Endemic Jeffrey Pine Beetle Associates: Beetle/Mite Fungal Dissemination Strategies and Interactions That May Influence Beetle Population Levels
Abstract
1. Introduction
2. Materials and Methods
2.1. Bark Beetle Sample Collection
2.2. Isolation of Fungi from Beetle Structures and Mites
2.3. Single Fungal Strain Isolations
2.4. Morphological Determination
2.5. Molecular Data
2.6. Sequences Alignment and Phylogenetic Analyses
2.7. Prevalence of Fungi in Beetle Maxillae and Pellet, and in Mites
2.8. Fungal Growth Rates across a Temperature Gradient and Interactions with Yeast
3. Results
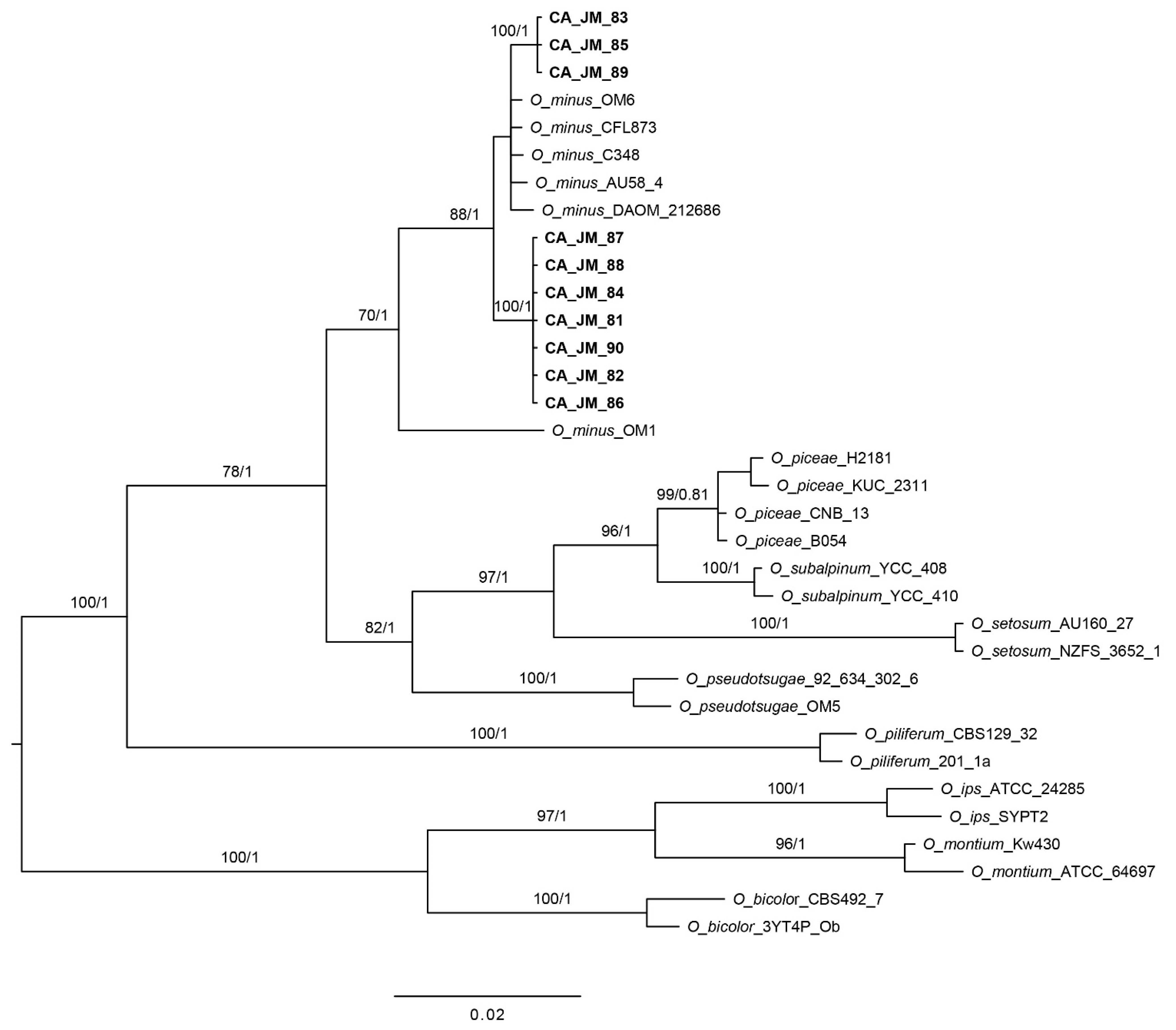
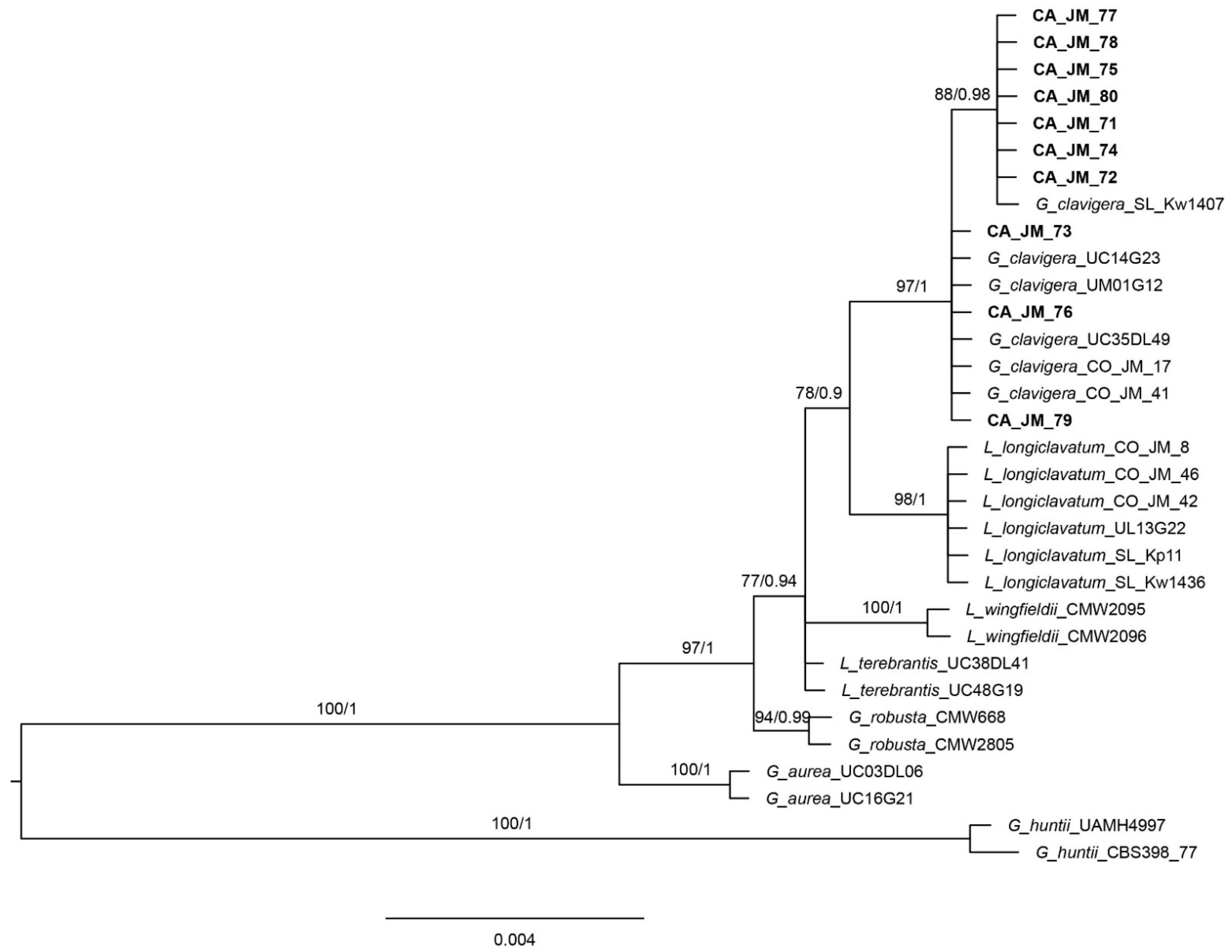
3.1. Phylogenetic Analyses
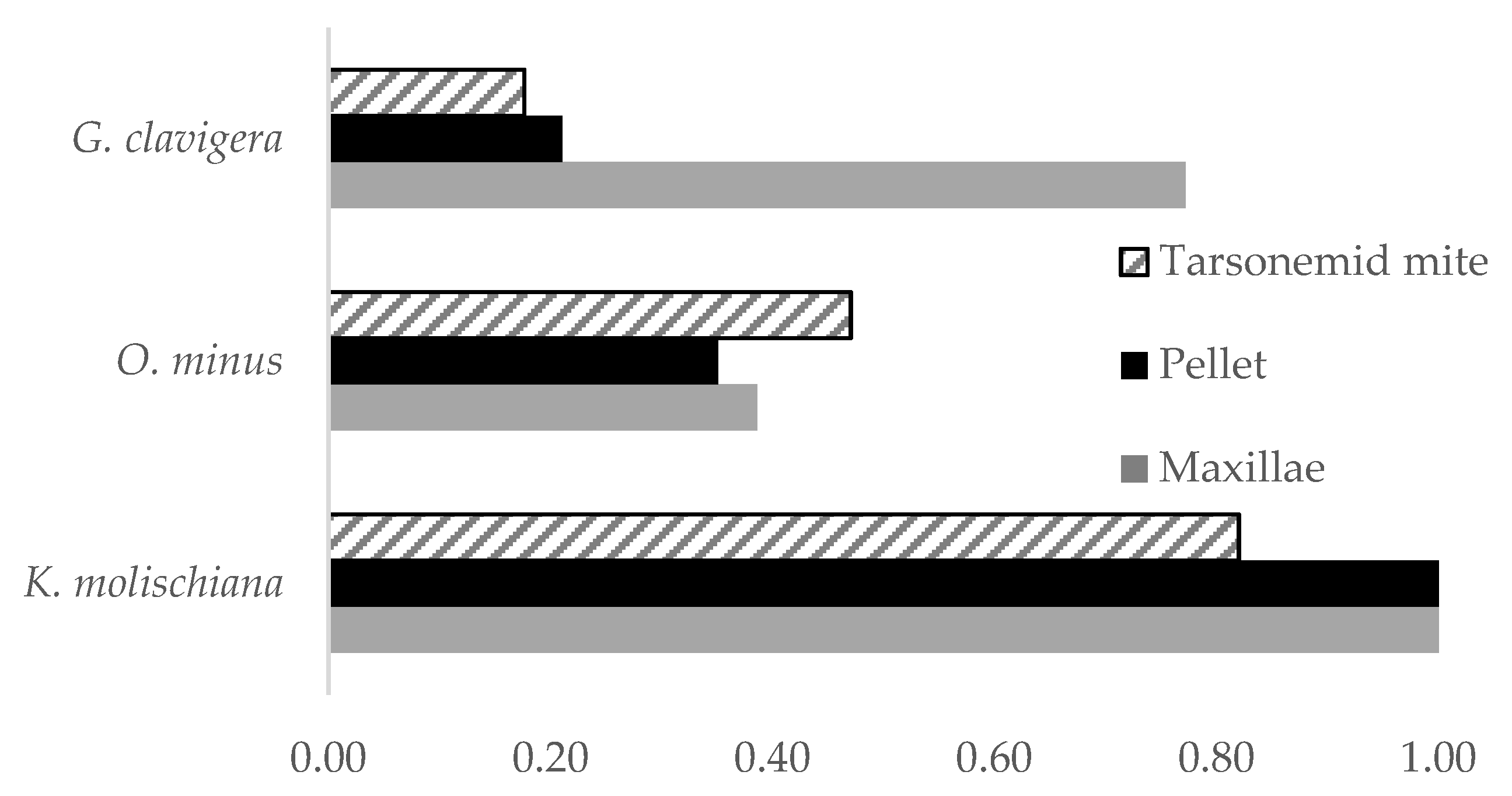
3.2. Prevalence of Fungal Associates on Beetles and Mites
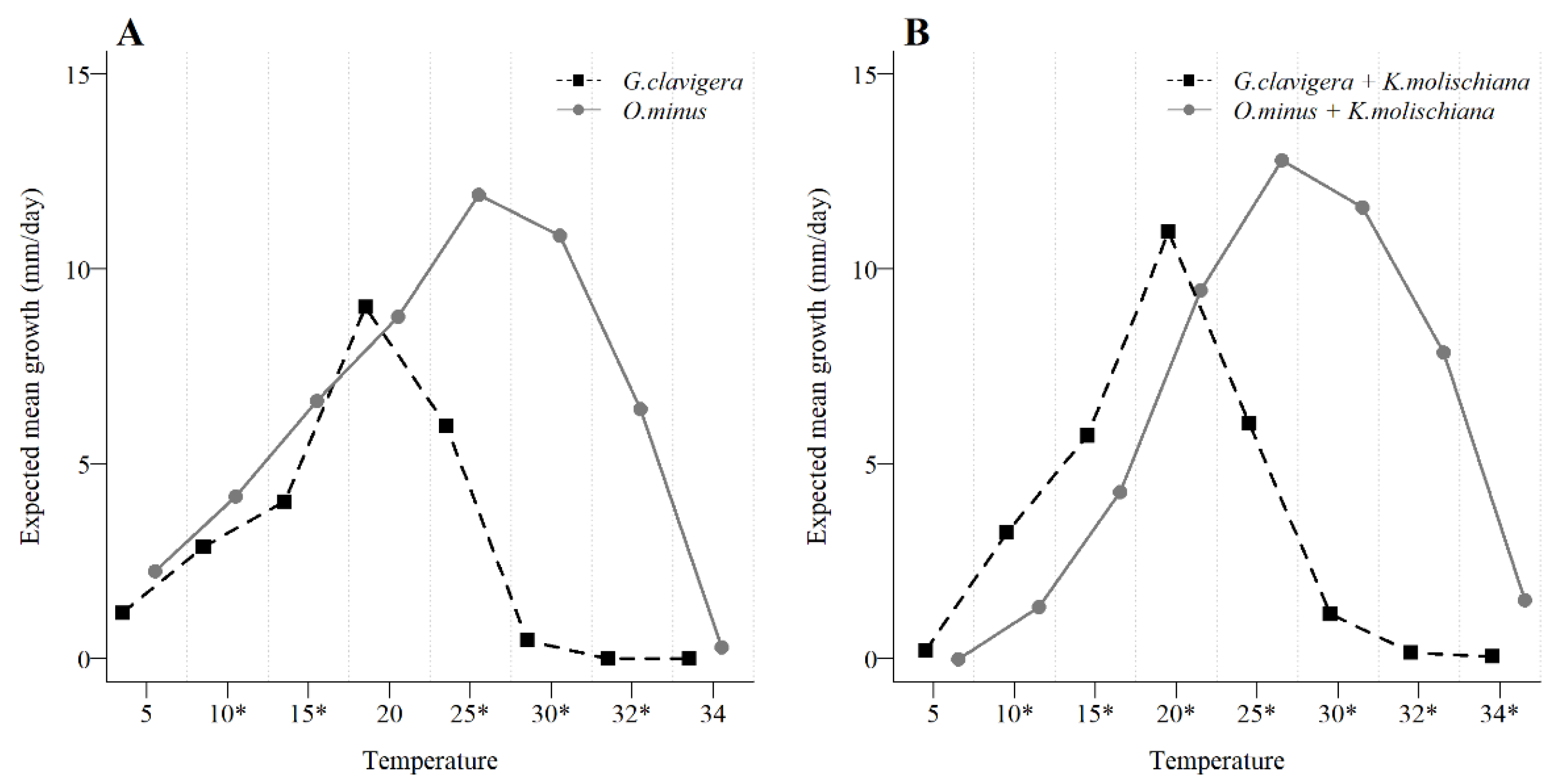
3.3. Yeast Interaction with Blue-Stain Fungal Growth across a Temperature Gradient
