Unraveling the Underlying Heavy Metal Detoxification Mechanisms of Bacillus Species
Abstract
1. Introduction to Heavy Metals
2. Methods of Heavy Metal Remediation
3. Bioremediation—An Environmentally-Friendly Approach for the Removal of Heavy Metals
4. Bacillus spp. as Potential Agents for Heavy Metal Removal and Their Overall Significance
5. Bacillus Species and Heavy Metals
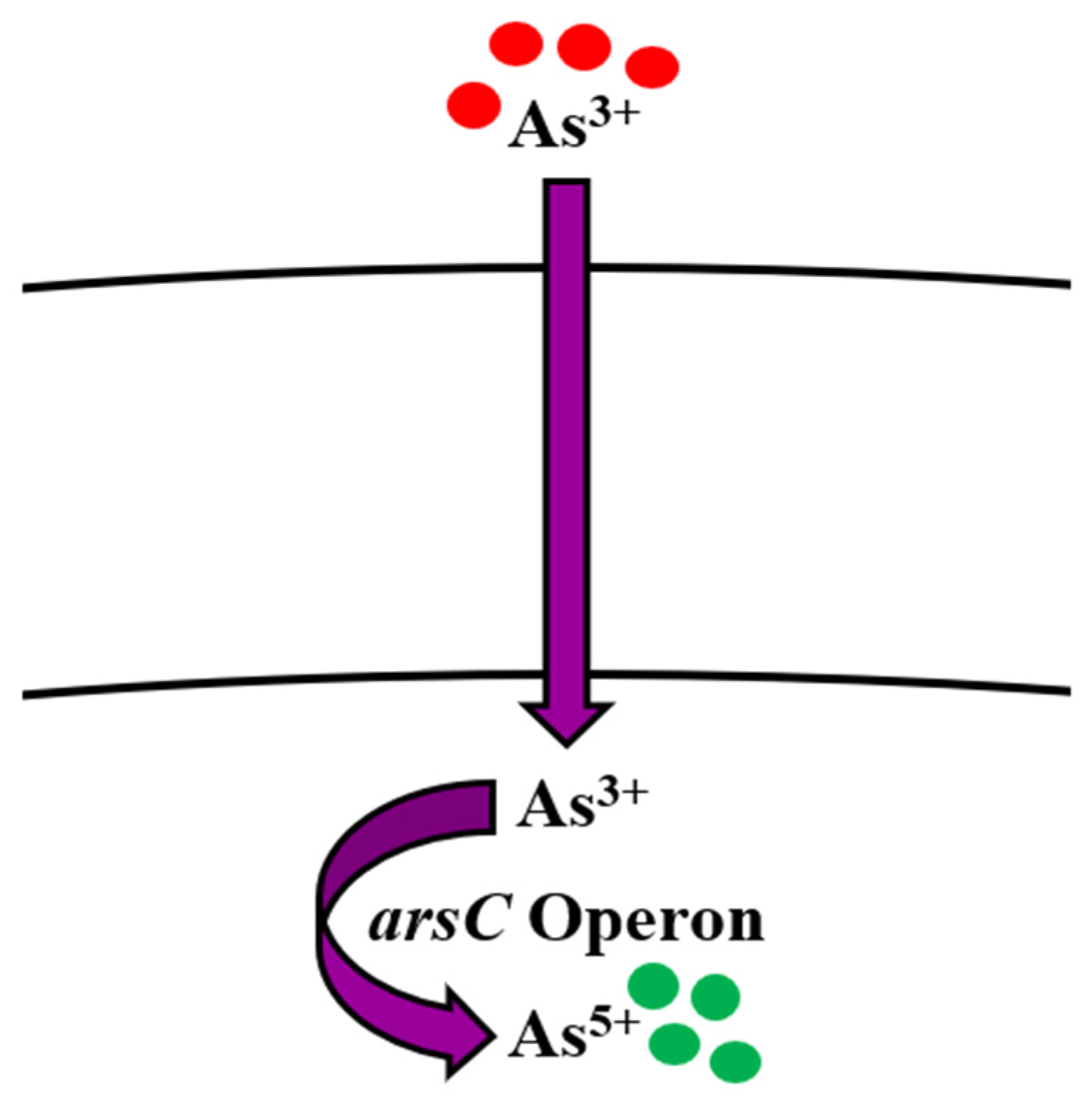
5.1. Arsenic
Bioregulation of Arsenic by Bacillus spp.
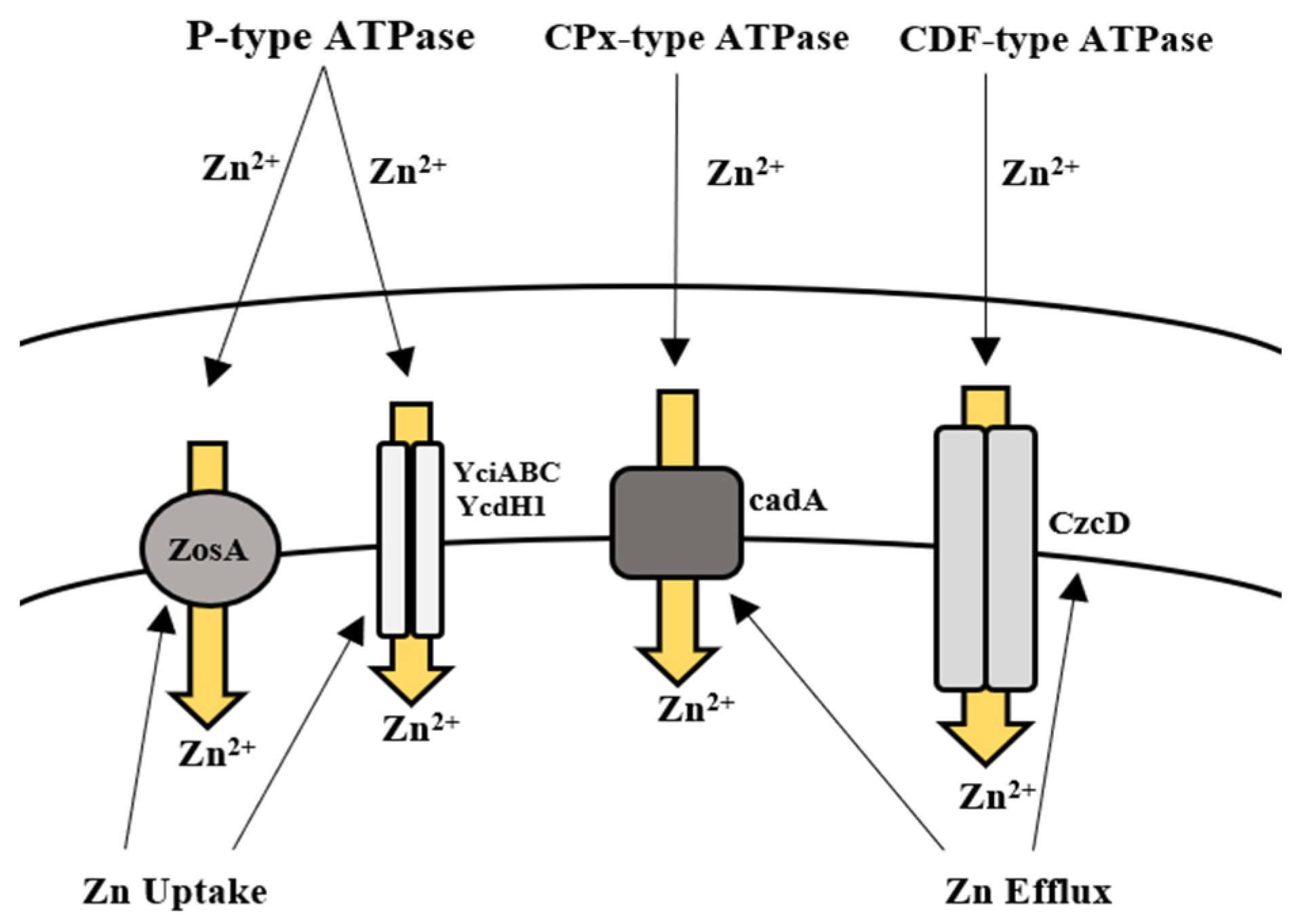
5.2. Zinc
Bioregulation of Zinc by Bacillus spp.
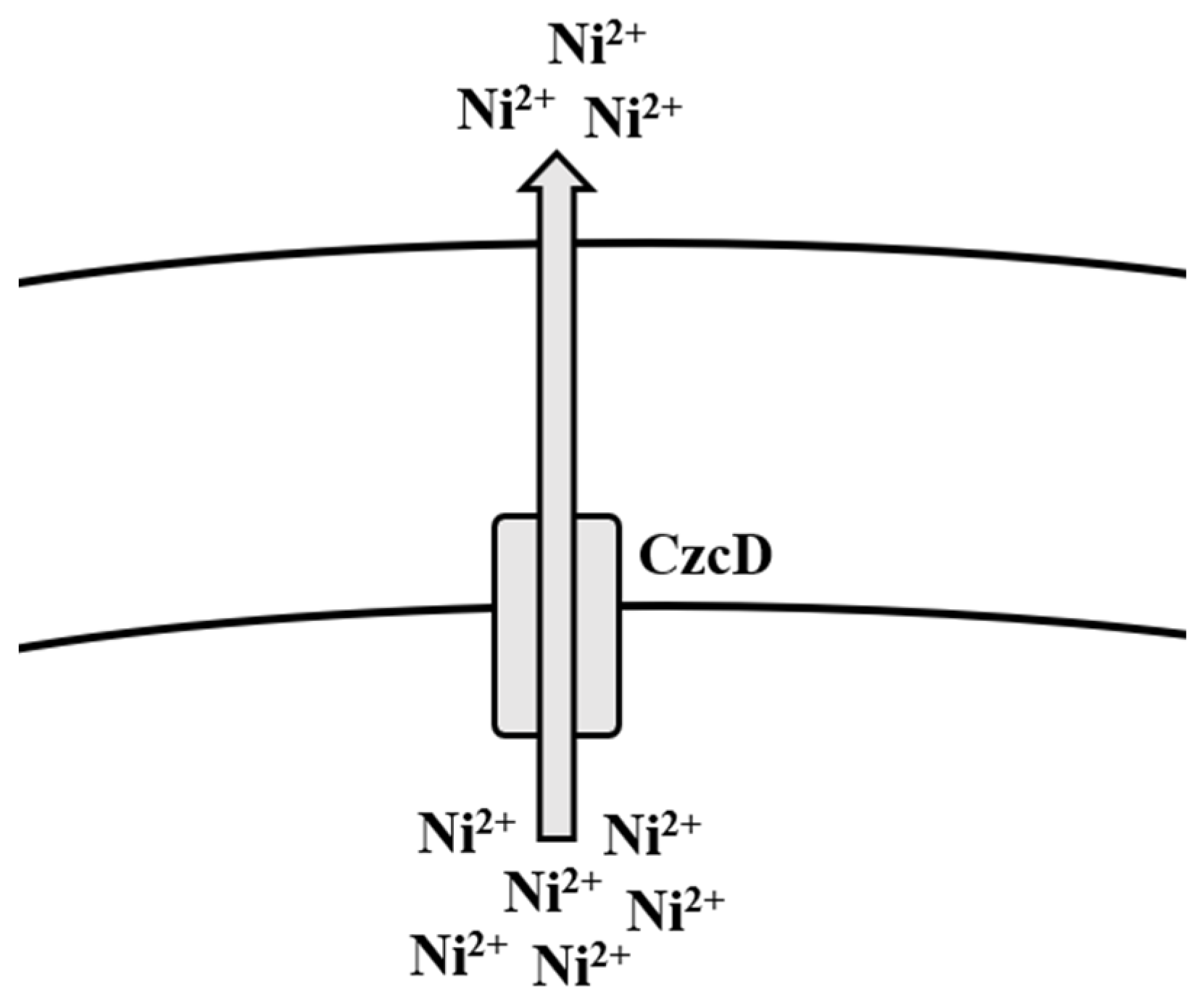
5.3. Nickel
Bioregulation of Nickel by Bacillus spp.
5.4. Cadmium
Bioregulation of Cadmium by Bacillus spp.
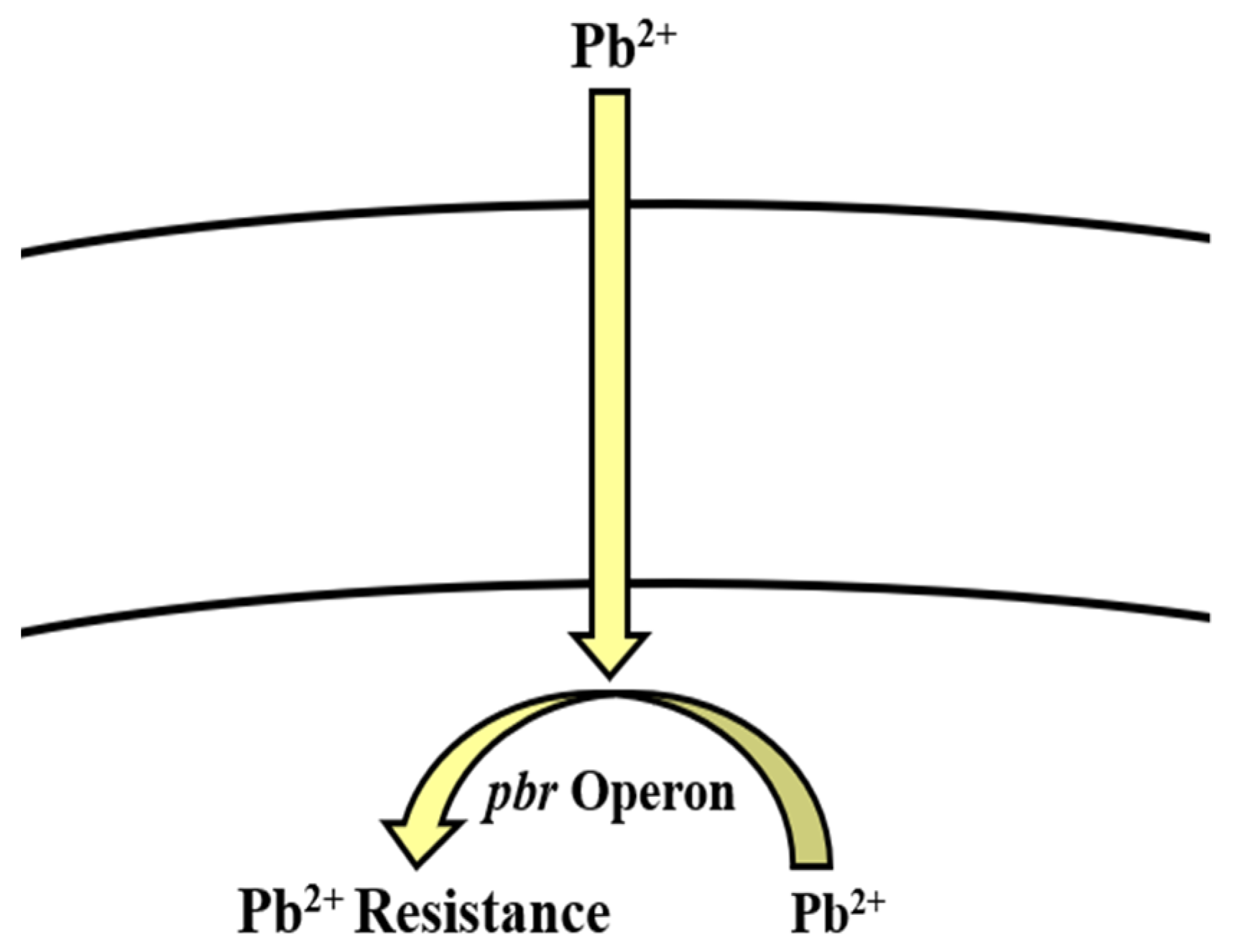
5.5. Lead
Bioregulation of Lead by Bacillus spp.
