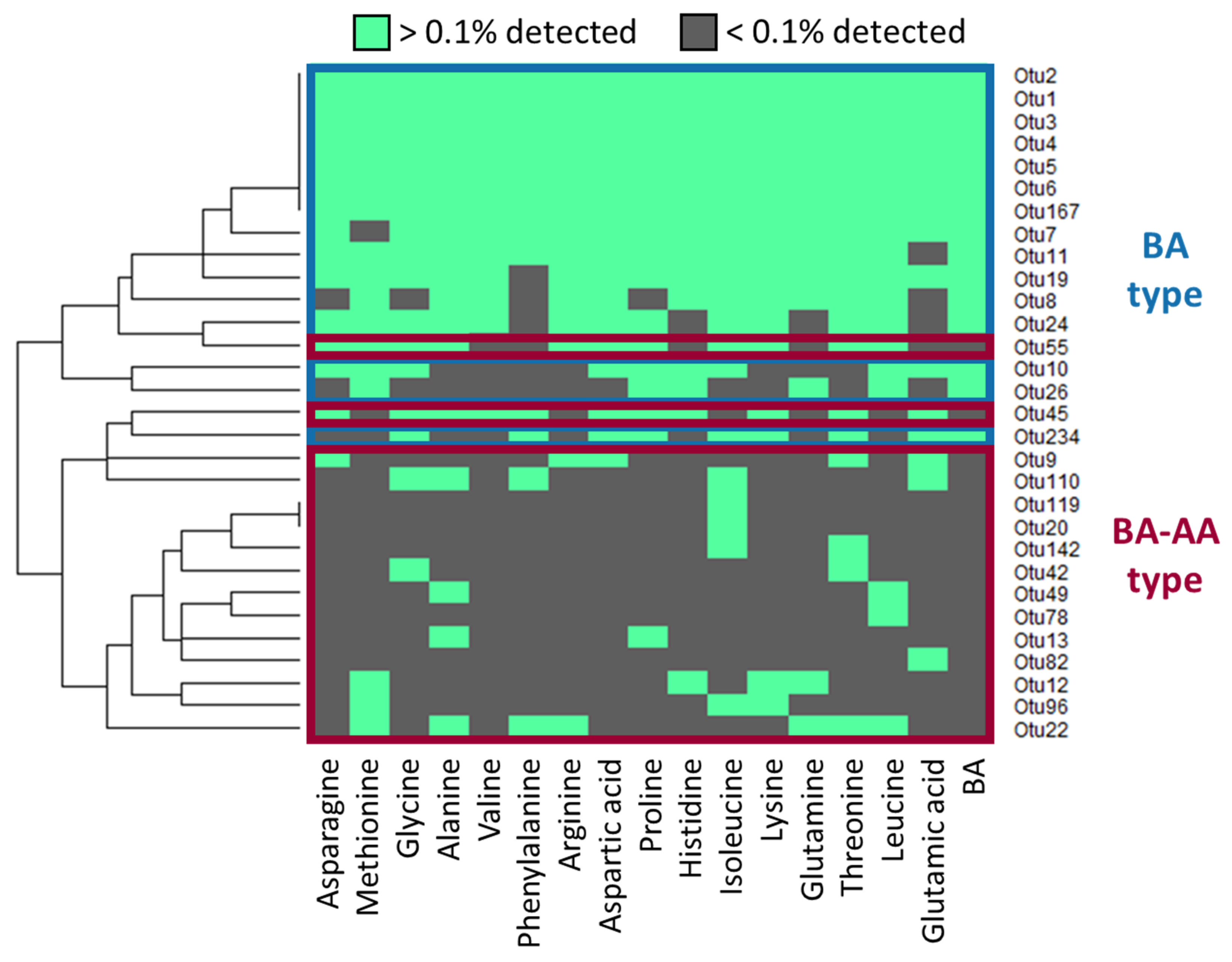
3.3. Classification of Germinating Bacteria by Responsiveness to AAs and BAs
3.4. Phylogenetic Analysis of Bacterial Species Germinated by the Combination of BAs and/or AAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Lora, V.H.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Tarashi, K.A. The Role of Gut Microbiota in the Development and Function of Intestinal T cells. J. Intest. Microbiol. 2015, 29, 1–7. [Google Scholar] [CrossRef]
- Fraune, S.; Bosch, T.C.G. Why bacteria matter in animal development and evolution. BioEssays 2010, 32, 571–580. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Köller, Y.; McCoy, K.D. The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol. 2015, 36, 460–470. [Google Scholar] [CrossRef]
- Khan, M.T.; Nieuwdorp, M.; Bäckhed, F. Microbial modulation of insulin sensitivity. Cell Metab. 2014, 20, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Shi, Z.J.; Seshadri, R.; Pollard, K.S.; Kyrpides, N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature 2019, 568, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y. Genome diversity of spore-forming firmicutes. In The Bacterial Spore: From Molecules to Systems; ASM Press: Washington, DC, USA, 2016; pp. 1–18. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Gerding, D.N.; Muto, C.A.; Owens, R.C., Jr. Measures to Control and Prevent Clostridium difficile Infection. Clin. Infect. Dis. 2008, 46, S43–S49. [Google Scholar] [CrossRef] [PubMed]
- Lawley, T.D.; Clare, S.; Walker, A.W.; Goulding, D.; Stabler, R.A.; Croucher, N.; Mastroeni, P.; Scott, P.; Raisen, C.; Mottram, L.; et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 2009, 77, 3661–3669. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.B.; Allen, C.A.; Shrestha, R.; Sorg, J.A. Bile Acid Recognition by the Clostridium difficile Germinant Receptor, CspC, Is Important for Establishing Infection. PLoS Pathog. 2013, 9, e1003356. [Google Scholar] [CrossRef]
- Moir, A. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J. Bacteriol. 1981, 146, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef]
- Zwicker, B.L.; Agellon, L.B. Transport and biological activities of bile acids. Int. J. Biochem. Cell Biol. 2013, 45, 1389–1398. [Google Scholar] [CrossRef]
- Russell, D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef]
- Hamilton, J.P.; Xie, G.; Raufman, J.P.; Hogan, S.; Griffin, T.L.; Packard, C.A.; Chatfield, D.A.; Hagey, L.R.; Steinbach, J.H.; Hofmann, A.F. Human cecal bile acids: Concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, 256–263. [Google Scholar] [CrossRef]
- Hofmann, A.F. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 1999, 159, 2647–2658. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef]
- Wilson, K.H.; Kennedy, M.J.; Fekety, F.R. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 1982, 15, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Sorg, J.A.; Sonenshein, A.L. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 2010, 192, 4983–4990. [Google Scholar] [CrossRef]
