Effects of Lactobacillus salivarius LN12 in Combination with Amoxicillin and Clarithromycin on Helicobacter pylori Biofilm In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain, Medium and Growth Conditions
2.2. MIC and FIC of Antibiotics
2.3. Construction of H. pylori Biofilm
2.4. H. pylori Biofilm Resistance to Antibiotics
2.5. Zone of Inhibition on H. pylori
2.6. MIC of LN12 CFS
2.7. Different Treatments on the Effectiveness of LN12 CFS
2.8. The Effect of LN12 CFS Combined with Antibiotics on the Biomass of H. pylori Biofilm
2.9. The Effect of LN12 CFS Combined with Antibiotics on the Morphology and Structure of H. pylori Biofilm
2.10. The Effect of LN12 CFS Combined with Antibiotics on the Survival Rate of H. pylori Biofilm
2.11. Effect of LN12 CFS Combined with Antibiotics on EPS of H. pylori Biofilm
2.12. The Effect of LN12 CFS Combined with Antibiotics on the Expression of Virulence Genes of H. pylori Biofilm
2.13. Statistical Analysis
3. Results
3.1. MIC of Antibiotics and Lactobacillus salivarius LN12 Cell-Free Supernatant (CFS)
3.2. FIC of Amoxicillin and Clarithromycin for H. pylori
3.3. H. pylori Biofilm
3.4. MBEC of Amoxicillin and Clarithromycin for H. pylori 3192
3.5. Zone of Inhibition on H. pylori
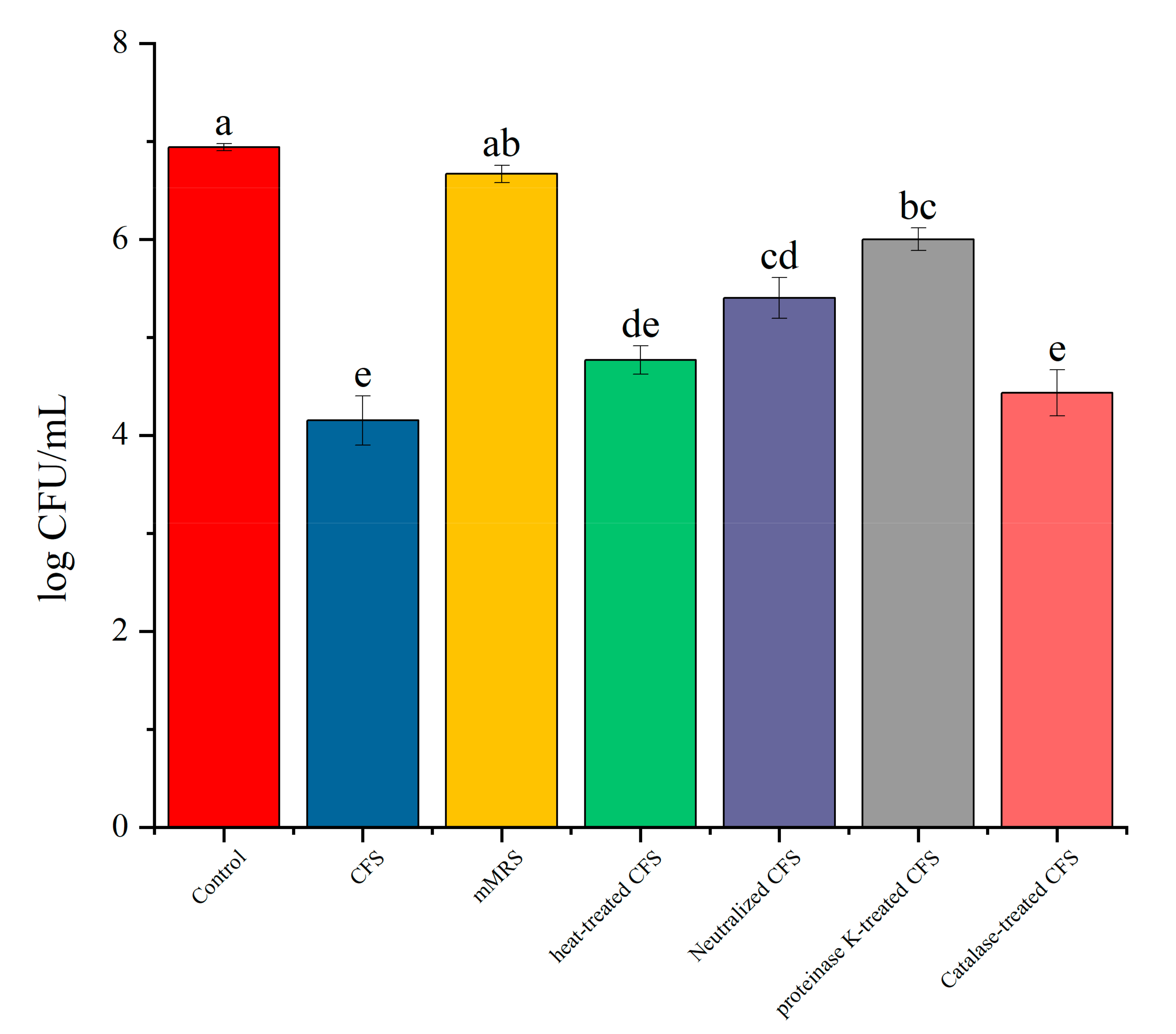
3.6. The Impact of Different Treatments of CFS on the Survival Rate of Biofilm
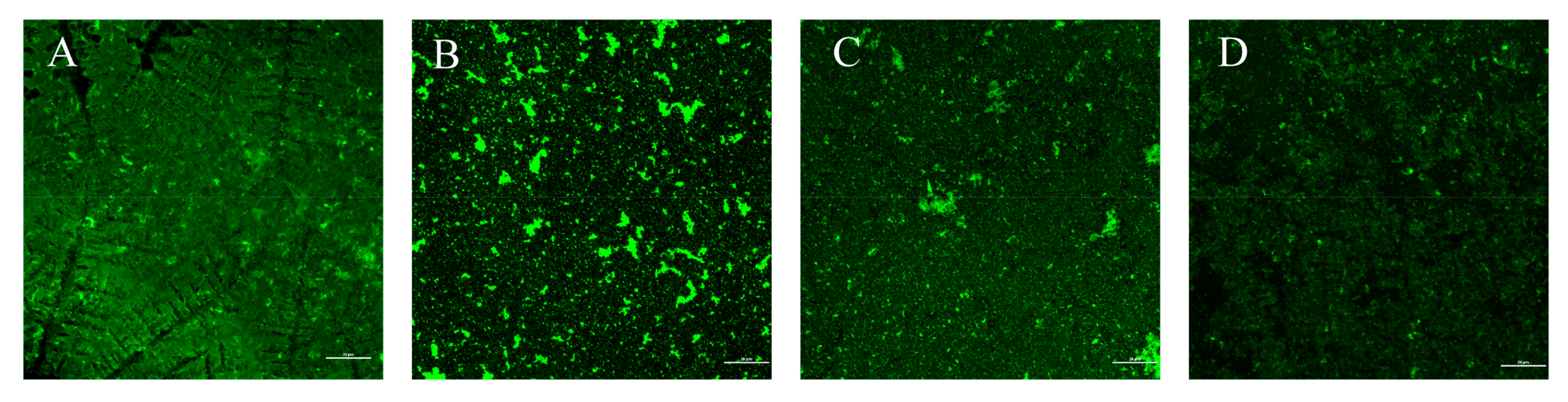
3.7. The Effect of LN12 CFS with Antibiotics on the Morphology of H. pylori Biofilm
3.8. The Effect of LN12 CFS with Antibiotics on H. pylori Biofilm Biomass
3.9. The Effect of LN12 CFS with Antibiotics on the Survival Rate of H. pylori Biofilm
3.10. Effect of LN12 CFS Combined with Antibiotics on EPS of H. pylori Biofilm
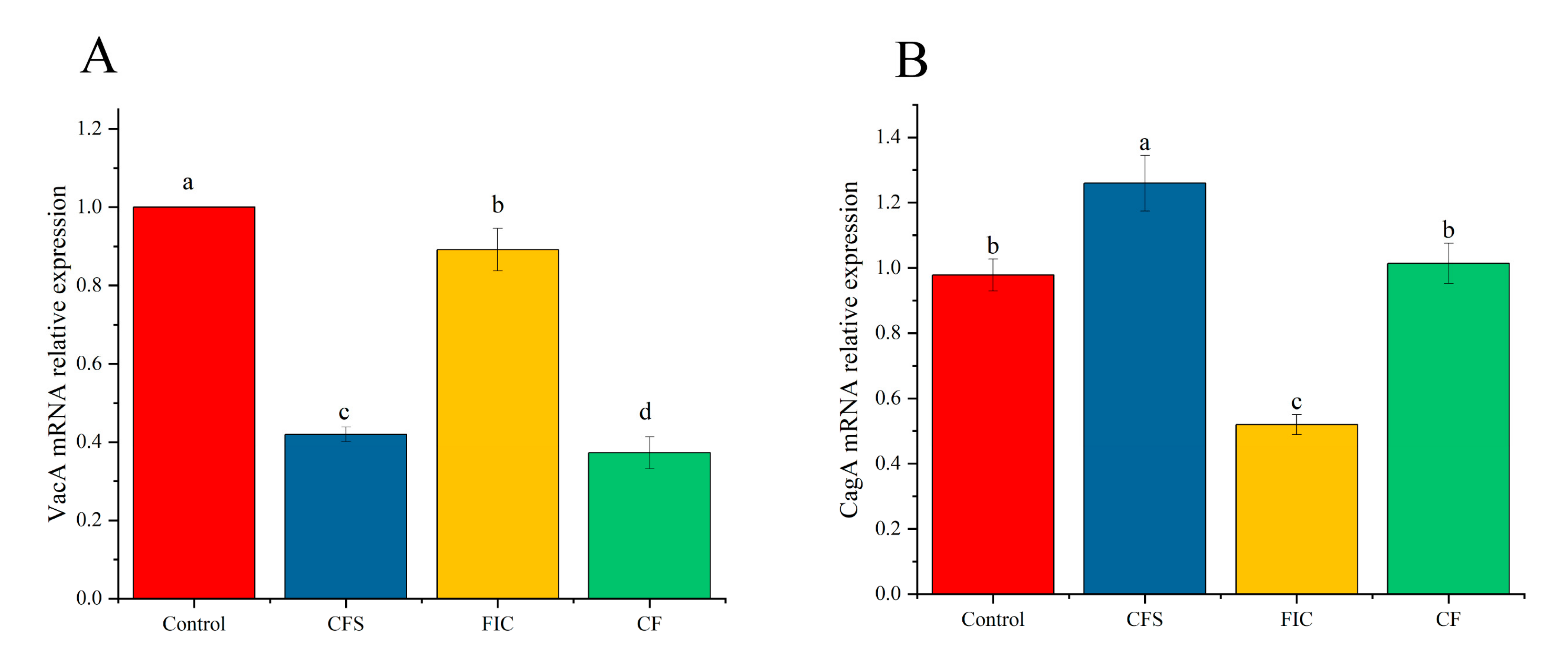
3.11. The Effect of LN12 CFS Combined with Antibiotics on the Expression of Virulence Genes of H. pylori Biofilm
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crowe, S.E. Helicobacter pylori Infection. N. Engl. J. Med. 2019, 380, 1158–1165. [Google Scholar] [CrossRef] [PubMed]
- Hathroubi, S.; Zerebinski, J.; Clarke, A.; Ottemann, K.M. Helicobacter pylori Biofilm Confers Antibiotic Tolerance in Part via A Protein-Dependent Mechanism. Antibiotics 2020, 9, 355. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P.; Kuipers, E.J.; Axon, A.T.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.Y.; et al. Management of Helicobacter pylori infection—The Maastricht V/Florence Consensus Report. Gut 2017, 66, 6–30. [Google Scholar] [CrossRef] [Green Version]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef] [Green Version]