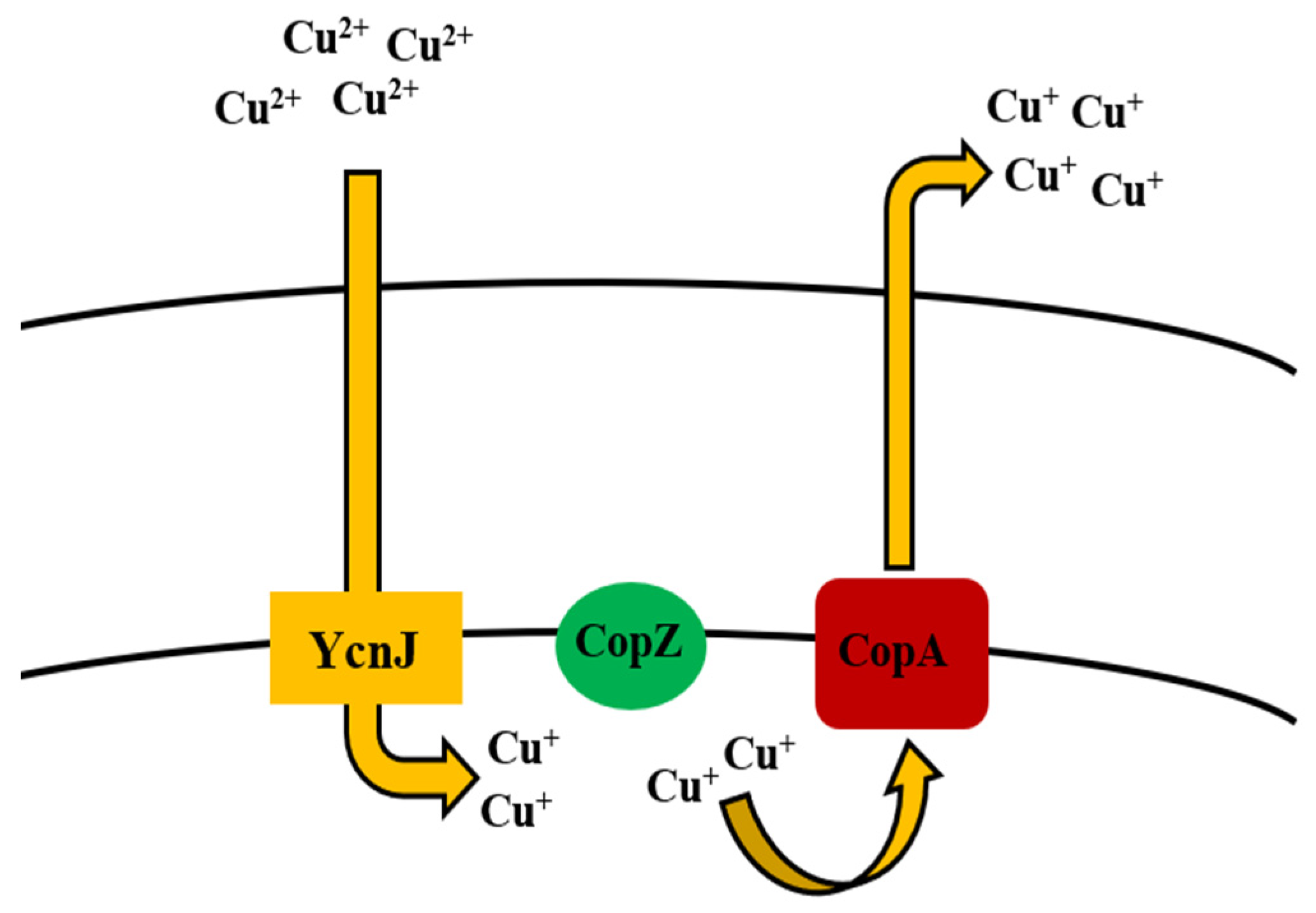
5.6. Copper
Bioregulation of Copper by Bacillus spp.
5.7. Chromium
Bioregulation of Chromium by Bacillus spp.
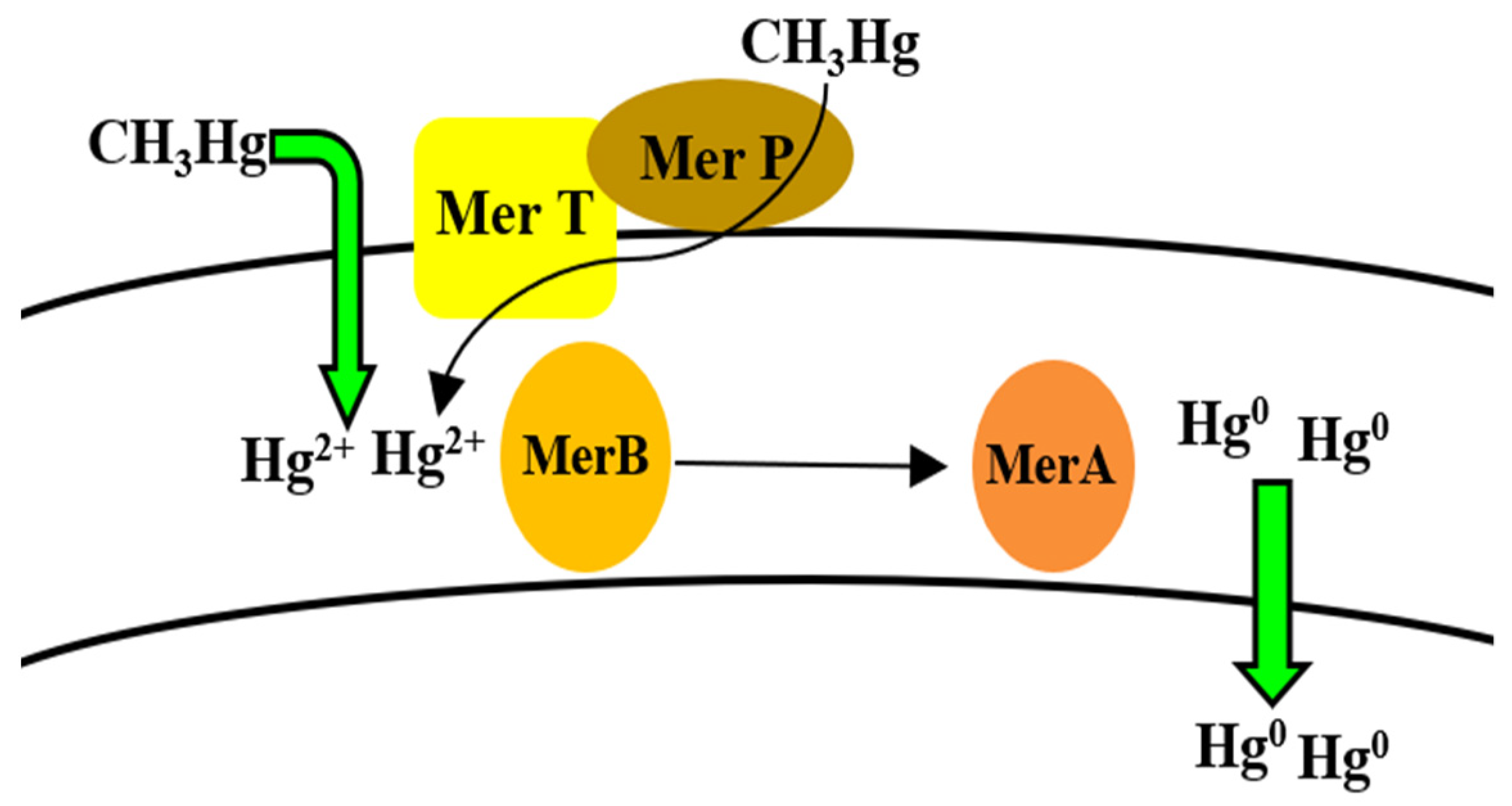
5.8. Mercury
Bioregulation of Mercury by Bacillus spp.
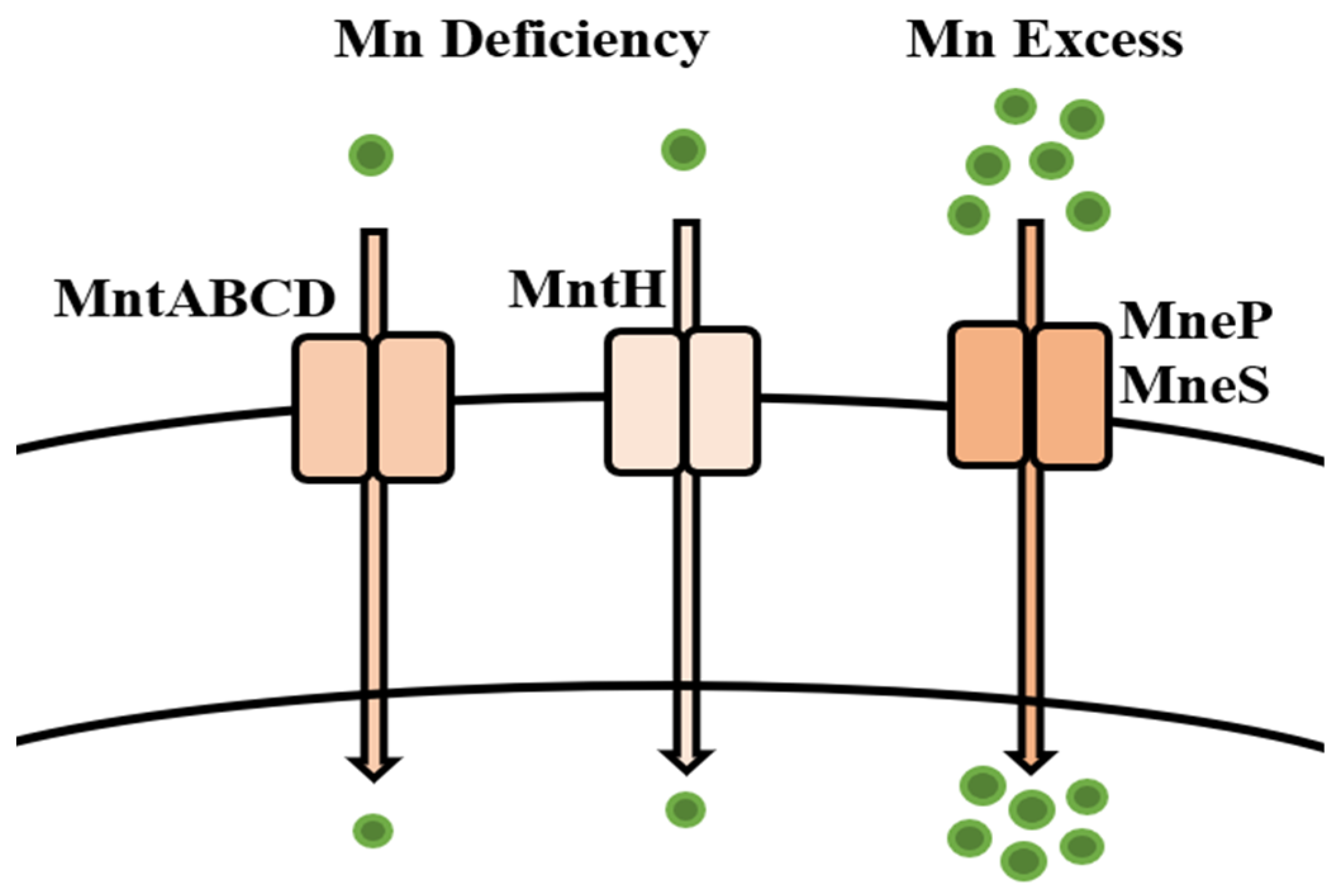
5.9. Manganese
Bioregulation of Manganese by Bacillus spp.
5.10. Molybdenum
Bioregulation of Molybdenum by Bacillus spp.
5.11. Gold
Bioregulation of Gold by Bacillus spp.
5.12. Silver
Bioregulation of Silver by Bacillus spp.