- Sorg, J.A.; Sonenshein, A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Onizuka, S.; Mishima, R.; Nakayama, J. Cultural isolation of spore-forming bacteria in human feces using bile acids. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Pelczar, P.L.; Igarashi, T.; Setlow, B.; Setlow, P. Role of GerD in germination of Bacillus subtilis spores. J. Bacteriol. 2007, 189, 1090–1098. [Google Scholar] [CrossRef]
- Atluri, S.; Ragkousi, K.; Cortezzo, D.E.; Setlow, P. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 2006, 188, 28–36. [Google Scholar] [CrossRef]
- Bhattacharjee, D.; Francis, M.B.; Ding, X.; McAllister, K.N.; Shrestha, R.; Sorg, J.A. Reexamining the germination phenotypes of several Clostridium difficile strains suggests another role for the CspC germinant receptor. J. Bacteriol. 2016, 198, 777–786. [Google Scholar] [CrossRef]
- Jr, P.E.C.; Kaiser, A.M.; Mccolm, S.A.; Bauer, J.M.; Vincent, B.; Aronoff, D.M.; Hanna, P.C. Variation in germination of Clostridium difficile clinical isolates correlates to disease severity. Anaerobe 2016, 33, 64–70. [Google Scholar] [CrossRef]
- Heeg, D.; Burns, D.A.; Cartman, S.T.; Minton, N.P. Spores of clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS ONE 2012, 7, e32381. [Google Scholar] [CrossRef]
- Shen, A.; Edwards, A.N.; Sarker, M.R.; Paredes-Sabja, D. Sporulation and germination in clostridial pathogens. In Gram-Positive Pathogens; ASM Press: Washington, DC, USA, 2019; pp. 903–926. [Google Scholar] [CrossRef]
- Adams, C.M.; Eckenroth, B.E.; Putnam, E.E.; Doublié, S.; Shen, A. Structural and Functional Analysis of the CspB Protease Required for Clostridium Spore Germination. PLoS Pathog. 2013, 9, e1003165. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.B.; Allen, C.A.; Sorg, J.A. Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J. Bacteriol. 2015, 197, 2276–2283. [Google Scholar] [CrossRef] [PubMed]
- Kochan, T.J.; Foley, M.H.; Shoshiev, M.S.; Somers, M.J.; Carlson, P.E.; Hanna, P.C. Updates to Clostridium difficile spore germination. J. Bacteriol. 2018, 200, 1–12. [Google Scholar] [CrossRef]
- Masayama, A.; Hamasaki, K.; Urakami, K.; Shimamoto, S.; Kato, S.; Makino, S.; Yoshimura, T.; Moriyama, M.; Moriyama, R. Expression of germination-related enzymes, CspA, CspB, CspC, SleC, and SleM, of Clostridium perfringens S40 in the mother cell compartment of sporulating cells. Genes Genet. Syst. 2006, 81, 227–234. [Google Scholar] [CrossRef]
- Kevorkian, Y.; Shirley, D.J.; Shen, A.; Genetics, M. Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie 2016, 122, 243–254. [Google Scholar] [CrossRef]
- Setlow, P.; Wang, S.; Li, Y.Q. Germination of Spores of the Orders Bacillales and Clostridiales. Annu. Rev. Microbiol. 2017, 71, 459–477. [Google Scholar] [CrossRef]
- Paredes-Sabja, D.; Setlow, P.; Sarker, M.R. Germination of spores of Bacillales and Clostridiales species: Mechanisms and proteins involved. Trends Microbiol. 2011, 19, 85–94. [Google Scholar] [CrossRef]