- Graham, D.Y.; Fischbach, L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut 2010, 59, 1143–1153. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, Y.; Lu, N.H. Recent progress in Helicobacter pylori treatment. Chin. Med. J. 2020, 133, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Odenbreit, S.; Faller, G.; Haas, R. Role of the alpAB proteins and lipopolysaccharide in adhesion of Helicobacter pylori to human gastric tissue. Int. J. Med. Microbiol. 2002, 292, 247–256. [Google Scholar] [CrossRef]
- Kao, C.Y.; Sheu, B.S.; Wu, J.J. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed. J. 2016, 39, 14–23. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.; Mao, X.H.; Li, J.X.; Tong, W.D.; Wang, B.; Zhang, Y.J.; Guo, G.; Zhao, Z.J.; Li, L.; Wu, D.L.; et al. Efficacy, safety, and immunogenicity of an oral recombinant Helicobacter pylori vaccine in children in China: A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2015, 386, 1457–1464. [Google Scholar] [CrossRef]
- Winter, J.A.; Letley, D.P.; Cook, K.W.; Rhead, J.L.; Zaitoun, A.A.; Ingram, R.J.; Amilon, K.R.; Croxall, N.J.; Kaye, P.V.; Robinson, K.; et al. A role for the vacuolating cytotoxin, VacA, in colonization and Helicobacter pylori-induced metaplasia in the stomach. J. Infect. Dis. 2014, 210, 954–963. [Google Scholar] [CrossRef] [Green Version]
- Yonezawa, H.; Osaki, T.; Hojo, F.; Kamiya, S. Effect of Helicobacter pylori biofilm formation on susceptibility to amoxicillin, metronidazole and clarithromycin. Microb. Pathog. 2019, 132, 100–108. [Google Scholar] [CrossRef]
- Hathroubi, S.; Servetas, S.L.; Windham, I.; Merrell, D.S.; Ottemann, K.M. Helicobacter pylori Biofilm Formation and Its Potential Role in Pathogenesis. Microbiol. Mol. Biol. Rev. 2018, 82, e00001-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzyzek, P.; Grande, R.; Migdal, P.; Paluch, E.; Gosciniak, G. Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori. Pathogens 2020, 9, 1062. [Google Scholar] [CrossRef] [PubMed]
- Attaran, B.; Falsafi, T.; Ghorbanmehr, N. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J. Gastroenterol. 2017, 23, 1163–1170. [Google Scholar] [CrossRef]