6. Conclusions
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gautam, P.K.; Gautam, R.K.; Banerjee, S.; Chattopadhyaya, M.C.; Pandey, J.D. Heavy Metals in the Environment: Fate, Transport, Toxicity and Remediation Technologies. In Heavy Metals: Sources, Toxicity and Remediation Techniques; Pathania, D., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2016; pp. 1–27. [Google Scholar]
- Masindi, V.; Muedi, K.L. Environmental Contamination by Heavy Metals. In Heavy Metals; Saleh, H.E.-D., Aglan, R.F., Eds.; IntechOpen Publishers: London, UK, 2018; pp. 115–132. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E. What are heavy metals? Long-standing controversy over the scientific use of the term “heavy metals”-proposal of a comprehensive definition. Toxicol. Environ. Chem. 2018, 100, 6–19. [Google Scholar] [CrossRef]
- Presentato, A.; Piacenza, E.; Turner, R.J.; Zannoni, D.; Cappelletti, M. Processing of Metals and Metalloids by Actinobacteria: Cell Resistance Mechanisms and Synthesis of Metal(loid)-Based Nanostructures. Microorganisms 2020, 8, 2027. [Google Scholar] [CrossRef]
- Duffus, J.H. Heavy metals—A meaningless term? Pure Appl. Chem. 2002, 74, 793–807. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Exp. Suppl. 2012, 101, 133–164. [Google Scholar] [CrossRef]
- Martin, Y.E.; Johnson, E.A. Biogeosciences survey: Studying interactions of the biosphere with the lithosphere, hydrosphere and atmosphere. Prog. Phys. Geogr. 2012, 36, 833–852. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E.; Ilahi, I. Environmental chemistry and ecotoxicology of hazardous heavy metals: Environmental persistence, toxicity, and bioaccumulation. J. Chem. 2019, 2019, 730305. [Google Scholar] [CrossRef]
- Nagajyoti, P.C.; Lee, K.D.; Sreekanth, T.V.M. Heavy metals, occurrence and toxicity for plants: A review. Environ. Chem. Lett. 2010, 8, 199–216. [Google Scholar] [CrossRef]
- He, Z.L.; Yang, X.E.; Stoffella, P.J. Trace elements in agroecosystems and impacts on the environment. J. Trace Elem. Med. Biol. 2005, 19, 125–140. [Google Scholar] [CrossRef]
- Muchuweti, M.; Birkett, J.W.; Chinyanga, E.; Zvauya, R.; Scrimshaw, M.D.; Lester, J.N. Heavy metal content of vegetables irrigated with mixtures of wastewater and sewage sludge in Zimbabwe: Implications for human health. Agric. Ecosyst. Environ. 2006, 112, 41–48. [Google Scholar] [CrossRef]
- Gupta, N.; Khan, D.K.; Santra, S.C. Heavy metal accumulation in vegetables grown in a long-term wastewater-irrigated agricultural land of tropical India. Environ. Monit. Assess. 2012, 184, 6673–6682. [Google Scholar] [CrossRef]
- Selvi, A.; Rajasekar, A.; Theerthagiri, J.; Ananthaselvam, A.; Sathishkumar, K.; Madhavan, J.; Rahman, P.K.S.M. Integrated remediation processes toward heavy metal removal/recovery from various environments-A review. Front. Environ. Sci. 2019, 7, 66–81. [Google Scholar] [CrossRef]
- Briffa, J.; Sinagra, E.; Blundell, R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 2020, 6, 1–26. [Google Scholar] [CrossRef]
- Türkmen, M.; Türkmen, A.; Tepe, Y.; Töre, Y.; Ateş, A. Determination of metals in fish species from Aegean and Mediterranean seas. Food Chem. 2009, 113, 233–237. [Google Scholar] [CrossRef]
- Ramírez, R. The gastropod Osilinus atrata as a bioindicator of Cd, Cu, Pb and Zn contamination in the coastal waters of the Canary Islands. Chem. Ecol. 2013, 29, 208–220. [Google Scholar] [CrossRef]
- Rahim, M.; Ullah, I.; Khan, A.; Haris, M.R.H.M. Health risk from heavy metals via consumption of food crops in the vicinity of District Shangla. J. Chem. Soc. Pak. 2016, 38, 177–185. [Google Scholar]
- Appenroth, K.-J. What are “heavy metals” in Plant Sciences? Acta Physiol. Plant. 2010, 32, 615–619. [Google Scholar] [CrossRef]
- Chalkiadaki, O.; Dassenakis, M.; Lydakis-Simantiris, N. Bioconcentration of Cd and Ni in various tissues of two marine bivalves living in different habitats and exposed to heavily polluted seawater. Chem. Ecol. 2014, 30, 726–742. [Google Scholar] [CrossRef]
- Valko, M.; Morris, H.; Cronin, M. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005, 12, 1161–1208. [Google Scholar] [CrossRef]
- Engwa, G.A.; Ferdinand, P.U.; Nwalo, F.N.; Unachukwu, N.M. Mechanism and Health Effects of Heavy Metal Toxicity in Humans. In Poisoning in the Modern World—New Tricks for an Old Dog? Karcioglu, O., Arslan, B., Eds.; IntechOpen Publishers: London, UK, 2019; pp. 1–24. [Google Scholar] [CrossRef]
- Shahid, M.; Arshad, M.; Kaemmerer, M.; Pinelli, E. Long-term field metal extraction by pelargonium: Phytoextraction efficiency in relation to plant maturity. Int. J. Phytoremed. 2012, 14, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Minnikova, T.V.; Denisova, T.V.; Mandzhieva, S.S.; Kolesnikov, S.I.; Minkina, T.M.; Chaplygin, V.A.; Burachevskaya, M.V.; Sushkova, S.N.; Bauer, T.V. Assessing the effect of heavy metals from the Novocherkassk power station emissions on the biological activity of soils in the adjacent areas. J. Geochem. Explor. 2017, 174, 70–78. [Google Scholar] [CrossRef]
- Murtaza, G.; Murtaza, B.; Niazi, K.N.; Sabir, M. Soil Contaminants: Sources, Effects, and Approaches for Remediation. In Improvement of Crops in the Era of Climatic Changes; Ahmad, P., Wani, R.M., Azooz, M.M., Phan Tran, L.-S., Eds.; Springer: New York, NY, USA, 2014; pp. 171–196. [Google Scholar] [CrossRef]
- Shi, W.; Liu, C.; Ding, D.; Lei, Z.; Yang, Y.; Feng, C.; Zhang, Z. Immobilization of heavy metals in sewage sludge by using subcritical water technology. Bioresour. Technol. 2013, 137, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Li, J.; Xie, H.; Yu, C. Review on remediation technologies of soil contaminated by heavy metals. Proceed. Environ. Sci. 2012, 16, 722–729. [Google Scholar] [CrossRef]
- Zhou, D.M.; Hao, X.Z.; Xue, Y. Advances in remediation technologies of contaminated soils. Ecol. Environ. Sci. 2004, 13, 234–242. [Google Scholar]
- Elicker, C.; Filho, P.S.; Castagno, K. Electroremediation of heavy metals in sewage sludge. Braz. J. Chem. Eng. 2014, 31, 365–371. [Google Scholar] [CrossRef]
- Navarro, A.; Cardellach, E.; Cañadas, I.; Rodríguez, J. Solar thermal vitrification of mining contaminated soils. Int. J. Min. Process. 2013, 119, 65–74. [Google Scholar] [CrossRef]
- Khalid, S.; Shahid, M.; Niazi, N.K.; Murtaza, B.; Bibi, I.; Dumate, C. A comparison of technologies for remediation of heavy metal contaminated soils. J. Geochem. Explor. 2017, 182, 247–268. [Google Scholar] [CrossRef]
- Albers, C.N.; Jacobsen, O.S.; Flores, E.M.M.; Johnsen, A.R. Arctic and subarctic natural soils emit chloroform and brominated analogues by alkaline hydrolysis of trihaloacetyl compounds. Environ. Sci. Technol. 2017, 51, 6131–6138. [Google Scholar] [CrossRef]
- Alghanmi, S.I.; Al Sulami, A.F.; El-Zayat, T.A.; Alhogbi, B.G.; Salam, M.A. Acid leaching of heavy metals from contaminated soil collected from Jeddah, Saudi Arabia: Kinetic and thermodynamics studies. Int. Soil Water Conserv. Res. 2015, 3, 196–208. [Google Scholar] [CrossRef]
- Lee, M.; Paik, I.S.; Do, W.; Kim, I.; Lee, Y.; Lee, S. Soil washing of As-contaminated stream sediments in the vicinity of an abandoned mine in Korea. Environ. Geochem. Health 2007, 29, 319–329. [Google Scholar] [CrossRef]
- Li, Z.; Wang, L.; Meng, J.; Liu, X.; Xu, J.; Wang, F.; Brookes, P. Zeolite-supported nanoscale zero-valent iron: New findings on simultaneous adsorption of Cd(II), Pb(II), and As(III) in aqueous solution and soil. J. Hazard. Mater. 2018, 344, 1–11. [Google Scholar] [CrossRef]
- Venkatachalam, P.; Jayaraj, M.; Manikandan, R.; Geetha, N.; Rene, E.R.; Sharma, N.C.; Sahi, S.V. Zinc oxide nanoparticles (ZnONPs) alleviate heavy metal-induced toxicity in Leucaena leucocephala seedlings: A physiochemical analysis. Plant. Physiol. Biochem. 2017, 110, 59–69. [Google Scholar] [CrossRef]
- Venegas, A.; Rigol, A.; Vidal, M. Viability of organic wastes and biochars as amendments for the remediation of heavy metal-contaminated soils. Chemosphere 2015, 119, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.H.; Ismail, I.M.; Mostafa, T.M.; Sulaymon, A.H. Biosorption of heavy metals: A review. J. Chem. Sci. Technol. 2014, 3, 74–102. [Google Scholar]
- Mustapha, M.U.; Halimoon, N. Microorganisms and biosorption of heavy metals in the environment: A review paper. J. Microb. Biochem. Technol. 2015, 7, 253–256. [Google Scholar] [CrossRef]
- Kang, S.Y.; Lee, J.U.; Kim, K.M. Biosorption of Cr(III) and Cr(VI) onto the cell surface of Pseudomonas aeruginosa. Biochem. Eng. J. 2007, 36, 54–58. [Google Scholar] [CrossRef]
- Akhtar, K.; Akhtar, M.W.; Khalid, A.M. Removal and recovery of uranium from aqueous solutions by Trichoderma harzianum. Water Res. 2007, 41, 1366–1378. [Google Scholar] [CrossRef]
- Ozer, A.; Ozer, D. Comparative study of the biosorption of Pb(II), Ni(II) and Cr(VI) ions onto S. cerevisiae: Determination of biosorption heats. J. Hazard. Mater. 2003, 100, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Mosa, K.A.; Saadoun, I.; Kumar, K.; Helmy, M.; Dhankher, O.P. Potential biotechnological strategies for the cleanup of heavy metals and metalloids. Front. Plant. Sci. 2016, 7, 303. [Google Scholar] [CrossRef] [PubMed]
- Kapahi, M.; Sachdeva, S. Bioremediation options for heavy metal pollution. J. Health Pollut. 2019, 9, 191203. [Google Scholar] [CrossRef] [PubMed]
- Abioye, O.P.; Oyewole, O.A.; Oyeleke, S.B.; Adeyemi, M.O.; Orukotan, A.A. Biosorption of lead, chromium and cadmium in tannery effluent using indigenous microorganisms. Braz. J. Biol. Sci. 2018, 5, 25–32. [Google Scholar] [CrossRef]
- Tarekegn, M.M.; Salilih, F.Z.; Ishetu, A.I. Microbes used as a tool for bioremediation of heavy metal from the environment. Cog. Food Agric. 2020, 6. [Google Scholar] [CrossRef]
- Kotb, E. Purification and partial characterization of serine fibrinolytic enzyme from Bacillus megaterium KSK-07 isolated from kishk, a traditional Egyptian fermented food. Appl. Biochem. Microbiol. 2015, 51, 34–43. [Google Scholar] [CrossRef]
- Elshaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus as potential probiotics: Status, concerns, and future perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef]
- Setlow, P.; Wang, S.; Li, Y.-Q. Germination of spores of the orders Bacillales and Clostridiales. Annu. Rev. Microbiol. 2017, 71, 459–477. [Google Scholar] [CrossRef]
- Cai, D.; Rao, Y.; Zhan, Y.; Wang, Q.; Chen, S. Engineering Bacillus for efficient production of heterologous protein: Current progress, challenge and prospect. J. Appl. Microbiol. 2019, 126, 1632–1642. [Google Scholar] [CrossRef] [PubMed]
- Wolken, W.A.M.; Tramper, J.; van der Werf, M.J. What Can Spores Do for Us? Trends Biotechnol. 2003, 21, 338–345. [Google Scholar] [CrossRef]
- Sanders, M.E.; Morelli, L.; Tompkins, T.A. Sporeformers as human probiotics: Bacillus, Sporolactobacillus and Brevibacillus. Comp. Rev. Food Sci. Food Saf. 2003, 2, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.A.; Duc, L.H.; Cutting, S.M. The Use of Bacterial Spore Formers as Probiotics. FEMS Microbiol. Rev. 2005, 29, 813–835. [Google Scholar] [CrossRef]
- Lee, N.-K.; Kim, W.-S.; Paik, H.-D. Bacillus strains as human probiotics: Characterization, safety, microbiome, and probiotic carrier. Food Sci. Biotechnol. 2019, 28, 1297–1305. [Google Scholar] [CrossRef]
- Ouattara, H.G.; Reverchon, S.; Niamke, S.L.; Nasser, W. Regulation of the synthesis of pulp degrading enzymes in Bacillus isolated from cocoa fermentation. Food Microbiol. 2017, 63, 255–262. [Google Scholar] [CrossRef]
- Tanaka, K.; Takanaka, S.; Yoshida, K. A second-generation Bacillus cell factory for rare inositol production. Bioengineered 2014, 5, 331–334. [Google Scholar] [CrossRef]
- Takano, H. The regulatory mechanism underlying light-inducible production of carotenoids in nonphototrophic bacteria. Biosci. Biotechnol. Biochem. 2016, 80, 1264–1273. [Google Scholar] [CrossRef]