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Lim, L.; Madilao, L.; Lah, L.; Bohlmann, J.; Breuil, C. Gene discovery for enzymes involved in limonene modification or utilization by the mountain pine beetle-associated pathogen Grosmannia clavigera. Appl. Environ. Microbiol. 2014, 80, 4566–4576. [Google Scholar] [CrossRef]
- Hunt, D.W.A.; Borden, J.H. Conversion of verbenols to verbenone by yeasts isolated from Dendroctonus ponderosae (Coleoptera: Scolytidae). J. Chem. Ecol. 1990, 16, 1385–1397. [Google Scholar] [CrossRef]
- Barras, S.J. Antagonism between Dendroctonus frontalis and the fungus Ceratocystis minor. Ann. Entomol. Soc. Am. 1970, 63, 1187–1190. [Google Scholar] [CrossRef]
- Hofstetter, R.W.; Cronin, J.T.; Klepzig, K.D.; Moser, J.C.; Ayres, M.P. Antagonisms, mutualisms and commensalisms affect outbreak dynamics of the southern pine beetle. Oecologia 2006, 147, 679–691. [Google Scholar] [CrossRef]
- Mercado, J.E.; Ortiz-Santana, B.; Kay, S.L. Fungal Frequency and Mite Load Trends Interact with a Declining Mountain Pine Beetle Population. Forests 2018, 9, 484. [Google Scholar] [CrossRef]
- Bridges, J.R.; Moser, J.C. Role of two phoretic mites in transmission of bluestain fungus, Ceratocystis minor. Ecol. Entomol. 1983, 8, 9–12. [Google Scholar] [CrossRef]