- Fauzia, K.A.; Miftahussurur, M.; Syam, A.F.; Waskito, L.A.; Doohan, D.; Rezkitha, Y.A.A.; Matsumoto, T.; Tuan, V.P.; Akada, J.; Yonezawa, H.; et al. Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates. Toxins 2020, 12, 473. [Google Scholar] [CrossRef]
- Tayseer, I.; Aburjai, T.; Abu-Qatouseh, L.; AL-Karabieh, N.; Ahmed, W.; Al-Samydai, A. In vitro Anti-Helicobacter pylori Activity of Capsaicin. J. Pure Appl. Microbiol. 2020, 14, 279–286. [Google Scholar] [CrossRef] [Green Version]
- Francavilla, R.; Polimeno, L.; Demichina, A.; Maurogiovanni, G.; Principi, B.; Scaccianoce, G.; Ierardi, E.; Russo, F.; Riezzo, G.; Di Leo, A.; et al. Lactobacillus reuteri Strain Combination In Helicobacter pylori Infection A Randomized, Double-Blind, Placebo-Controlled Study. J. Clin. Gastroenterol. 2014, 48, 407–413. [Google Scholar] [CrossRef]
- Cárdenas, P.A.; Garcés, D.; Prado-Vivar, B.; Flores, N.; Fornasini, M.; Cohen, H.; Salvador, I.; Cargua, O.; Baldeón, M.E. Effect of Saccharomyces boulardii CNCM I-745 as complementary treatment of Helicobacter pylori infection on gut microbiome. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1365–1372. [Google Scholar] [CrossRef] [PubMed]
- Lü, M.; Yu, S.; Deng, J.; Yan, Q.; Yang, C.; Xia, G.; Zhou, X. Efficacy of Probiotic Supplementation Therapy for Helicobacter pylori Eradication: A Meta-Analysis of Randomized Controlled Trials. PLoS ONE 2016, 11, e0163743. [Google Scholar] [CrossRef]
- Barzegari, A.; Kheyrolahzadeh, K.; Khatibi, S.M.H.; Sharifi, S.; Memar, M.Y.; Vahed, S.Z. The Battle of Probiotics and Their Derivatives Against Biofilms. Infect. Drug Resist. 2020, 13, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.; Wang, B.; Zhou, X.; Wang, F.; Xie, Y.; Zheng, H.; Lv, N. Efficacy and safety of probiotics as adjuvant agents for Helicobacter pylori infection: A meta-analysis. Exp. Ther. Med. 2015, 9, 707–716. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Li, P.; Shen, Y.; Zou, Y.; Yuan, G.; Hu, H. Rhamnolipid-involved antibiotics combinations improve the eradication of Helicobacter pylori biofilm in vitro: A comparison with conventional triple therapy. Microb. Pathog. 2019, 131, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Fiorini, G.; Zullo, A.; Saracino, I.M.; Pavoni, M.; Vaira, D. Antibiotic resistance pattern of Helicobacter pylori strains isolated in Italy during 2010–2016. Scand. J. Gastroenterol. 2018, 53, 661–664. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, H. Effect of Bifidobacterium breve in Combination with Different Antibiotics on Clostridium difficile. Front. Microbiol. 2018, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, X.; Ling, F.; Wang, H.; Zhang, P.; Shao, S. Atractylodes lancea volatile oils attenuated Helicobacter pylori NCTC11637 growth and biofilm. Microb. Pathog. 2019, 135, 103641. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Kurata, S.; Fukuda, M.; Kawakami, H.; Ochiai, K.; Hanawa, T.; Kamiya, S. Outer Membrane Vesicles of Helicobacter pylori TK1402 are Involved in Biofilm Formation. BMC Microbiol. 2009, 9, 197. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.H.; Tsai, W.H.; Wu, H.Y.; Chen, C.Y.; Yeh, W.L.; Chen, Y.H.; Hsu, H.Y.; Chen, W.W.; Chen, Y.W.; Chang, W.W.; et al. Probiotic Lactobacillus spp. Act Against Helicobacter pylori-induced Inflammation. J. Clin. Med. 2019, 8, 90. [Google Scholar] [CrossRef] [Green Version]
- Piotrowski, M.; Karpiński, P.; Pituch, H.; van Belkum, A.; Obuch-Woszczatyński, P. Antimicrobial effects of Manuka honey on in vitro biofilm formation by Clostridium difficile. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1661–1664. [Google Scholar] [CrossRef]
- Ji, J.; Yang, H. In Vitro Effects of Lactobacillus plantarum LN66 and Antibiotics Used Alone or in Combination on Helicobacter pylori Mature Biofilm. Microorganisms 2021, 9, 424. [Google Scholar] [CrossRef]
- Hirasawa, M.; Kurita-Ochia, T. Probiotic Potential of Lactobacilli Isolated from Saliva of Periodontally Healthy Individuals. Oral Health Prev. Dent. 2020, 18, 563–570. [Google Scholar] [CrossRef]
- Cornacchione, L.P.; Klein, B.A.; Duncan, M.J.; Hu, L.T. Interspecies Inhibition of Porphyromonas gingivalis by Yogurt-Derived Lactobacillus delbrueckii Requires Active Pyruvate Oxidase. Appl. Environ. Microbiol. 2019, 85, e01271-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Chen, X.; Shen, Y.; Li, H.; Zou, Y.; Yuan, G.; Hu, P.; Hu, H. Mucus penetration enhanced lipid polymer nanoparticles improve the eradication rate of Helicobacter pylori biofilm. J. Control. Release 2019, 300, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.T.; Liou, J.M.; ElOmar, E.M.; Wu, J.Y.; Leow, A.H.R.; Goh, K.L.; Das, R.; Lu, H.; Lin, J.T.; Tu, Y.K.; et al. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2017, 2, 707–715. [Google Scholar] [CrossRef]