- Chikere, C.B.; Okpokwasili, G.C.; Chikere, B.O. Bacterial diversity in a tropical crude oil-polluted soil undergoing bioremediation. Afr. J. Biotechnol. 2009, 8, 2535–2540. [Google Scholar] [CrossRef]
- Nwinyi, O.C.; Kanu, I.A.; Tunde, A.; Ajanaku, K.O. Characterization of diesel degrading bacterial species from contaminated tropical ecosystem. Braz. Arch. Biol. Technol. 2014, 57, 789–796. [Google Scholar] [CrossRef]
- Dey, U.; Chatterjee, S.; Mondal, N.K. Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotech. Rep. 2016, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Matschullat, J. Arsenic in the geosphere—A review. Sci. Total Environ. 2000, 249, 297–312. [Google Scholar] [CrossRef]
- WHO. Environmental Health Criteria: Arsenic and Arsenic Compounds; WHO: Geneva, Switzerland, 2001; p. 224. [Google Scholar]
- Oritz-Escobar, M.E.; Hue, N.V.; Cutler, W.G. Recent developments on arsenic: Contamination and remediation. In Recent Research Developments in Bioenergetics; Transworld Research Network: Trivandrum, India, 2006; Volume 4, pp. 1–32. [Google Scholar]
- Wilcox, D.E. Arsenic. Can This Toxic Metalloid Sustain Life? In Interrelations between Essential Metal Ions and Human Diseases. Metal Ions in Life Sciences; Sigel, A., Sigel, H., Sigel, R., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 475–498. [Google Scholar] [CrossRef]
- Mandal, B.K.; Suzuki, K.T. Arsenic around the world: A review. Talanta 2002, 58, 201–235. [Google Scholar] [CrossRef]
- Hue, N.V. Bioremediation of Arsenic Toxicity. In Arsenic Toxicity: Prevention and Treatment; Chakrabarty, N., Ed.; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Cervantes, C.; Ji, G.; Ramírez, J.L.; Silver, S. Resistance to arsenic compounds in microorganisms. FEMS Microbiol. Rev. 1994, 15, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Ratnaike, R.N. Acute and chronic arsenic toxicity. Postgrad. Med. J. 2003, 79, 391–396. [Google Scholar] [CrossRef]
- Lim, K.T.; Shukor, M.Y.; Wasoh, H. Physical, chemical, and biological methods for the removal of arsenic compounds. Biomed. Res. Int. 2014, 2014, 503784. [Google Scholar] [CrossRef] [PubMed]
- Elangovan, D.; Chalakh, M.L. Arsenic pollution in West Bengal. Tech. Dig. 2006, 9, 31–35. [Google Scholar]
- Smedley, P.L.; Kinniburgh, D.G. A review of the source, behavior and distribution of arsenic in natural waters. Appl. Geochem. 2002, 17, 517–568. [Google Scholar] [CrossRef]
- Ghosh, S.; Mohapatra, B.; Satyanrayana, T.; Sar, P. Molecular and taxonomic characterization of arsenic (As) transforming Bacillus sp. strain IIIJ3–1 isolated from As-contaminated groundwater of Brahmaputra river basin, India. BMC Microbiol. 2020, 20, 256–275. [Google Scholar] [CrossRef]
- Mateos, L.M.; Ordonez, E.; Letek, M.; Gil, J.A. Corynebacterium glutamicum as a model bacteria for the bioremediation of arsenic. Int. Microbiol. 2006, 9, 2007–2015. [Google Scholar]
- Zhu, Y.G.; Yoshinaga, M.; Zhao, F.J.; Rosen, B.P. Earth abides arsenic biotransformations. Ann. Rev. Earth Planet. Sci. 2014, 42, 443–467. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Kazy, S.K.; Gupta, A.K.; Pal, T.; Sar, P. Diversity, metabolic properties and arsenic mobilization potential of indigenous bacteria in arsenic contaminated groundwater of West Bengal, India. PLoS ONE 2015, 19, e0118735. [Google Scholar] [CrossRef]
- Khan, A.H.; Rasul, S.B.; Munir, A.; Habibuddowla, M.; Alauddin, M.; Newaz, S.S.; Hussan, A. Appraisal of a simple arsenic removal method for groundwater of Bangladesh. J. Environ. Sci. Health 2000, 35, 1021–1041. [Google Scholar] [CrossRef]
- Banerjee, S.; Datta, S.; Chattyopadhyay, D.; Sarkar, P. Arsenic accumulating and transforming bacteria isolated from contaminated soil for potential use in bioremediation. J. Environ. Sci. Health Part A 2011, 46, 1736–1747. [Google Scholar] [CrossRef]
- Smith, A.H.; Lingas, E.O.; Rahman, M. Contamination of drinking water by arsenic in Bangladesh: A public health emergency. Bull. World Health Org. 2000, 78, 1093–1103. [Google Scholar] [PubMed]
- Rhine, E.D.; Phelps, C.D.; Young, L.Y. Anaerobic arsenic oxidation by novel denitrifying isolates. Environ. Microbiol. 2006, 8, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Simeonova, D.D.; Micheva, K.; Muller, D.A.E.; Lagarde, F.; Lett, M.C.; Groudeva, V.I.; Lieremont, D. Arsenite oxidation in batch reactors with alginate-immobilized ULPAs1 strain. Biotechnol. Bioeng. 2005, 91, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Kamaluddin, S.P.; Arunkumar, K.R.; Avudainayagam, S.; Ramasamy, K. Bioremediation of chromium contaminated environments. IndianJ. Exp. Biol. 2003, 41, 972–985. [Google Scholar]
- Anyanwu, C.U.; Ugwu, C.E. Incidence of arsenic resistant bacteria isolated from a sewage treatment plant. Int. J. Basic Appl. Sci. 2010, 10, 64–78. [Google Scholar]
- Liao, V.H.-C.; Chu, Y.-J.; Su, Y.-C.; Hsiao, S.-Y.; Wei, C.-C.; Liu, C.-W.; Liao, C.-M.; Shen, W.-C.; Chang, F.-J. Arsenite-oxidizing and arsenate-reducing bacteria associated with arsenic-rich groundwater in Taiwan. J. Contam. Hydrol. 2011, 123, 20–29. [Google Scholar] [CrossRef]
- Miyatke, M.; Hayashi, S. Characteristics of arsenic removal from aqueous solution by Bacillus megaterium strain UM-123. J. Environ. Biotechnol. 2009, 9, 123–129. [Google Scholar]
- Mohamed, E.A.H.; Farag, A.G. Arsenic removal from aqueous solutions by different Bacillus and Lysinibacillus species. Bioremediat. J. 2015, 19, 269–276. [Google Scholar] [CrossRef]
- Singh, N.; Gupta, S.; Marwa, N.; Pandey, V.; Verma, P.C.; Rathaur, S.; Singh, N. Arsenic mediated modifications in Bacillus aryabhattai and their biotechnological application for arsenic bioremediation. Chemosphere 2016, 164, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Miyatake, M.; Hayashi, S. Characteristics of arsenic removal by Bacillus cereus strain W2. Resour. Process. 2011, 58, 101–107. [Google Scholar] [CrossRef][Green Version]
- Blum, J.S.; Bindi, A.B.; Buzzelli, J.; Stolz, J.F.; Oremland, R.S. Bacillus arsenicoselenatis, sp. nov. and Bacillus selenitireducens sp. nov.: Two haloalkaliphiles from mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 1998, 171, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.R.; Cook, G.M. Isolation and characterization of arsenate-reducing bacteria from arsenic contaminated sites in New Zealand. Curr. Microbiol. 2004, 48, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Suresh, K.; Parabagaran, S.R.; Sengupta, S.; Shivaji, S. Bacillus indicus sp. nov. an arsenic-resistant bacterium isolated from an aquifer in West Bengal, India. Int. J. Syst. Evol. Microbiol. 2004, 54, 1369–1375. [Google Scholar] [CrossRef]
- Shivaji, S.; Suresh, K.; Chaturvedi, P.; Dube, S.; Sengupta, S. Bacillus arsenicus sp. nov., an arsenic-resistant bacterium isolated from a siderite concretion in West Bengal, India. Int. J. Syst. Evol. Microbiol. 2005, 55, 1123–1127. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, G.; Blanch, A.R.; Tamames, J.; Rosselló-Mora, R. Complete genome sequence of Bacillus toyonensis BCT-7112T, the active ingredient of the feed additive preparation Toyocerin. Genome Announc. 2013, 1, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Podder, M.S.; Majumder, C.B. Biosorptive performance of Bacillus arsenicus MTCC 4380 biofilm supported on sawdust/MnFe2O4 composite for the removal of As(III) and As(V). Water Conser. Sci. Eng. 2016, 1, 103–125. [Google Scholar] [CrossRef]
- Andreoni, V.; Zanchi, R.; Cavalca, L.; Corsini, A.; Romagnoli, C.; Canzi, E. Arsenite oxidation in Ancylobacter dichloromethanicus As3–1b strain: Detection of genes involved inn arsenite oxidation and CO2 fixation. Curr. Microbiol. 2012, 65, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Santini, J.M.; Stolz, J.F.; Macy, J.M. Isolation of a new arsenate-respiring bacterium-physiological and phylogenetic studies. Geomicrobiol. J. 2002, 19, 41–52. [Google Scholar] [CrossRef]
- Rosen, B.P. Biochemistry of arsenic detoxification. FEBS Lett. 2002, 529, 86–92. [Google Scholar] [CrossRef]
- Liu, Z.; Shen, J.; Carbrey, J.M.; Mukhopadhyay, R.; Agre, P.; Rosen, B.P. Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP 9. Proc. Natl. Acad. Sci. USA 2002, 99, 6053–6058. [Google Scholar] [CrossRef]
- Oremland, R.S.; Stolz, J.F. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 2005, 13, 45–49. [Google Scholar] [CrossRef]
- Villegas-Torres, M.F.; Bedoya-Reina, O.C.; Salazar, C.; Vives-Florez, M.J.; Dussan, J. Horizontal arsC gene transfer among microorganisms isolated from arsenic polluted soil. Int. Biodeter. Biodegrad. 2011, 65, 147–152. [Google Scholar] [CrossRef]
- Rosen, B.P. Families of arsenic transporters. Trends Microbiol. 1999, 7, 207–212. [Google Scholar] [CrossRef]
- Yang, H.C.; Fu, H.L.; Lin, Y.F.; Rosen, B.P. Pathways of arsenic uptake and efflux. Curr. Top. Membr. 2012, 69, 325–358. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Rosen, B.P.; Phung, L.T.; Silver, S. Microbial arsenic: From geocycles to genes and enzymes. FEMS Microbiol. Rev. 2002, 26, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Radaev, S.; Rosen, B.P.; Gatti, D.L. Structure of the ArsA ATPase: The catalytic subunit of a heavy metal resistance pump. EMBO J. 2000, 19, 4838–4845. [Google Scholar] [CrossRef] [PubMed]
- Afkar, E. Localization of the dissimilatory arsenate reductase in Sulfurospiriullum barnesii strain Ses-2. Am. J. Agric. Biol. Sci. 2012, 7, 97–105. [Google Scholar] [CrossRef]
- Musingarimi, W.; Tuffin, M.; Cowan, D. Characterisation of the arsenic resistance genes in Bacillus sp. UWC isolated from maturing fly ash acid mine drainage Neutralised soilds. S. Afr. J. Sci. 2010, 106, 59–63. [Google Scholar] [CrossRef]
- Wierzba, S. Biosorption of lead(II), zinc(II) and nickel(II) from industrial wastewater by Stenotrophomonas maltophilia and Bacillus subtilis. Pol. J. Chem. Technol. 2015, 17. [Google Scholar] [CrossRef]
- Singh, P.P.; Chopra, A.K. Removal of Zn2+ and Pb2+ using new isolates of Bacillus spp. PPS03 and Bacillus subtilis PPS04 from Paper mill effluents using indigenously designed Bench-top Bioreactor. J. Appl. Nat. Sci. 2014, 6, 47–56. [Google Scholar] [CrossRef]
- Kamika, I.; Momba, M.N. Assessing the resistance and bioremediation ability of selected bacterial and protozoan species to heavy metals in metal-rich industrial wastewater. BMC Microbiol. 2013, 13, 28. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.H.; Hassan, S.H.; Oh, S.E. Comparative study of biosorption of Zn2+ by Pseudomonas aeruginosa and Bacillus cereus. Int. Biodeter. Biodegrad. 2010, 64, 734–741. [Google Scholar] [CrossRef]
- Green-Ruiz, C.; Rodriguez-Tirado, V.; Gomez-Gil, B. Cadmium and zinc removal from aqueous solutions by Bacillus jeotgali: pH, salinity and temperature effects. Bioresour. Technol. 2008, 99, 3864–3870. [Google Scholar] [CrossRef] [PubMed]
- Sabae, S.Z.; Hazaa, M.; Hallim, S.A.; Awny, N.M.; Daboor, S.M. Bioremediation of Zn+2, Cu+2 and Fe+2 using Bacillus subtilis D215 and Pseudomonas putida biovar A D225. Biosci. Res. 2006, 3, 189–204. [Google Scholar]
- Salehizadeh, H.; Shojaosadati, S.A. Removal of metal ions from aqueous solution by polysaccharide produced from Bacillus firmus. Water Res. 2003, 37, 4231–4235. [Google Scholar] [CrossRef]
- Khan, M.; Ijaz, M.; Chotana, G.A.; Murtaza, G.; Malik, A.; Shamim, S. Bacillus altitudinis MT422188: A potential agent for zinc bioremediation. Bioremediat. J. 2021. [Google Scholar] [CrossRef]
- Mardiyono; Sajidan; Masykuri, M.; Setyono, P. Bioremediation of nickel heavy metals in electroplating industrial liquid waste with Bacillus subtilis. AIP Conf. Proc. 2019, 2202. [Google Scholar] [CrossRef]
- Al-Gheethi, A.; Mohamed, R.; Noman, E.; Ismail, N.; Kadir, O.A. Removal of heavy metal ions from aqueous solutions using Bacillus subtilis biomass pre-treated by supercritical carbon dioxide. Clean Soil Air Water 2017, 45, 1700356. [Google Scholar] [CrossRef]