- Mercado, J.E.; Hofstetter, R.W.; Reboletti, D.M.; Negrón, J.F. Phoretic symbionts of the mountain pine beetle (Dendroctonus ponderosae Hopkins). For. Sci. 2014, 60, 512–526. [Google Scholar] [CrossRef]
- Rivera, F.N.; González, E.; Gómez, Z.; López, N.; Hernández-Rodríguez, C.; Berkov, A.; Zuñiga, G. Gut-associated yeast in bark beetles of the genus Dendroctonus Erichson (Coleoptera: Curculionidae: Scolytinae). Biol. J. Linn. Soc. 2009, 98, 325–342. [Google Scholar] [CrossRef][Green Version]
- Davis, T.S. The ecology of yeasts in the bark beetle holobiont: A century of research revisited. Microb. Ecol. 2015, 69, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.S.; Six, D.L. Temporal variation in mycophagy and prevalence of fungi associated with developmental stages of Dendroctonus ponderosae (Coleoptera: Curculionidae). Environ. Entomol. 2014, 36, 64–72. [Google Scholar] [CrossRef]
- Farmer, L.J. The Phloem-Yeast Complex during Infestations of the Mountain Pine Beetle in Lodgepole Pine. Ph.D. Thesis, University of Utah, Salt Lake City, UT, USA, 1965. [Google Scholar]
- Wood, S.L. The Bark and Ambrosia Beetles of North and Central America (Coleoptera: Scolytidae), a Taxonomic Monograph; Great Basin Nat. (Mem. 6); Brigham John University: Provo, UT, USA, 1982; pp. 150–203. [Google Scholar]
- Kelley, S.T.; Farrell, B.D. Is specialization a dead end? The phylogeny of host use in Dendroctonus bark beetles (Scolytidae). Evolution 1998, 52, 1731–1743. [Google Scholar] [CrossRef]
- Reeve, J.D.; Anderson, F.E.; Kelley, S.T. Ancestral state reconstruction for Dendroctonus bark beetles: Evolution of a tree killer. Environ. Entomol. 2012, 41, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Victor, J.; Zuñiga, G. Phylogeny of Dendroctonus bark beetles (Coleoptera: Curculionidae: Scolytinae) inferred from morphological and molecular data. Syst. Entomol. 2016, 41, 162–177. [Google Scholar] [CrossRef]
- Whitney, H.S.; Farris, S.H. Maxillary mycangium in the mountain pine beetle. Science 1970, 167, 54–55. [Google Scholar] [CrossRef]