- Hu, Y.; Zhu, Y.; Lu, N.-H. Primary Antibiotic Resistance of Helicobacter pylori in China. Dig. Dis. Sci. 2017, 62, 1146–1154. [Google Scholar] [CrossRef]
- Xie, C.; Lu, N.-H. Review: Clinical management of Helicobacter pylori infection in China. Helicobacter 2015, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Fighting biofilms with lantibiotics and other groups of bacteriocins. NPJ Biofilms Microbiomes 2018, 4, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hathroubi, S.; Zerebinski, J.; Ottemann, K.M. Helicobacter pylori Biofilm Involves a Multigene Stress-Biased Response, Including a Structural Role for Flagella. mBio 2018, 9, e01973-18. [Google Scholar] [CrossRef] [Green Version]
- Goldman, R.C.; Zakula, D.; Flamm, R.; Beyer, J.; Capobianco, J. Tight binding of clarithromycin, its 14-(R)-hydroxy metabolite, and erythromycin to Helicobacter pylori ribosomes. Antimicrob. Agents Chemother. 1994, 38, 1496–1500. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.Z.; Xie, Y.; Lu, H.; Cheng, H.; Zeng, Z.R.; Zhou, L.Y.; Chen, Y.; Wang, J.B.; Du, Y.Q.; Lu, N.H.; et al. Fifth Chinese National Consensus Report on the management of Helicobacter pylori infection. Helicobacter 2018, 23, e12475. [Google Scholar] [CrossRef]
- Gupta, P.; Sarkar, S.; Das, B.; Bhattacharjee, S.; Tribedi, P. Biofilm, pathogenesis and prevention-a journey to break the wall: A review. Arch. Microbiol. 2016, 198, 1–15. [Google Scholar] [CrossRef]
- Tao, Z.H.; Han, J.X.; Fang, J.Y. Helicobacter pylori infection and eradication: Exploring their impacts on the gastrointestinal microbiota. Helicobacter 2020, 25, e12754. [Google Scholar] [CrossRef]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef]
- Yao, X.; Smolka, A.J. Gastric Parietal Cell Physiology and Helicobacter pylori–Induced Disease. Gastroenterology 2019, 156, 2158–2173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peek, R.M., Jr.; Fiske, C.; Wilson, K.T. Role of innate immunity in Helicobacter pylori-induced gastric malignancy. Physiol. Rev. 2010, 90, 831–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, C.; Schütte, K.; Koch, N.; Vilchez-Vargas, R.; Wos-Oxley, M.L.; Oxley, A.P.A.; Vital, M.; Malfertheiner, P.; Pieper, D.H. The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut 2018, 67, 216–225. [Google Scholar] [CrossRef] [Green Version]