- Taran, M.; Sisakhtnezhad, S.; Azin, T. Biological removal of nickel (II) by Bacillus KL1 in different conditions: Optimization by Taguchi statistical approach. Pol. J. Chem. Technol. 2015, 17, 29–32. [Google Scholar] [CrossRef]
- Das, P.; Sinha, S.; Mukherjee, S.K. Nickel Bioremediation potential of Bacillus thuringiensis KUNi1 and some environmental factors in nickel removal. Bioremediat. J. 2014, 18, 169–177. [Google Scholar] [CrossRef]
- Oves, M.; Khan, M.S.; Zaidi, A. Biosorption of heavy metals by Bacillus thuringiensis strain OSM29 originating from industrial effluent contaminated north Indian soil. Saudi J. Biol. Sci. 2013, 20, 121–129. [Google Scholar] [CrossRef]
- Öztürk, A. Removal of nickel from aqueous solution by the bacterium Bacillus thuringiensis. J. Hazard. Mater. 2007, 147, 518–523. [Google Scholar] [CrossRef]
- Priyalaxmi, R.; Murugan, A.; Raja, P.; Raj, K.D. Bioremediation of cadmium by Bacillus safensis (JX126862), a marine bacterium isolated from mangrove sediments. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 326–335. [Google Scholar]
- Basha, S.A.; Rajaganesh, K. Microbial bioremediation of heavy metals from textile industry dye effluents using isolated bacterial strains. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 785–794. [Google Scholar]
- Kim, S.Y.; Jin, M.R.; Chung, C.H.; Yun, Y.-S.; Jahng, K.Y.; Yu, K.-Y. Biosorption of cationic basic dye and cadmium by the novel biosorbent Bacillus catenulatus JB-022 strain. J. Biosci. Bioeng. 2015, 119, 433–439. [Google Scholar] [CrossRef]
- El-Helow, E.R.; Sabry, S.A.; Amer, R.M. Cadmium biosorption by a cadmium resistant strain of Bacillus thuringiensis: Regulation and optimization of cell surface affinity for metal cations. BioMetals 2000, 13, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.; Goli, D. Bioremediation of lead by a halophilic bacteria Bacillus pumilus isolated from the mangrove regions of Karnataka. Int. J. Sci. Res. 2020, 9, 1337–1343. [Google Scholar] [CrossRef]
- Qiao, W.; Zhang, Y.; Xia, H.; Luo, Y.; Liu, S.; Wang, S.; Wang, W. Bioimmobilization of lead by Bacillus subtilis X3 biomass isolated from lead mine soil under promotion of multiple adsorption mechanisms. R. Soc. Open Sci. 2019, 6, 181701. [Google Scholar] [CrossRef]
- Pan, J.-H.; Liu, R.-X.; Tang, H.-X. Surface reaction of Bacillus cereus biomass and its biosorption for lead and copper ions. J. Environ. Sci. 2007, 19, 403–408. [Google Scholar] [CrossRef]
- Arifiyanto, A.; Apriyanti, F.D.; Purwaningsih, P.; Kalqutny, S.H.; Agustina, D.; Surtiningdih, T.; Shovitri, M.; Zulaika, E. Lead (Pb) bioaccumulation; genera Bacillus isolated S1 and SS19 as a case study. AIP Conf. Proc. 2017, 1854, 020003. [Google Scholar] [CrossRef]
- Cephidian, A.; Makhdoumi, A.; Mashreghi, M.; Mahmudy Gharaie, M.H. Removal of anthropogenic lead pollutions by a potent Bacillus species AS2 isolated from geogenic contaminated site. Int. J. Environ. Sci. Technol. 2016, 13, 2135–2142. [Google Scholar] [CrossRef]
- Raj, A.S.; Muthukumar, P.V.; Bharathiraja, B.; Priya, M. Comparative biosorption capacity of copper and chromium by Bacillus cereus. Int. J. Eng. Technol. 2018, 7, 442–444. [Google Scholar] [CrossRef]
- Rohini, B.; Jayalakshmi, S. Bioremediation potential of Bacillus cereus against copper and other heavy metals. Int. J. Adv. Res. Biol. Sci. 2015, 2, 200–209. [Google Scholar]
- Karakagh, R.M.; Chorom, M.; Motamedi, H.; Kalkhajeh, Y.K.Y.; Oustan, S. Biosorption of Cd and Ni by inactivated bacteria isolated from agricultural soil treated with sewage sludge. Ecohydrol. Hydrobiol. 2012, 12, 191–198. [Google Scholar] [CrossRef]
- Rodríguez-Tirado, V.; Green-Ruiz, C.; Gómez-Gil, B. Cu and Pb biosorption on Bacillus thioparans strain u3 in aqueous solution: Kinetic and equilibrium studies. Chem. Eng. J. 2012, 181, 352–359. [Google Scholar] [CrossRef]
- da Costa, A.C.A.; Duta, F.P. Bioaccumulation of copper, zinc, cadmium and lead by Bacillus sp., Bacillus cereus, Bacillus sphaericus and Bacillus subtilis. Braz. J. Microbiol. 2001, 32. [Google Scholar] [CrossRef]
- He, L.M.; Tebo, B.M. Surface charge properties of and Cu(II) Adsorption by Spores of the Marine Bacillus sp. Strain SG-1. Appl. Environ. Microbiol. 1998, 64, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Ayangbenro, A.S.; Babalola, O.O. Genomic analysis of Bacillus cereus NWUAB01 and its heavy metal removal from polluted soil. Sci. Rep. 2020, 10, 19660–19671. [Google Scholar] [CrossRef]
- Nayak, A.K.; Panda, S.S.; Basu, A.; Dhal, N.K. Enhancement of toxic Cr (VI), Fe, and other heavy metals phytoremediation by the synergistic combination of native Bacillus cereus strain and Vetiveria zizanioides L. Int. J. Phytoremediat. 2018, 20, 682–691. [Google Scholar] [CrossRef]
- Dadrasnia, A.; Wei, K.S.C.; Shahsavari, N.; Azirun, M.S.; Ismail, S. Biosorption potential of Bacillus salmalaya Strain 139SI for removal of Cr(VI) from aqueous solution. Int. J. Environ. Res. Public Health 2015, 12, 15321–15338. [Google Scholar] [CrossRef]
- Singh, N.; Verma, T.; Gaur, R. Detoxification of hexavalent chromium by an indigenous facultative anaerobic Bacillus cereus strain isolated from tannery effluent. Afr. J. Biotechnol. 2013, 12, 1091–1103. [Google Scholar]
- Samarth, D.P.; Chandekar, C.J.; Bhadekar, R. Biosorption of heavy metals from aqueous solution using Bacillus licheniformis. Int. J. Pure Appl. Sci. Technol. 2012, 10, 12–19. [Google Scholar]
- Chaturvedi, M.K. Studies on chromate removal by chromium-resistant Bacillus sp. isolated from tannery effluent. J. Environ. Prot. 2011, 2, 76. [Google Scholar] [CrossRef]
- Mishra, S.; Doble, M. Novel chromium tolerant microorganisms: Isolation, characterization and their biosorption capacity. Ecotoxicol. Environ. Saf. 2008, 71, 874–879. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, Y.; Zeng, G.; Li, X.; Xu, W.; Fan, T. Kinetic and equilibrium studies of Cr (VI) biosorption by dead Bacillus licheniformis biomass. World J. Microbiol. Biotechnol. 2007, 23, 43–48. [Google Scholar] [CrossRef]
- Şahin, Y.; Öztürk, A. Biosorption of chromium (VI) ions from aqueous solution by the bacterium Bacillus thuringiensis. Process. Biochem. 2005, 40, 1895–1901. [Google Scholar] [CrossRef]
- Zouboulis, A.; Loukidou, M.; Matis, K. Biosorption of toxic metals from aqueous solutions by bacteria strains isolated from metal-polluted soils. Process. Biochem. 2004, 39, 909–916. [Google Scholar] [CrossRef]
- Srinath, T.; Verma, T.; Ramteke, P.; Garg, S. Chromium (VI) biosorption and bioaccumulation by chromate resistant bacteria. Chemosphere 2002, 48, 427–435. [Google Scholar] [CrossRef]
- Saranya, K.; Sundaramanickam, A.; Shekhar, S.; Swaminathan, S. Biosorption of mercury by Bacillus thuringiensis (CASKS3) isolated from mangrove sediments of southeast coast India. Ind. J. Geo-Mar. Sci. 2019, 48, 143–150. [Google Scholar]
- Upadhyay, K.H.; Vaishnav, A.M.; Tipre, D.R.; Patel, B.C.; Dave, S.R. Kinetics and mechanisms of mercury biosorption by an exopolysaccharide producing marine isolate Bacillus licheniformis. 3 Biotech 2017, 7, 313. [Google Scholar] [CrossRef]
- Dash, H.R.; Das, S. Bioremediation of inorganic mercury through volatilization and biosorption by transgenic Bacillus cereus BW-03(pPW-05). Int. Biodeter. Biodegrad. 2015, 103, 179–185. [Google Scholar] [CrossRef]
- Muneer, B.; Iqbal, M.J.; Shakoori, F.R.; Shakoori, A.R. Tolerance and biosorption of mercury by microbial consortia: Potential use in bioremediation of wastewater. Pak. J. Zool. 2013, 45, 247–254. [Google Scholar]
- Sinha, A.; Khare, S.K. Mercury bioremediation by mercury accumulating Enterobacter sp. cells and its alginate immobilized application. Biodegradation 2012, 23, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Green-Ruiz, C. Mercury (II) removal from aqueous solutions by nonviable Bacillus sp. from a tropical estuary. Bioresour. Technol. 2006, 97, 1907–1911. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhao, Y.; Xu, Z.; Ding, Y.; Zhou, X.; Dong, M. A high Mn(II)-tolerance strain, Bacillus thuringiensis HM7, isolated from manganese ore and its biosorption characteristics. PeerJ 2020, 8, e8589. [Google Scholar] [CrossRef]
- Zhenggang, X.; Yi, D.; Huimin, H.; Liang, W.; Yunlin, Z.; Guiyan, Y. Biosorption characteristics of Mn (II) by Bacillus cereus Strain HM-5 isolated from soil contaminated by manganese ore. Pol. J. Environ. Stud. 2019, 463–472. [Google Scholar] [CrossRef]
- Hasan, H.A.; Abdullah, S.R.S.; Kofli, N.T.; Kamaruddin, S.K. Biosorption of manganese in drinking water by isolated bacteria. J. Appl. Sci. 2010, 10, 2653–2657. [Google Scholar] [CrossRef]
- Adnan, A.S.M.; Abu Zeid, I.M.; Ahmad, S.A.; Halmi, M.I.E.; Abdullah, S.R.S.; Masdor, N.A.; Shukor, M.S.; Shukor, M.Y. A molybdenum-reducing Bacillus sp. strain Zeid 14 in soils from Sudan that could grow on amides and acetonitrile. Malays. J. Soil Sci. 2016, 20, 111–134. [Google Scholar]
- Othman, A.R.; Bakar, N.A.; Halmi, M.I.; Johari, W.L.; Ahmad, S.A.; Jirangon, H.; Syed, M.A.; Shukor, M.Y. Kinetics of molybdenum reduction to molybdenum blue by Bacillus sp. strain A.rzi. Biomed. Res. Int. 2013, 2013, 371058. [Google Scholar] [CrossRef]
- Sun, D.; Li, X.; Zhang, G. Biosorption of Ag(I) from aqueous solution by Bacillus licheniformis strain R08. Appl. Mech. Mater. 2013, 295–298, 129–134. [Google Scholar] [CrossRef]
- Gaballa, A.; Wang, T.; Ye, R.W.; Helmann, J.D. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 2002, 184, 6508–6514. [Google Scholar] [CrossRef]
- Mikhaylina, A.; Ksibe, A.Z.; Scanlan, D.J.; Blindauer, C.A. Bacterial zinc uptake regulator proteins and their regulons. Biochem. Soc. Trans. 2018, 46, 983–1001. [Google Scholar] [CrossRef]
- Moore, C.M.; Gaballa, A.; Hui, M.; Ye, R.W.; Helmann, J.D. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol. Microbiol. 2005, 57, 27–40. [Google Scholar] [CrossRef]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Solovieva, I.M.; Entian, K.-D. Investigation of the yvgW Bacillus subtilis chromosomal gene involved in Cd2+ ion resistance. FEMS Microbiol. Lett. 2002, 208, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, Y.; Bao, H.; Zhang, L.; Wang, R.; Zhou, X. Plasmid-borne cadmium resistant determinants are associated with the susceptibility of Listeria monocytogenes to bacteriophage. Microbiol. Res. 2015, 172, 1–6. [Google Scholar] [CrossRef]
- Hoch, J.A. Regulation of the phsophorelay and the initiation of sporulation in Bacillus subtilis. Ann. Rev. Microbiol. 1993, 47, 441–465. [Google Scholar] [CrossRef]
- Herzberg, M.; Bauer, L.; Kirsten, A.; Nies, D.H. Interplay between seven secondary metal uptake system is required for full metal resistance of Cupriavidus metallidurans. Metallomics 2016, 8, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ding, Z.; Ji, Y.; Zhao, J.; Liu, X.; Tian, J.; Wu, N.; Fan, Y. An operon consisting of a P-type ATPase gene and a transcriptional regulator gene responsible for cadmium resistances in Bacillus vietamensis 151–6 and Bacillus marisflavi 151–25. BMC Microbiol. 2020, 20, 18–30. [Google Scholar] [CrossRef]
- Hynninen, A.; Touze, T.; Pitkanen, L.; Lecreulx, D.M.; Virta, M. An efflux transporter PbrA and a phosphatase PbrB cooperate in lead-resistance mechanism in bacteria. Mol. Microbiol. 2009, 74, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Silver, S. Bacterial resistance to toxic metal ions—A review. Gene 1996, 179, 9–19. [Google Scholar] [CrossRef]
- Silver, S.; Phung, L.T. A bacterial view of periodic table: Genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 2005, 32, 587–605. [Google Scholar] [CrossRef]
- Gaballa, A.; Helmann, J.D. Bacillus subtilis CPx-type ATPases: Characterization of Cd, Zn, Co and Cu efflux systems. Biometals 2003, 16, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Smaldone, G.T.; Helmann, J.D. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 2007, 153, 4123–4128. [Google Scholar] [CrossRef] [PubMed]