- Six, D.L.; Paine, T.D. Ophiostoma clavigerum is the mycangial fungus of the Jeffrey pine beetle, Dendroctonus jeffreyi. Mycologia 1997, 89, 858–866. [Google Scholar] [CrossRef]
- Mathre, D.E. Survey of Ceratocystis spp. associated with bark beetles in California. Contrib. Boyce Thompson Inst. 1964, 22, 353–362. [Google Scholar] [CrossRef]
- Alamouti, S.M.; Wang, V.; Diguistini, S.; Six, D.L.; Bohlmann, J.; Hamelin, R.C.; Feau, N.; Breuil, C. Gene genealogies reveal cryptic species and host preferences for the pine fungal pathogen Grosmannia clavigera. Mol. Ecol. 2011, 20, 2581–2602. [Google Scholar] [CrossRef]
- Shifrine, M.; Phaff, H.J. The association of yeasts with certain bark beetles. Mycologia 1956, 48, 41–55. [Google Scholar] [CrossRef]
- Bleiker, K.P.; Potter, S.E.; Lauzon, C.R.; Six, D.L. Transport of fungal symbionts by mountain pine beetles. Can. Entomol. 2009, 141, 503–514. [Google Scholar] [CrossRef]
- California Forest Pest Council. Available online: https://caforestpestcouncil.org/tag/pest-condition-reports/ (accessed on 9 January 2020).