- Howerton, A.; Ramirez, N.; Abel-Santos, E. Mapping interactions between germinants and Clostridium difficile spores. J. Bacteriol. 2011, 193, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.; Lockless, S.W.; Sorg, J.A. A Clostridium difficile alanine racemase affects spore germination and accommodates serine as a substrate. J. Biol. Chem. 2017, 292, 10735–10742. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.; Sorg, J.A. Hierarchical recognition of amino acid co-germinants during Clostridioides difficile spore germination. Anaerobe 2018, 49, 41–47. [Google Scholar] [CrossRef]
- Tanaka, M.; Korenori, Y.; Washio, M.; Kobayashi, T.; Momoda, R.; Kiyohara, C.; Kuroda, A.; Saito, Y.; Sonomoto, K.; Nakayama, J. Signatures in the gut microbiota of Japanese infants who developed food allergies in early childhood. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Fisher, N.; Hanna, P. Characterization of Bacillus anthracis germinant receptors in vitro. J. Bacteriol. 2005, 187, 8055–8062. [Google Scholar] [CrossRef][Green Version]
- Luu, H.; Akoachere, M.; Patra, M.; Abel-Santos, E. Cooperativity and interference of germination pathways in Bacillus anthracis spores. J. Bacteriol. 2011, 193, 4192–4198. [Google Scholar] [CrossRef] [PubMed]
- Banawas, S.; Paredes-Sabja, D.; Korza, G.; Li, Y.; Hao, B.; Setlow, P.; Sarker, M.R. The Clostridium perfringens germinant receptor protein GerKC is located in the spore inner membrane and is crucial for spore germination. J. Bacteriol. 2013, 195, 5084–5091. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, D.; McAllister, K.N.; Sorg, J.A. Germinants and their receptors in clostridia. J. Bacteriol. 2016, 198, 2767–2775. [Google Scholar] [CrossRef] [PubMed]
- Bouillaut, L.; Dubois, T.; Sonenshein, A.L.; Dupuy, B. Integration of metabolism and virulence in Clostridium difficile. Res. Microbiol. 2015, 166, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Elsholz, A.K.W.; Turgay, K.; Michalik, S.; Hessling, B.; Gronau, K.; Oertel, D.; Mäder, U.; Bernhardt, J.; Becher, D.; Hecker, M.; et al. Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2012, 109, 7451–7456. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onizuka, S.; Tanaka, M.; Mishima, R.; Nakayama, J. Cultivation of Spore-Forming Gut Microbes Using a Combination of Bile Acids and Amino Acids. Microorganisms 2021, 9, 1651. https://doi.org/10.3390/microorganisms9081651
Onizuka S, Tanaka M, Mishima R, Nakayama J. Cultivation of Spore-Forming Gut Microbes Using a Combination of Bile Acids and Amino Acids. Microorganisms. 2021; 9(8):1651. https://doi.org/10.3390/microorganisms9081651
Chicago/Turabian StyleOnizuka, Sakura, Masaru Tanaka, Riko Mishima, and Jiro Nakayama. 2021. "Cultivation of Spore-Forming Gut Microbes Using a Combination of Bile Acids and Amino Acids" Microorganisms 9, no. 8: 1651. https://doi.org/10.3390/microorganisms9081651
APA StyleOnizuka, S., Tanaka, M., Mishima, R., & Nakayama, J. (2021). Cultivation of Spore-Forming Gut Microbes Using a Combination of Bile Acids and Amino Acids. Microorganisms, 9(8), 1651. https://doi.org/10.3390/microorganisms9081651