- Odermatt, A.; Suter, H.; Krapf, R.; Solioz, M. An ATPase operon involved in copper resistance by Enterococcus hirae. Ann. N. Y. Acad. Sci. 1992, 671, 484–486. [Google Scholar] [CrossRef] [PubMed]
- Chillappagari, S.; Miethke, M.; Trip, H.; Kuipers, O.P.; Marahiel, M.A. Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J. Bacteriol. 2009, 191, 2362–2370. [Google Scholar] [CrossRef]
- Dhal, B.; Thatoi, H.; Das, N.; Pandey, B.D. Reduction of hexavalent chromium by Bacillus sp. isolated from chromite mine soils and characterization of reduced product. J. Chem. Technol. Biotechnol. 2010, 85, 1471–1479. [Google Scholar] [CrossRef]
- Cervantes, C.; Campos-Garcia, J.; Devars, S.; Gutierrez-Corona, F.; Loza-Tavera, H.; Torres-Guzman, J.C.; Moreno-Sanchez, R. Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 2001, 25, 335–347. [Google Scholar] [CrossRef]
- Aguilar-Barajas, E.; Paluscio, E.; Cervantes, C.; Rensing, C. Expression of chromate resistance genes from Shewanella sp. strain ANA-3 in Escherichia coli. FEMS Microbiol. Lett. 2008, 285, 97–100. [Google Scholar] [CrossRef]
- Permina, E.A.; Kazakov, A.E.; Kalinina, O.V.; Gelfand, M.S. Comparative genomics of regulation of heavy metal resistance in Eubacteria. BMC Microbiol. 2006, 6, 49. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dash, H.R.; Chakraborty, J. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl. Microbiol. Biotechnol. 2016, 100, 2967–2984. [Google Scholar] [CrossRef]
- Bogdanova, E.S.; Bass, I.A.; Minakhin, L.S.; Petrova, M.A.; Mindlin, S.Z.; Volodin, A.A.; Kalyaeva, E.S.; Tiedje, J.M.; Hobman, J.L.; Brown, N.L.; et al. Horizontal spread of mer operons among Gram-positive bacteria in natural environments. Microbiology 1998, 144, 609–620. [Google Scholar] [CrossRef]
- Narita, M.; Chiba, K.; Nishizawa, H.; Ishii, H.; Huang, C.-C.; Kawabata, Z.; Silver, S.; Endo, G. Diversity of mercury resistance determinants among Bacillus strains isolated from sediment of Minamata Bay. FEMS Microbiol. Lett. 2003, 223, 73–82. [Google Scholar] [CrossRef]
- Medina, J.A.C.; Farias, J.E.; Hernndez, A.C.; Martinez, R.G.; Valdes, S.S.; Silva, G.H.; Jones, G.H.; Campos-Guillen, J. Isolation and characterization of mercury resistant Bacillus sp. from soils with an extensive history as substrates for mercury Extraction in Mexico. Geomicrobiol. J. 2013, 30, 454–461. [Google Scholar] [CrossRef]
- Guedon, E.; Helmann, J.D. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol. Microbiol. 2003, 48, 495–506. [Google Scholar] [CrossRef]
- Fisher, S.; Buxbaum, L.; Toth, K.; Eisenstadt, E.; Silver, S. Regulation of manganese accumulation and exchange in Bacillus subtilis W23. J. Bacteriol. 1973, 113, 1373–1380. [Google Scholar] [CrossRef]
- Glasfeld, A.; Guedon, E.; Helmann, J.D.; Brennan, R.G. Structure of the manganese bound manganese transport regulator of Bacillus subtilis. Nat. Struct. Biol. 2003, 10, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Guedon, E.; Moore, C.M.; Que, Q.; Wang, T.; Ye, R.W.; Helmann, J.D. The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and sigmaB regulons. Mol. Microbiol. 2003, 49, 1477–1491. [Google Scholar] [CrossRef] [PubMed]
- Meaden, G.T. The general physical properties of manganese metal. Met. Rev. 1968, 13, 97–114. [Google Scholar] [CrossRef]
- Langenhoff, A.A.M.; Bronwers-Ceiler, D.L.; Engelberting, J.H.L.; Quist, J.J.; Wolkenfelt, J.P.N.; Zehnder, A.J.B.; Schraa, G. Microbial reduction of manganese coupled to toluene oxidation. FEMS Microbiol. Ecol. 1997, 22, 119–127. [Google Scholar] [CrossRef]
- Zhongxing, S.; Guimin, W.; Yonggen, J. Relationship between environmental manganese exposure and children’s neurological behavior. Shanghai J. Prev. Med. 2017, 29, 288–291. [Google Scholar]
- Zeng, X.-C.; Chao, S.-H.; Zhu, M.-L.; Fan, X.-T.; Jiang, Y.-X.; Cao, H.-B. The contents of five trace elements in Panaxnotoginseng and the associated health risk. China Environ. Sci. 2016, 36, 293. [Google Scholar]
- Merchant, A.T.; Spatafora, G.A. A role for the DtxR family of metalloregulators in gram-positive pathogenesis. Mol. Oral Microbiol. 2014, 29, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Shin, J.H.; Pinochet-Barros, A.; Su, T.T.; Helmann, J.D. Bacillus subtilis MntR coordinates the transcriptional regulation of manganese uptake and efflux systems. Mol. Microbiol. 2016, 103, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Reith, F.; Lengke, M.F.; Falconer, D.; Craw, D.; Southam, G. The geomicrobiology of gold. ISME J. 2007, 1, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Karamushka, V.I.; Ulberg, Z.R.; Gruzina, T.G.; Podolska, V.I.; Pertsov, N.V. Study of the role of surface structural components of microorganisms in heterocoagulation with colloidal gold particles. Prikl. Biokhim. Microbiol. 1987, 23, 697–702. [Google Scholar]
- Silver, S. Bacterial silver resistance: Molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 2003, 27, 341–353. [Google Scholar] [CrossRef]
- Woods, E.J.; Cochrane, C.A.; Percival, S.L. Prevalence of silver resistance genes in bacteria isolated from human and horse wounds. Vet. Microbiol. 2009, 138, 325–329. [Google Scholar] [CrossRef]
- Elkrewi, E.; Randall, C.P.; Ooi, N.; Cottell, J.L.; O’Neill, A.J. Cryptic silver resistance is prevalent and readily activated in certain Gram-negative pathogens. J. Antimicrob. Chemother. 2017, 72, 3043–3046. [Google Scholar] [CrossRef]
- Loh, J.V.; Percival, S.L.; Woods, E.J.; Williams, N.J.; Cochrane, C.A. Silver resistance in MRSA isolated from wound and nasal sources in humans and animals. Int. Wound J. 2009, 6, 32–38. [Google Scholar] [CrossRef]
- Randall, C.P.; Gupta, A.; Jackson, N.; Busse, D.; O’Neill, A.J. Silver resistance in Gram-negative bacteria: A dissection of endogenous and exogenous mechanisms. J. Antimicrob. Chemother. 2015, 70, 1037–1046. [Google Scholar] [CrossRef]
- Kędziora, A.; Wernecki, M.; Korzekwa, K.; Speruda, M.; Gerasymchuk, Y.; Łukowiak, A.; Bugla-Płoskońska, G. Consequences of long-term bacteria’s exposure to silver nanoformulations with different physicochemical properties. Int. J. Nanomed. 2020, 15, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Jensen, W.B. The place of Zinc, Cadmium, and Mercury in the Periodic Table. J. Chem. Ed. 2003, 80, 952–961. [Google Scholar] [CrossRef]
- Wuana, R.A.; Okieimen, F.E. Heavy Metals in contaminated soils: A review of sources, chemistry, risks and best available strategies for remediation. Int. Sch. Res. Not. 2011, 2011, 402647. [Google Scholar] [CrossRef]
- Cerasi, M.; Ammendola, S.; Battistoni, A. Competition for zinc binding in the host-pathogen interaction. Front. Cell. Infect. Microbiol. 2013, 3, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.T.; Karim, M.M. Metallothionein: A potential link in the regulation of zinc in nutritional immunity. Biol. Trace Elem. Res. 2018, 182, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chandrangsu, P.; Rensing, C.; Helmann, J.D. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017, 15, 338–350. [Google Scholar] [CrossRef]
- Zinicovscaia, I.; Yushin, N.; Grozdov, D.; Abdusamadzoda, D.; Safonov, A.; Rodlovskaya, E. Zinc-containing effluent treatment using Shewanella xiamenensis biofilm formed on zeolite. Materials 2021, 14, 1760. [Google Scholar] [CrossRef]
- Jain, D.; Kour, R.; Bhojiya, A.A.; Meena, R.H.; Singh, A.; Mohanty, S.R.; Rajpurohit, D.; Ameta, K.D. Zinc tolerant plant growth promoting bacteria alleviates phytotoxic effects of zinc on maize through zinc immobilization. Sci. Rep. 2020, 10, 1385–1398. [Google Scholar] [CrossRef]
- Zhou, C.; Gong, X.; Han, J.; Guo, R. Removal of Pb(II) and Zn(II) from aqueous solutions by raw crab shell: A comparative study. Water Environ. Res. 2016, 88, 374–383. [Google Scholar] [CrossRef]
- Rodrigues, A.C.D.; do Amaral Sobrinho, N.M.B.; dos Santos, F.S.; dos Santos, A.M.; Pereira, A.C.C.; Lima, E.S.A. Biosorption of toxic metals by water lettuce (Pistia stratiotes) biomass. Water Air Soil Pollut. 2017, 228, 156. [Google Scholar] [CrossRef]
- Silver, S. Plasmid-determined metal resistance mechanisms: Range and overview. Plasmid 1991, 27, 1–3. [Google Scholar] [CrossRef]
- Krishna, M.P.; Varghese, R.; Babu, V.A.; Jyothy, S.; Hatha, A.A.M. Bioremediation of Zinc Using Bacillus sp. Isolated from Metal Contaminated Industrial Zone. In Prospects in Bioscience: Addressing the Issues; Sabu, A., Augustine, A., Eds.; Springer India: Delhi, India, 2013; pp. 11–18. [Google Scholar] [CrossRef]
- Das, K.K.; Das, S.N.; Dhundasi, S.A. Nickel, its adverse health effects & oxidative stress. Ind. J. Med. Res. 2008, 128, 412–425. [Google Scholar]
- Schaumlöffel, D. Nickel species: Analysis and toxic effects. J. Trace Elem. Med. Biol. 2012, 26, 1–6. [Google Scholar] [CrossRef]
- Das, K.; Reddy, R.; Bagoji, I.; Das, S.; Bagali, S.; Mullur, L.; Khodnapur, J.; Biradar, M. Primary concept of nickel toxicity—An overview. J. Basic Clin. Physiol. Pharm. 2019, 30, 141–152. [Google Scholar] [CrossRef]
- Cempel, M.; Nikel, G. Nickel: A review of its sources and environmental toxicology. Pol. J. Environ. Stud. 2006, 15, 375–382. [Google Scholar]
- Beattie, H.; Keen, C.; Coldwell, M.; Tan, E.; Morton, J.; McAlinden, J.; Smith, P. The use of bio-monitoring to assess exposure in the electroplating industry. J. Expo. Sci. Environ. Epidemiol. 2017, 27, 47–55. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer (IARC). Nickel and Nickel Compounds Monograph; IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; WHO Press: Geneva, Switzerland, 2017; pp. 169–218. [Google Scholar]
- Buxton, S.; Garman, E.; Heim, K.E.; Lyons-Darden, T.; Schlekat, C.E.; Taylor, M.D.; Oller, A.R. Concise review of nickel human health toxicology and ecotoxicology. Inorganics 2019, 7, 89. [Google Scholar] [CrossRef]
- Boer, J.L.; Mulrooney, S.B.; Hausinger, R.P. Nickel-dependent metalloenzymes. Arch. Biochem. Biophys. 2014, 544, 142–152. [Google Scholar] [CrossRef]
- Genchi, G.; Carocci, A.; Lauria, G.; Sinicropi, M.S.; Catalano, A. Nickel: Human health and environmental toxicology. Int. J. Environ. Res. Public Health 2020, 17, 679. [Google Scholar] [CrossRef] [PubMed]
- Cagnetta, G.; Huang, J.; Yu, G. A mini-review on mechanochemical treatment of contaminated soil: From laboratory to large-scale. Crit. Rev. Environ. Sci. Technol. 2018, 48, 723–771. [Google Scholar] [CrossRef]
- Moghbeli, M.R.; Khajeh, A.; Alikhani, M. Nanosilica reinforced ion-exchange polyHIPE type membrane for removal of nickel ions: Preparation, characterization and adsorption studies. Chem. Eng. J. 2017, 309, 552–562. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Cui, X.; Lou, Z.; Shan, W.; Xiong, Y.; Fan, Y. Recovery of silver from nickel electrolyte using corn stalk-based sulfur-bearing adsorbent. Hydrometallurgy 2018, 176, 192–200. [Google Scholar] [CrossRef]
- Vakili, M.; Rafatullah, M.; Yuan, J.; Zwain, H.M.; Mojiri, A.; Gholami, Z.; Gholami, F.; Wang, W.; Giwa, A.S.; Yu, Y.; et al. Nickel ion removal from aqueous solutions through the adsorption process: A review. Rev. Chem. Eng. 2020, 20190047. [Google Scholar] [CrossRef]
- Njokua, K.L.; Akinyede, O.R.; Obidi, O.F. Microbial remediation of heavy metals contaminated media by Bacillus megaterium and Rhizopus stolonifer. Sci. Afr. 2020, 10, e00545. [Google Scholar] [CrossRef]
- Krom, B.P.; Huttinga, H.; Warner, J.B.; Lolkema, J.S. Impact of the Mg2+-citrate transporter CitM on heavy metal toxicity in Bacillus subtilis. Arch. Microbiol. 2002, 178, 370–375. [Google Scholar] [CrossRef][Green Version]
- Sinicropi, M.S.; Amantea, D.; Caruso, A.; Saturnino, C. Chemical and biological properties of toxic metals and use of chelating agents for the pharmacological treatment of metal poisoning. Arch. Toxicol. 2010, 84, 501–520. [Google Scholar] [CrossRef] [PubMed]
- Lokhande, B.; Patil, P.S.; Uplane, M.D. Studies on cadmium oxide sprayed thin films deposited through non-aqueous medium. Mater. Chem. Phys. 2004, 84, 238–242. [Google Scholar] [CrossRef]