- Solheim, H.; Krokene, P. Growth and virulence of mountain pine beetle associated blue-stain fungi, Ophiostoma clavigerum and Ophiostoma montium. Can. J. Bot. 1998, 76, 561–566. [Google Scholar] [CrossRef]
- Upadhyay, H.P. A Monograph of Ceratocystis and Ceratocystiopsis; University of Georgia Press: Athens, GA, USA, 1981; 176p. [Google Scholar]
- Lee, S.; Kim, J.J.; Breuil, C. Leptographium longiclavatum sp. nov., a new species associated with the mountain pine beetle, Dendroctonus ponderosae. Mycol. Res. 2005, 109, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef]
- Kim, J.-J.; Kim, S.H.; Lee, S.; Breuil, C. Distinguishing Ophiostoma ips and Ophiostoma montium, two bark beetle-associated sapstain fungi. FEMS Microbiol. Lett. 2003, 222, 187–192. [Google Scholar] [CrossRef]
- Gorton, C.; Kim, S.H.; Henricot, B.; Webber, J.; Breuil, C. Phylogenetic analysis of the bluestain fungus Ophiostoma minus based on partial ITS rDNA and beta-tubulin gene sequences. Mycol. Res. 2004, 108, 759–765. [Google Scholar] [CrossRef]
- Zipfel, R.D.; De Beer, Z.W.; Jacobs, K.; Wingfield, B.D.; Winfield, M.J. Multi-gene phylogenies define Ceratocystiopsis and Grosmannia distinct from Ophiostoma. Stud. Mycol. 2006, 55, 75–97. [Google Scholar] [CrossRef]
- Roe, A.D.; Rice, A.V.; Bromilow, S.E.; Cooke, J.E.K.; Sperling, F.A.H. Multilocus species identification and fungal DNA barcoding: Insights from blue stain fungal symbionts of the mountain pine beetle. Mol. Ecol. Resour. 2010, 10, 946–959. [Google Scholar] [CrossRef]
- Six, D.L.; De Beer, Z.W.; Duong, T.A.; Carroll, A.L.; Wingfield, M.J. Fungal associates of the lodgepole pine beetle, Dendroctonus murrayanae. Antonie Van Leuwenhoek 2011, 100, 231–244. [Google Scholar] [CrossRef][Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIP-RES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (eds. GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4.4. Institute of Evolutionary Biology, University of Edinburgh. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 11 March 2021).
- R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 9 March 2020).
- Péter, G.; Dlauchy, D.; Tornai-Lehoczki, J.; Kurtzman, C.P. Kuraishia molischiana sp. nov., the teleomorph of Candida molischiana. Antonie van Leeuwenhoek 2005, 88, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Lombardero, M.J.; Ayres, M.P.; Hofstetter, R.W.; Moser, J.C.; Lepzig, K.D. Strong indirect interactions of Tarsonemus mites (Acarina: Tarsonemidae) and Dendroctonus frontalis (Coleoptera: Scolytidae). Oikos 2003, 102, 243–252. [Google Scholar] [CrossRef]
- Myrholm, C.L.; Langor, D.W. Assessment of the impact of symbiont Ophiostomatales (Fungi) on mountain pine beetle (Coleoptera: Curculionidae) performance on a jack pine (Pinaceae) diet using a novel in vitro rearing method. Can. Entomol. 2016, 148, 68–82. [Google Scholar] [CrossRef]