- Oh, B.; Kim, B.-S.; Kim, J.W.; Kim, J.S.; Koh, S.-J.; Kim, B.G.; Lee, K.L.; Chun, J. The Effect of Probiotics on Gut Microbiota during the Helicobacter pylori Eradication: Randomized Controlled Trial. Helicobacter 2016, 21, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.; Kim, J.W.; Kim, B.-S. Changes in the Functional Potential of the Gut Microbiome Following Probiotic Supplementation during Helicobacter Pylori Treatment. Helicobacter 2016, 21, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, N.; Li, P.; Gu, Q. Probiotic therapy in Helicobacter pylori infection: A potential strategy against a serious pathogen? Appl. Microbiol. Biotechnol. 2019, 103, 1573–1588. [Google Scholar] [CrossRef]
- Homan, M.; Orel, R. Are probiotics useful in Helicobacter pylori eradication? World J. Gastroenterol. 2015, 21, 10644–10653. [Google Scholar] [CrossRef] [PubMed]
- Fontes, L.E.S.; Martimbianco, A.L.C.; Zanin, C.; Riera, R. N-acetylcysteine as an adjuvant therapy for Helicobacter pylori eradication. Cochrane Database Syst. Rev. 2019, 2. [Google Scholar] [CrossRef]
- Ryan, K.A.; Daly, P.; Li, Y.; Hooton, C.; O’Toole, P.W. Strain-specific inhibition of Helicobacter pylori by Lactobacillus salivarius and other Lactobacilli. J. Antimicrob. Chemother. 2008, 61, 831–834. [Google Scholar] [CrossRef] [Green Version]
- Ryan, K.A.; O’Hara, A.M.; van Pijkeren, J.-P.; Douillard, F.P.; O’Toole, P.W. Lactobacillus salivarius modulates cytokine induction and virulence factor gene expression in Helicobacter pylori. J. Med. Microbiol. 2009, 58, 996–1005. [Google Scholar] [CrossRef] [Green Version]
- Salama, N.R.; Hartung, M.L.; Müller, A. Life in the human stomach: Persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 2013, 11, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Ding, Y.; Xu, H.; Shen, C.; Chen, X.; Li, C. Effects of sodium butyrate supplementation on inflammation, gut microbiota, and short-chain fatty acids in Helicobacter pylori-infected mice. Helicobacter 2021, 26, e12785. [Google Scholar] [CrossRef]
- Brown, A.; Fernández, I.S.; Gordiyenko, Y.; Ramakrishnan, V. Ribosome-dependent activation of stringent control. Nature 2016, 534, 277–280. [Google Scholar] [CrossRef] [Green Version]