- Yuan, Z.; Luo, T.; Liu, X.; Hua, H.; Zhuang, Y.; Zhang, X.; Zhang, L.; Zhang, Y.; Xu, W.; Ren, R. Tracing anthropogenic cadmium emissions: From sources to pollution. Sci. Total Environ. 2019, 676, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, A. Cadmium toxicity: Effects on human reproduction and fertility. Rev. Environ. Health 2019, 34, 327–338. [Google Scholar] [CrossRef]
- Parsons, C.; Lee, S.; Kathariou, S. Dissemination and conservation of cadmium and arsenic resistance determinants in Listeria and other Gram-positive bacteria. Mol. Microbiol. 2020, 113, 560–569. [Google Scholar] [CrossRef]
- Endo, G.; Silver, S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 1995, 177, 4437–4441. [Google Scholar] [CrossRef]
- Yoon, K.P.; Silver, S. A second gene in the Staphylococcus aureus cad A cadmium resistance determinant of plasmid pI258. J. Bacteriol. 1991, 173, 7636–7642. [Google Scholar] [CrossRef]
- Silver, S.; Phung, L.T. Bacterial plasmid-mediated heavy metal resistances: New surprises. Annu. Rev. Microbiol. 1996, 50, 753–897. [Google Scholar] [CrossRef]
- Lang, W.K.; Glassey, K.; Archibald, A.R. Influence of phosphate supply on teichoic acid and teichuronic acid content of Bacillus subtilis cell walls. J. Bacteriol. 1982, 151, 367–375. [Google Scholar] [CrossRef]
- Abbas, S.Z.; Rafatullah, M.; Hossain, K.; Ismail, N.; Tajarudin, H.A.; Khalil, H.P.S.A. A review on mechanism and future perspectives of cadmium-resistant bacteria. Int. J. Environ. Sci. Technol. 2017, 15, 243–262. [Google Scholar] [CrossRef]
- Kumari, W.M.N.H.; Thiruchittampalam, S.; Weerasinghe, M.S.S.; Chandrasekharan, N.V.; Wijayarathna, C.D. Characterization of a Bacillus megaterium strain with metal bioremediation potential and in silico discovery of novel cadmium binding motifs in the regulator, CadC. Appl. Microbiol. Biotechnol. 2021, 105, 2573–2586. [Google Scholar] [CrossRef]
- Peraferrer, C.; Martinez, M.; Poch, J.; Villaescusa, I. Toxicity of metal-ethylene diaminetetraacetic acid solution as a function of chemical speciation: An approach for toxicity assessment. Arch. Environ. Contam. Toxicol. 2012, 63, 484–494. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rukhsana, F.; Butterly, C.R.; Baldock, J.A.; Xu, J.M.; Tang, C. Model organic compounds differ in priming effects on alkalinity release in soils through carbon and nitrogen mineralization. Soil Biol. Biochem. 2012, 51, 35–43. [Google Scholar] [CrossRef]
- Blanco, A. Immobilization of Nonviable Cyanobacteria and Their Use for Heavy Metal Adsorption from Water in Environmental Biotechnology and Cleaner Bioprocesses; Oluguin, E.J., Sanchez, E.J., Hernandez, E., Eds.; Taylor and Francis: Philadelphia, PA, USA, 2000; p. 135. [Google Scholar]
- Qin, W.; Zhao, J.; Yu, X.; Liu, Z.; Chu, Z.; Tian, J.; Wu, N. Improving cadmium resistance in Escherichia coli through continuous genome evolution. Front. Microbiol. 2019, 10, 278. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, S.; Kilinc, E.; Poli, A.; Nicolaus, B.; Guven, K. Biosorption of Cd, Cu, Ni, Mn and Zn from aqueous solutions by thermophilic bacteria, Geobacillus toebii subsp. Decanicus and Geobacillus thermoleovorans sub. sp. Stromboliensis: Equilibrium, kinetic and thermodynamic studies. Chem. Eng. J. 2009, 152, 195–206. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.-R.; Zhang, L.-M.; He, J.-Z. Sorption mechanism and distribution of cadmium by different microbial species. J. Environ. Manag. 2019, 237, 552–559. [Google Scholar] [CrossRef]
- Chen, M.; Li, Y.; Zhang, L.; Wang, J.; Zhenf, C.; Zhang, X. Analysis of gene expression provides insights into the mechanism of cadmium tolerance in Acidthiobacillus ferrooxidans. Curr. Microbiol. 2015, 70, 290–297. [Google Scholar] [CrossRef]
- Kushwaha, A.; Hans, N.; Kumar, S.; Rani, R. A critical review on speciation, mobilization and toxicity of lead in soil-microbe-plant system and bioremediation strategies. Ecotoxicol. Environ. Saf. 2018, 147, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Pacyna, E.G.; Pacyna, J.M.; Fudala, J.; Strzelecka-Jastrzab, E.; Hlawiczka, S.; Panasiuk, D.; Nitter, S.; Pregger, T.; Pfeiffer, H.; Friedrich, R. Current and future emissions of selected heavy metals to the atmosphere from anthropogenic sources in Europe. Atmos. Environ. 2007, 41, 8557–8566. [Google Scholar] [CrossRef]
- Varghese, R.K.M.P.; Arun, B.V.; Hatha, M.A.A. Biological removal of lead by Bacillus sp. obtained from metal contaminated industrial area. J. Microbiol. Biotechnol. 2012, 2, 756–770. [Google Scholar]
- Lindsay, W.L. Chemical Equilibria in Soils; John Wiley and Sons: Chichester, UK, 1979. [Google Scholar]
- Rigoletto, M.; Calza, P.; Gaggero, E.; Malandrino, M.; Fabbri, D. Bioremediation methods for the recovery of lead-contaminated soils: A review. Appl. Sci. 2020, 10, 3528. [Google Scholar] [CrossRef]
- Silver, S.; Ji, G. Newer systems for bacterial resistances to toxic heavy metals. Environ. Health Perspect. 1994, 102, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, A.; Asmuss, M.; Ehleben, I.; Herzer, U.; Kostelac, D.; Pelzer, A.; Schwerdtle, T.; Burkle, A. Interference by toxic metal ions with DNA repair processes and cell cycle control: Molecular mechanisms. Environ. Health Perspect. 2002, 110, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Crichton, R.R.; Pierre, J.L. Old iron, young copper: From Mars to Venus. BioMetals 2001, 14, 99–112. [Google Scholar] [CrossRef]
- Osman, D.; Cavet, J.S. Copper Homeostasis in Bacteria. Adv. Appl. Microbiol. 2008, 65, 217–247. [Google Scholar] [CrossRef]
- Zhao, S.L.; Liu, Q.; Qi, Y.T.; Duo, L. Responses of root growth and protective enzymes to copper stress in turfgrass. Acta Biol. Crac. Bot. 2010, 52, 7–11. [Google Scholar] [CrossRef]
- Ma, Z.; Cowart, D.M.; Scott, R.A.; Giedroc, D.P. Molecular insights into the metal selectivity of the Cu(I)-sensing repressor CsoR from Bacillus Subtilis. Biochemisty 2009, 48, 3325–3334. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.; Liu, L.; Wang, Q.; Saleem, M.H.; Ullah, B.; Peng, S. Copper environmental toxicology, recent advances, and future outlook: A review. Environ. Sci. Pollut. Res. 2019, 26, 18003–18016. [Google Scholar] [CrossRef] [PubMed]
- Babak, L.; Šupinova, P.; Zichova, M.; Burdychova, R.; Vitova, E. Biosorption of Cu, Zn and Pb by thermophilic bacteria—Effect of biomass concentration on biosorption capacity. Acta Univ. Agric. Silvic. Mendel. Brunesis 2012, 60, 9–18. [Google Scholar] [CrossRef]
- Liu, Y.-G.; Liao, T.; He, Z.-B.; Li, T.-T.; Wang, H.; Hu, X.-J.; Guo, Y.-M.; He, Y. Biosorption of copper(II) from aqueous solution by Bacillus subtilis cells immobilized into chitosan beads. Trans. Nonferr. Met. Soc. 2013, 23, 1804–1814. [Google Scholar] [CrossRef]
- Changela, A.; Chen, K.; Xue, Y.; Holschen, J.; Outten, C.E.; O’Halloran, T.V.; Mondragon, A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 2003, 301, 1383–1387. [Google Scholar] [CrossRef]
- Liang, M.; Xu, S.; Zhu, Y.; Chen, X.; Deng, Z.; Yan, L.; He, H. Preparation and characterization of Fe-Mn binary oxide/mulberry stem biochar composite adsorbent and adsorption of Cr(VI) from aqueous solution. Int. J. Environ. Res. Public Health 2020, 17, 676. [Google Scholar] [CrossRef]
- Goswami, L.; Mukhopadhyay, R.; Bhattacharya, S.S.; Das, P.; Goswami, R. Detoxification of chromium-rich tannery industry sludge by Eudrillus eugeniae: Insight on compost quality fortification and microbial enrichment. Bioresour. Technol. 2018, 266, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.A.; Apel, W.A.; Petersen, J.N.; Peyton, B.M. Effect of carbon and energy source on bacterial chromate reduction. Bioremediat. J. 2002, 6, 205–215. [Google Scholar] [CrossRef]
- Daulton, T.L.; Little, B.J.; Jones-Meehan, J.; Blom, D.A.; Allard, L.F. Microbial reduction of chromium from the hexavalent to divalent state. Geochim. Cosmochim. Acta 2007, 71, 556–565. [Google Scholar] [CrossRef]
- Aranda-García, E.; Cristiani-Urbina, E. Effect of pH on hexavalent and total chromium removal from aqueous solutions by avocado shell using batch and continuous systems. Environ. Sci. Pollut. Res. Int. 2019, 26, 3157–3173. [Google Scholar] [CrossRef]
- Módenes, A.N.; de Oliveira, A.P.; Espinoza-Quiñones, F.R.; Trigueros, D.E.G.; Kroumov, A.D.; Bergamasco, R. Study of the involved sorption mechanisms of Cr(VI) and Cr(III) species onto dried Salvinia auriculata biomass. Chemosphere 2017, 172, 373–383. [Google Scholar] [CrossRef]
- Bibi, I.; Niazi, N.K.; Choppala, G.; Burton, E.D. Chromium (VI) removal by siderite (FeCO3) in anoxic aqueous solutions: An X-ray absorption spectroscopy investigation. Sci. Total Environ. 2018, 640, 1424–1431. [Google Scholar] [CrossRef]
- Viti, C. Response of microbial communities to different doses of chromate in soil microcosms. J. Appl. Soil Ecol. 2006, 34, 125–139. [Google Scholar] [CrossRef]
- Tanga, X.; Huang, Y.; Li, Y.; Wang, L.; Pei, X.; Zhou, D.; He, P.; Hughes, S.S. Study on detoxification and removal mechanisms of hexavalent chromium by microorganisms. Ecotoxicol. Environ. Saf. 2021, 208, 111699. [Google Scholar] [CrossRef]
- Fernandez, M.; Paisio, C.E.; Perotti, R.; Pereira, P.P.; Agostini, E.; Gonzalez, P.S. Laboratory and field microcosms as useful experimental systems to study the bioaugmentation treatment of tannery effluents. J. Environ. Manag. 2019, 234, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Ahemad, M. Bacterial mechanisms for Cr(VI) resistance and reduction: An overview and recent advances. Folia Microbiol. 2014, 59, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Hossan, S.; Hossain, S.; Islam, M.R.; Kabir, M.H.; Ali, S.; Islam, M.S.; Imran, K.M.; Moniruzzaman, M.; Mou, T.J.; Parvez, A.K.; et al. Bioremediation of hexavalent chromium by chromium resistant bacteria reduces phytotoxicity. Int. J. Environ. Res. Public Health 2020, 17, 6013. [Google Scholar] [CrossRef]
- Congeevaram, S.; Dhanarani, S.; Park, J.; Dexilin, M.; Thamaraiselvi, K. Biosorption of chromium and nickel by heavy metal resistant fungal and bacterial isolates. J. Hazard. Mater. 2007, 146, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bhatia, D.; Singh, R.; Rani, S.; Bishnoi, N.R. Sorption of heavy metals from electroplating effluent using immobilized biomass Trichoderma viride in a continuous packed-bed column. Int. Biodeter. Biodegrad. 2011, 65, 1133–1139. [Google Scholar] [CrossRef]
- Vijayaraghavan, K.; Yun, Y.S. Bacterial biosorbents and biosorption. Biotechnol. Adv. 2008, 26, 266–291. [Google Scholar] [CrossRef] [PubMed]
- Habashi, F. Mercury, Physical and Chemical Properties. In Encyclopedia of Metalloproteins; Kretsinger, R.H., Uversky, V.N., Permyakov, E.A., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Driscoll, C.T.; Mason, R.P.; Chan, H.M.; Jacob, D.J.; Pirrone, N. Mercury as a global pollutant: Sources, pathways, and effects. Environ. Sci. Technol. 2013, 47, 4967–4983. [Google Scholar] [CrossRef] [PubMed]
- Velásquez-Riaño, M.; Benavides-Otaya, H.D. Bioremediation techniques applied to aqueous media contaminated with mercury. Crit. Rev. Biotechnol. 2016, 36, 1124–1130. [Google Scholar] [CrossRef]
- Beckers, F.; Rinklebe, J. Cycling of mercury in the environment: Sources, fate, and human health implications: A review. Crit. Rev. Environ. Sci. Technol. 2017, 47, 693–794. [Google Scholar] [CrossRef]
- Wagner-Dobler, I. Pilot plant for bioremediation of mercury containing industrial wastewater. Appl. Microbiol. Biotechnol. 2003, 62, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Amit Jamwal, R.; Mishra, N.; Singh, D.K. Recent developments in environmental mercury bioremediation and its toxicity: A review. Environ. Nanotechnol. Monit. Manag. 2020, 13, 100283. [Google Scholar] [CrossRef]
- Imam, S.S.A.; Rajpoot, I.K.; Gajjar, B.; Sachdeva, A. Comparative study of heavy metal bioremediation in soil by Bacillus subtilis and Saccharomyces cerevisiae. Ind. J. Sci. Technol. 2016, 9. [Google Scholar] [CrossRef]