- Paine, T.D.; Hanlon, C.C. Influence of oleoresin constituents from Pinus ponderosa and Pinus jeffreyi on growth of mycangial fungi from Dendroctonus ponderosae and Dendroctonus jeffreyi. J. Chem. Ecol. 1994, 20, 2551–2563. [Google Scholar] [CrossRef] [PubMed]
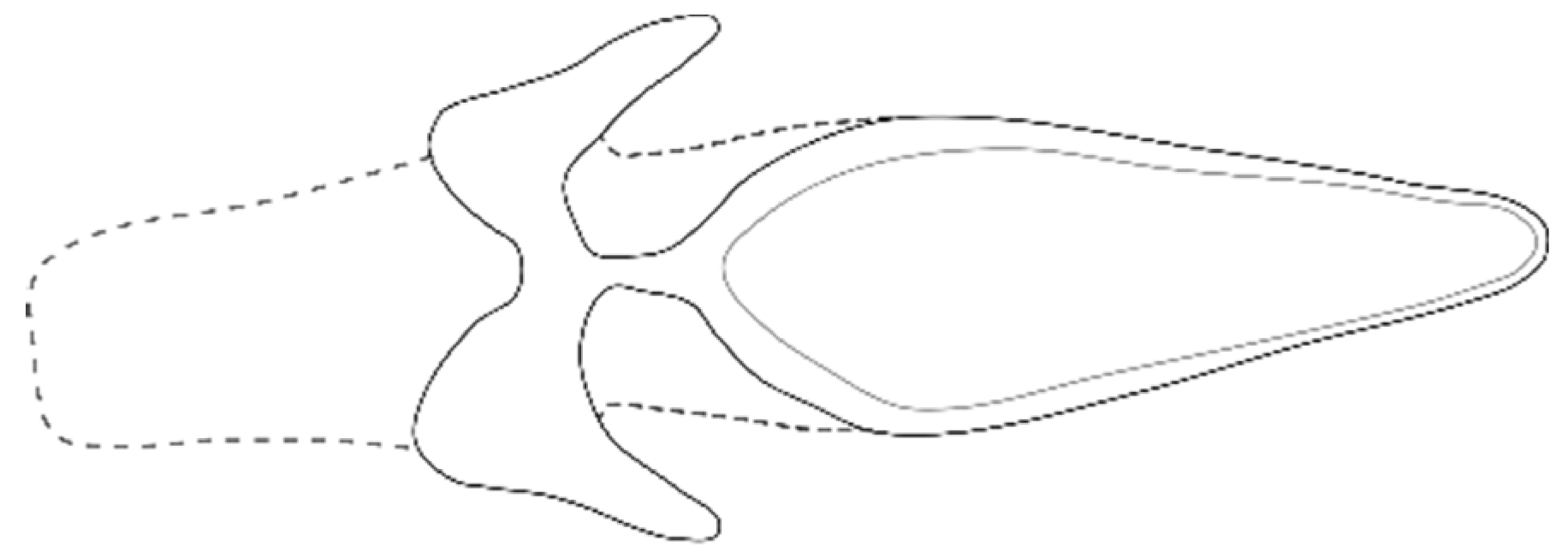

| Species | BeetleStructure | Locality | Reference |
|---|---|---|---|
| Ophiostoma ips/montium | unreported | Stanislaus NF, CA | [18] |
| Grosmannia clavigera | Maxillae | S. Bernandino Mnts., CA and NV | [17] |
| Leptographium longiclavatum | Maxillae | Monitor Pass, Toiyabe NF, CA | [19] |
| Candida silvicola | Gut | Northern California | [20] |
| Diutina rugosa | Gut | Northern California | [20] |
| Kuraishia capsulata | Gut | Northern California | [20] |
| Nakazawaea holstii | Gut | Northern California | [20] |
| Priceomyces haplophilus | Gut | Northern California | [20] |
| Spencerozyma crocea | Gut | Northern California | [20] |
| Species | Isolate | Location | ITS-LSU | β-tubulin |
|---|---|---|---|---|
| Grosmannia aurea | UC03DL06 | Canada-AB | GU370267 | GU370181 |
| UC16G21 | Canada-AB | GU370293 | GU370207 | |
| G. clavigera | CA-JM-71 | USA-CA | MZ297376 | MZ296739 |
| CA-JM-72 | USA-CA | MZ297377 | MZ296740 | |
| CA-JM-73 | USA-CA | MZ297378 | MZ296741 | |
| CA-JM-74 | USA-CA | MZ297379 | MZ296742 | |
| CA-JM-75 | USA-CA | MZ297380 | MZ296743 | |
| CA-JM-76 | USA-CA | MZ297381 | MZ296744 | |
| CA-JM-77 | USA-CA | MZ297382 | MZ296745 | |
| CA-JM-78 | USA-CA | MZ297383 | MZ296746 | |
| CA-JM-79 | USA-CA | MZ297384 | MZ296747 | |
| CA-JM-80 | USA-CA | MZ297385 | MZ296748 | |
| CO-JM-17 | USA-CO | KY940832 | KY940842 | |
| CO-JM-41 | USA-CO | KY940833 | KY940843 | |
| SL-Kw1407 | Canada-BC | AY544615 | AY263195 | |
| UC14G23 | Canada-AB | GU370266 | GU370180 | |
| UC35DL49 | Canada-AB | GU370288 | GU370202 | |
| UM01G12 | Canada-AB | GU370274 | GU370188 | |
| G. huntii | CBS398.77 | Canada | AY707208 | AY707194 |
| UAMH4997 | Canada | AY544617 | AY349023 | |
| G. robusta | CMW 668 | USA-ID | AY553397 | AY534945 |
| CMW 2805 | USA-ID | AY553396 | AY534944 | |
| Leptographium longiclavatum | CO-JM-8 | USA-CO | KY940824 | KY940834 |
| CO-JM-42 | USA-CO | KY940826 | KY940836 | |
| CO-JM-46 | USA-CO | KY940830 | KY940840 | |
| SL-Kp11 | Canada-BC | AY816687 | AY816712 | |
| SL-Kw1436 | Canada-BC | AY816686 | AY288934 | |
| UL13G22 | Canada-BC | GU370300 | GU370214 | |
| L. terebrantis | UC38DL41 | Canada-AB | GU370295 | GU370209 |
| UC48G19 | Canada-AB | GU370272 | GU370186 | |
| L. wingfieldii | CMW 2095 | France | AY553400 | AY534948 |
| CMW 2096 | France | AY553398 | AY534946 | |
| Ophiostoma bicolor | 3YT4P-Ob | Canada | DQ268606 | DQ268637 |
| CBS492.7 | USA-CO | DQ268604 | DQ268635 | |
| O. ips | ATCC-24285 | Canada | AY194937 | AY194949 |
| SYPT2 | Canada | AY194940 | AY194955 | |
| O. minus | AU58.4 | Canada-BC | — | AY548743 |
| C348 | Canada | AY542496 | AY542507 | |
| CFL873 | Canada | KC305144 | KC336019 | |
| DAOM-212686 | Canada | — | AY305690 | |
| OM1 | UK | AY542494 | AY542505 | |
| OM6 | USA-NE | AY542498 | AY542509 | |
| CA-JM-81 | USA-CA | MZ297386 | MZ296749 | |
| CA-JM-82 | USA-CA | MZ297387 | MZ296750 | |
| CA-JM-83 | USA-CA | MZ297388 | MZ298931 | |
| CA-JM-84 | USA-CA | MZ297389 | MZ296751 | |
| CA-JM-85 | USA-CA | MZ297390 | MZ298932 | |
| CA-JM-86 | USA-CA | MZ297391 | MZ296752 | |
| CA-JM-87 | USA-CA | MZ297392 | MZ296753 | |
| CA-JM-88 | USA-CA | MZ297393 | MZ296754 | |
| CA-JM-89 | USA-CA | MZ297394 | MZ298933 | |
| CA-JM-90 | USA-CA | MZ297395 | MZ296755 | |
| O. montium | ATCC-64697 | Canada | AY194941 | AY194957 |
| Kw430 | Canada | AY194948 | AY194964 | |
| O. piceae | B054 | Austria | HQ115733 | — |
| CNB-13 | Spain | AJ538341 | — | |
| H2181 | Japan | AF211844 | AB934362 | |
| KUC-2311 | Korea | — | DQ868379 | |
| O. piliferum | 201/1a | United Kingdom | — | AF221629 |
| CBS129.32 | Netherlands | — | AY305705 | |
| O. pseudotsugae | 92-634/302/6 | Canada-BC | AY542502 | AY542510 |
| OM5 | USA-ID | AY542500 | AY548744 | |
| O. setosum | AU160-27 | Canada | — | AY789160 |
| NZFS-3652-1 | New Zealand | — | AY789159 | |
| O. subalpinum | YCC-408 | Japan | — | AB200429 |
| YCC-410 | Japan | — | AB200430 |
| Fungal Species | Value | DF | Chisq | Pr (>Chisq) |
|---|---|---|---|---|
| Maxillae-Pellet: G. clavigera | 0.95 | 1 | 28.87 | <0.001 |
| Maxillae-Pellet: O. minus | 0.52 | 1 | 0.04 | 0.833 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mercado, J.E.; Ortiz-Santana, B.; Kay, S.L. Endemic Jeffrey Pine Beetle Associates: Beetle/Mite Fungal Dissemination Strategies and Interactions That May Influence Beetle Population Levels. Microorganisms 2021, 9, 1641. https://doi.org/10.3390/microorganisms9081641
Mercado JE, Ortiz-Santana B, Kay SL. Endemic Jeffrey Pine Beetle Associates: Beetle/Mite Fungal Dissemination Strategies and Interactions That May Influence Beetle Population Levels. Microorganisms. 2021; 9(8):1641. https://doi.org/10.3390/microorganisms9081641
Chicago/Turabian StyleMercado, Javier E., Beatriz Ortiz-Santana, and Shannon L. Kay. 2021. "Endemic Jeffrey Pine Beetle Associates: Beetle/Mite Fungal Dissemination Strategies and Interactions That May Influence Beetle Population Levels" Microorganisms 9, no. 8: 1641. https://doi.org/10.3390/microorganisms9081641
APA StyleMercado, J. E., Ortiz-Santana, B., & Kay, S. L. (2021). Endemic Jeffrey Pine Beetle Associates: Beetle/Mite Fungal Dissemination Strategies and Interactions That May Influence Beetle Population Levels. Microorganisms, 9(8), 1641. https://doi.org/10.3390/microorganisms9081641