- Yan-de, J.; Zhen-li, H.; Xiao-e, Y. Role of soil rhizobacteria in phytoremediation of heavy metal contaminated soils. J. Zhejiang Univ. Sci. 2007, 8, 192–207. [Google Scholar] [CrossRef]
- Stapleton, P.; Pike, R.; Mullany, P.; Lucas, V.; Roberts, G.; Rowbury, R.; Wilson, M.; Richards, H. Mercuric resistance genes in gram-positive oral bacteria. FEMS Microbiol. Lett. 2004, 236, 213–220. [Google Scholar] [CrossRef][Green Version]
- Das, A.P.; Sukla, L.B.; Pradhan, N.; Nayak, S. Manganese biomining: A review. Bioresour. Technol. 2011, 102, 7381–7387. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–664. [Google Scholar] [CrossRef]
- Ehrlich, H.L.; Newman, D.K. Geomicrobiology, 5th ed.; CRC/Taylor and Francis: Boca Raton, FL, USA, 2009. [Google Scholar] [CrossRef]
- Helmann, J.D. Specificity of metal sensing: Iron and manganese homeostasis in Bacillus subtilis. Biol. Chem. 2014, 289, 28112–28120. [Google Scholar] [CrossRef] [PubMed]
- Paruthiyil, S.; Pinochet-Barros, A.; Huang, X.; Helmann, J.D. Bacillus subtilis TerC family proteins help prevent manganese intoxication. J. Bacteriol. 2020, 202, e00624-19. [Google Scholar] [CrossRef] [PubMed]
- Barceloux, D.G. Molybdenum. Clin. Toxicol. 1999, 37, 231–237. [Google Scholar] [CrossRef]
- Morrison, S.J.; Mushovic, P.S.; Niesen, P.L. Early breakthrough of molybdenum and uranium in a permeable reactive barrier. Environ. Sci. Technol. 2006, 40, 2018–2024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Reardon, E.J. Removal of B, Cr, Mo, and Se from wastewater by incorporation into hydrocalumite and ettringite. Environ. Sci. Technol. 2003, 37, 2947–2952. [Google Scholar] [CrossRef]
- Grunden, A.M.; Shanmugam, K.T. Molybdate transport and regulation in bacteria. Arch. Microbiol. 1997, 168, 345–354. [Google Scholar] [CrossRef]
- Périnet, S.; Jeukens, J.; Kukavica-Ibrulj, I.; Ouellet, M.M.; Charette, S.J.; Levesque, R.C. Molybdate transporter ModABC is important for Pseudomonas aeruginosa chronic lung infection. BMC Res. Notes 2016, 9, 23. [Google Scholar] [CrossRef]
- Xia, Z.; Lei, L.; Zhang, H.Y.; Wei, H.L. Characterization of the ModABC molybdate transport system of Pseudomonas putida in nicotine degradation. Front. Microbiol. 2018, 9, 3030. [Google Scholar] [CrossRef]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef]
- Self, W.T.; Grunden, A.M.; Hasona, A.; Shanmugam, K.T. Molybdate transport. Res. Microbiol. 2001, 152, 311–321. [Google Scholar] [CrossRef]
- Zhong, Q.; Kobe, B.; Kappler, U. Molybdenum enzymes and how they support virulence in pathogenic bacteria. Front. Microbiol. 2020, 11, 3185. [Google Scholar] [CrossRef]
- Ge, X.; Thorgersen, M.P.; Poole, F.L.; Deutschbauer, A.M.; Chandonia, J.M.; Novichkov, P.S.; Gushgari-Doyle, S.; Lui, L.M.; Nielsen, T.; Chakraborty, R.; et al. Characterization of a metal-resistant Bacillus strain with a high molybdate affinity ModA from contaminated sediments at the oak ridge reservation. Front. Microbiol. 2020, 11, 2543. [Google Scholar] [CrossRef]
- Boyle, R.W. The geochemistry of gold and its deposits. Geol. Surv. Can. Bull. 1979, 280, 583. [Google Scholar]
- Karthikeyan, S.; Beveridge, T.J. Pseudomonas aeruginosa biofilms react with and precipitate toxic soluble gold. Environ. Microbiol. 2002, 4, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Bütof, L.; Wiesemann, N.; Herzberg, M.; Altzschner, M.; Holleitner, A.; Reith, F.; Nies, D.H. Synergistic gold-copper detoxification at the core of gold biomineralisation in Cupriavidus metallidurans. Metallomics 2018, 10, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Watterson, J.R. Preliminary evidence for the involvement of budding bacteria in the origin of Alaskan placer gold. Geology 1992, 20, 315–318. [Google Scholar] [CrossRef]
- Hough, R.M.; Butt, C.R.M.; Reddy, S.M.; Verrall, M. Gold nuggets: Supergene or hypogene? Aust. J. Earth Sci. 2007, 54, 959–964. [Google Scholar] [CrossRef]
- Nies, D.H. Microbial heavy metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef]
- Korobushkina, E.D.; Mineyev, G.G.; Praded, G.P. Mechanism of the microbiological process of dissolution of gold. Mikrobiologiia 1976, 45, 535–538. [Google Scholar]
- Korobushkina, E.D.; Karavaiko, G.I.; Korobushkin, I.M. Biochemistry of gold. In Environmental Biogeochemistry Ecological Bulletin; Hallberg, R., Ed.; Oikos Editorial Office: Stockholm, Sweden, 1983; pp. 325–333. [Google Scholar]
- Lengke, M.F.; Southam, G. The deposition of elemental gold from gold(I)-thiosulfate complex mediated by sulfate-reducing bacterial conditions. Econ. Geol. 2007, 102, 109–126. [Google Scholar] [CrossRef]
- Gee, A.R.; Dudeney, A.W.L. Adsorption and crystallization of gold on biological surfaces. In Biohydrometallurgy; Kelly, D.P., Norris, P.R., Eds.; Science & Technology Letters: London, UK, 1988; pp. 437–451. [Google Scholar]
- Ulberg, Z.R.; Karamushka, V.I.; Vidybida, A.K. Interaction of energized bacteria calls with particles of colloidal gold: Peculiarities and kinetic model of the process. Biochim. Biophys. Acta 1992, 1134, 89–95. [Google Scholar] [CrossRef]
- Agency for Toxic Substances and Disease Registry (ATSDR). Toxicological Profile for Silver; US Department of Health and Human Services, Public Health Service, Agency for Toxic Substances and Disease Registry (TP-90–24): Atlanta, GA, USA, 1990.
- Eisler, R. Silver Hazards to Fish, Wildlife and Invertebrates: A Synoptic Review; Department of the Interior, National Biological Service: Washington, DC, USA, 1997; pp. 1–44. [Google Scholar]
- Becker, R.O. Silver ions in the treatment of local infections. Met.-Based Drugs 1999, 6, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, L.J.; White, R.J.; Chipman, J.K. Silver and nanoparticles of silver in wound dressings: A review of efficacy and safety. J. Wound Care 2011, 20, 543–549. [Google Scholar] [CrossRef] [PubMed]


| Metal | Bacterial Strains | Initial Metal Concentration (%/mg/g/mM/ppm/mg/L) | Metal Uptake Ability (%/mg/g/mM/ppm/mg/L/mol/g) | Reference |
|---|---|---|---|---|
| Arsenic | Bacillus sp. KM02 | 100 ppm | 51.45% (As3+) | [59] |
| B. licheniformis B. polimyxa | 0–100 mM 0–20 mM | 100 ppm (As0) 100 ppm (As0) | [88] | |
| Bacillus sp. IIIJ3–1 | 350 smM (As5+) 10 mM (As3+) | 350 mM (As5+) 10 mM (As3+) | [71] | |
| B. barbaricus | - | 20 mM (As5+) 0.3 mM (As3+) | [91] | |
| B. indicus Sd/3T | 0 mM 0 mM | 20 mM (As5+) 30 mM (As3+) | [89] | |
| B. selenatiredreducens | 10 mM | 0 mM (As5+) 0.3 mM (As3+) | [87] | |
| B. arsenicus con a/3 | 20 mM 0.5 mM | 20 mM (As5+) 0.3 mM (As3+) | [90] | |
| B. cereus W2 | 50 mg/L | 1.870 mg/L (As3+) | [86] | |
| B. cereus EA5 B. fusiformis EA2 | 15 mg/L | 94.9% 99.7% | [84] | |
| B. arsenicus MTCC 4380 | 2000 mg/L 1800 mg/L | 89.462% (As5+) 83.043% (As3+) | [92] | |
| Zinc | B. subtilis | 178 mg/L | 49.7 mg/L | [105] |
| Bacillus sp. (KF710041) B. subtilis (KF710042) | - | 73.29% 78.15% | [106] | |
| B. licheniformis | - | 53% | [107] | |
| B. cereus | 0–200 mg/L | 66.6 mg/g | [108] | |
| B. jeotgali | 75 mg/l | 30% | [109] | |
| B. subtilis D215 | 100 mg/L | 63.73% | [110] | |
| B. firmus | 100 mg/L | 61.8% | [111] | |
| B. altitudinis | 100 mg/L | 87 mg/L | [112] | |
| Nickel | B. subtilis | 2.14 ppm | 85.61% | [113] |
| B. subtilis BM1 B. subtilis BM2 B. subtilis BM3 | 2–32 mg/L | 98.54% 99.2% 96.3% | [114] | |
| B. subtilis | 178 mg/L | 57.8 mg/g | [105] | |
| Bacillus sp. KL1 | 100 ppm | 55.06% | [115] | |
| B. thuringiensis KUNi1 | 0–7.5 mM | 82% | [116] | |
| B. thuringiensis OSM29 | 25–150 mg/L | 94% | [117] | |
| B. thuringiensis | 250 mg/L | 15.7% | [118] | |
| Cadmium | B. safensis |
40 ppm 60 ppm | 83.5% 98.10% | [119] |
| B. licheniformis | - | 98.34% | [120] | |
| B. catenulatus JB-022 | 150 mg/L | 66% | [121] | |
| B. thuringiensis DM55 | 0.25 mM | 79% | [122] | |
| Lead | B. pumilus MF472596 | 100–1000 ppm | 96% | [123] |
| B. subtilis X3 | 200–1400 mg/L | 590.49 mg/g | [124] | |
| B. cereus | 5–100 mg/L | 36.71 mg/g | [125] | |
| Bacillus S1 Bacillus SS19 | 75 and 100 mg/L 50 mg/mL | 53%, 51% 57% | [126] | |
| Bacillus sp. AS2 | 500 ppm | 74.5 mg/g (99.5 %) | [127] | |
| Copper | B. cereus | 100 ppm | 54% | [128] |
| B. cereus | 400 ppm | 48% | [129] | |
| B. thuringiensis OSM29 | 25 mg/L | 91.8% | [117] | |
| B. licheniformis | 5 gm/L | 32% | [130] | |
| B. thioparans | 40 mg/L | 27.3 mg/g | [131] | |
| B. subtilis D215 | 100 mg/L | 67.18% | [110] | |
| B. sphaericus B. cereus Bacillus sp. |
17.6 mg/L 44.0 mg/L 88.0 mg/L | 5.6 mol/g 5.9 mol/g 6.4 mol/g | [132] | |
| Bacillus sp. SG-1 | - | 60% | [133] | |
| Chromium | B. cereus NWUAB01 | 100 mg/L | 43% | [134] |
| B. cereus | 100 mg/L | 81% | [135] | |
| B. salmalaya 139SI | 50 ppm | 20.35 mg/g | [136] | |
| B. cereus FA-3 | 1000 μg/ml | 72% | [137] | |
| B. licheniformis | 15 mg/L | 95% | [138] | |
| Bacillus sp. B | 500–4500 mg/L | 47% | [139] | |
| B. marisflavi | 200 mg/L | 5.783% | [140] | |
| B. licheniformis | 300 mg/g | 69.4% | [141] | |
| B. thuringiensis | 250 mg/L | 83.3% | [142] | |
| B. licheniformis B. laterosporus | - | 62 mg/g 72.6 mg/g | [143] | |
| B. circulans B. megaterium | 0.96 mg/L | 34.5% 32% | [144] | |
| Mercury | B. thuringiensis CASKS3 | 200 mg/L 400 mg/L 600 mg/L | 62.4% 54% 40% | [145] |
| B. licheniformis | 50 mg/L | 70% | [146] | |
| B. cereus BW-03(pPW-05) | 5–50 ppm | 96.4% | [147] | |
| B. licheniformis | 100 μg/mL | 70% | [148] | |
| B. cereus | 5 mg/L | 104.1 mg/g | [149] | |
| Bacillus sp. | 1–10 mg/L | 7.9 mg/g | [150] | |
| Manganese | B. thuringiensis HM7 | 400 mg/L | 95.04% | [151] |
| B. cereus HM-5 | 600 mg/L | 67% | [152] | |
| Bacillus sp. | 13.3 mg/g | 55.56 mg/g | [153] | |
| Molybdenum | Bacillus sp. Zeid 14 | - | 200 mg/L | [154] |
| Bacillus sp. strain A.rzi | 0.1 mM | Not reported | [155] | |
| Silver | B. licheniformis R08 | 100 mg/L | 73.6 mg/g | [156] |
| Metal | Protein(s)/Gene(s) | Method(s) | Reference |
|---|---|---|---|
| Arsenic | ars operon (arsR, arsD, arsA, arsB, arsC) | Reduction (Detoxification) Efflux Cell membrane binding, Adsorption on cell surface Complexation by exopolysaccharides | [59,81,87,99,100,101,102] |
| Zinc | Zur ZosA ycdHI-yceA yciABC CadA CzcD | Physico-chemical adsorption Ion exchange Efflux Uptake | [108,157,158] |
| Nickel | CzcD CitM | Efflux | [159,160] |
| Cadmium | cad operon yvgW KinA | Efflux | [122,134,161,162,163,164,165] |
| Lead | pbr operon | Efflux | [166,167,168] |
| Copper | CueR copZA operon (CopA, CopZ, CopB) YcnJ | Efflux by chaperone Uptake | [169,170,171,172] |
| Chromium | ChrR | Efflux, Uptake Enzymatic reduction (Detoxification) | [173,174,175] |
| Mercury | mer operon (merR, merA, merB) MerR MerA MerB | Efflux Enzymatic reduction (Detoxification) | [176,177,178,179,180] |
| Manganese | mntABCD operon MntR MntH MneP MneS | Efflux Uptake | [181,182,183,184] |
| Molybdenum | modABC operon | Uptake | [148,185,186,187,188,189,190] |
| Gold | Not reported | Bioaccumulation | [191,192] |
| Silver | SilP sil genes | Efflux | [193,194,195,196,197,198] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alotaibi, B.S.; Khan, M.; Shamim, S. Unraveling the Underlying Heavy Metal Detoxification Mechanisms of Bacillus Species. Microorganisms 2021, 9, 1628. https://doi.org/10.3390/microorganisms9081628
Alotaibi BS, Khan M, Shamim S. Unraveling the Underlying Heavy Metal Detoxification Mechanisms of Bacillus Species. Microorganisms. 2021; 9(8):1628. https://doi.org/10.3390/microorganisms9081628
Chicago/Turabian StyleAlotaibi, Badriyah Shadid, Maryam Khan, and Saba Shamim. 2021. "Unraveling the Underlying Heavy Metal Detoxification Mechanisms of Bacillus Species" Microorganisms 9, no. 8: 1628. https://doi.org/10.3390/microorganisms9081628
APA StyleAlotaibi, B. S., Khan, M., & Shamim, S. (2021). Unraveling the Underlying Heavy Metal Detoxification Mechanisms of Bacillus Species. Microorganisms, 9(8), 1628. https://doi.org/10.3390/microorganisms9081628