- Xia, M.; Chen, H.; Liu, S. The synergy of resveratrol and alcohol against Helicobacter pylori and underlying anti-Helicobacter pylori mechanism of resveratrol. J. Appl. Microbiol. 2020, 128, 1179–1190. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, H.; Osaki, T.; Kamiya, S. Biofilm Formation by Helicobacter pylori and Its Involvement for Antibiotic Resistance. BioMed Res. Int. 2015, 2015, 914791. [Google Scholar] [CrossRef] [Green Version]
- Bessa, L.J.; Grande, R.; Di Iorio, D.; Di Giulio, M.; Di Campli, E.; Cellini, L. Helicobacter pylori free-living and biofilm modes of growth: Behavior in response to different culture media. APMIS 2013, 121, 549–560. [Google Scholar] [CrossRef]
- Franklin, M.J.; Chang, C.; Akiyama, T.; Bothner, B. New Technologies for Studying Biofilms. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [Green Version]








| Genes | Sequences (5′–3′) |
|---|---|
| cagA-F | TGGCTAAAGCAACGGGTGAT |
| cagA-R | CCGACTAGGGTTCCGTTCAC |
| vacA-F | CACTAACGCTGATGGCACGA |
| vacA-R | GGACAGATTGACACCGCCTT |
| 16s rRNA-F | GGCGACCTGCTGGAACATTACTG |
| 16s rRNA-R | CATCGTTTAGGGCGTGGACTACC |
| H. pylori Isolates | Minimal Inhibitory Concentration (mg/L) | ||||
|---|---|---|---|---|---|
| Amoxicillin | Clarithromycin | Levofloxacin | Metronidazole | Tetracycline | |
| SS1 | 0.094(S) | 0.032(S) | 0.094(S) | 0.032(S) | 0.032(S) |
| ATCC43504 | 0.016(S) | 0.032(S) | 0.19(S) | 256(R) | 0.19(S) |
| 3192 | 0.016(S) | 0.016(S) | 8(R) | 256(R) | 0.125(S) |
| 3750 | 0.094(S) | 24(R) | 32(R) | 256(R) | 0.38(S) |
| 3139 | 0.016(S) | 0.016(S) | 0.25(S) | 12(R) | 0.032(S) |
| 4386 | 0.25(R) | 16(R) | 0.5(S) | 256(R) | 0.75(S) |
| 2359 | 0.016(S) | 0.016(S) | 0.094(S) | 1(S) | 0.023(S) |
| 1758 | 0.023(S) | 2(R) | 32(R) | 24(R) | 0.023(S) |
| 3931 | 0.016(S) | 12(R) | 32(R) | 256(R) | 0.25(S) |
| 3179 | 0.016(S) | 0.023(S) | 32(R) | 256(R) | 0.25(S) |
| Strain | FIC s (MICCLR, MICAMX) | Effect |
|---|---|---|
| SS1 | 1.5 (0.5, 1) | Indifferent effect |
| ATCC43504 | 0.75 (0.25, 0.5) | Partially synergistic |
| 3192 | 2.125 (2, 0.125) | Antagonistic |
| 3750 | 1.06 (0.06, 1) | Indifferent effect |
| 3139 | 1.0 (0.5, 0.5) | Indifferent effect |
| 4386 | 1.06 (0.06, 1) | Indifferent effect |
| 2359 | 0.625 (0.125, 0.5) | Partially synergistic |
| 1758 | 1.0 (0.5, 0.5) | Indifferent effect |
| 3931 | 2.25 (0.25, 2) | Antagonistic |
| 3179 | 0.625 (0.125, 0.5) | Partially synergistic |
| Probiotic Strains | Diameter of Inhibition Zone/mm | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SS1 | 43504 | 3750 | 4386 | 3192 | 3931 | 3179 | 3139 | 2359 | 1758 | |
| LN66 | 6.8 ± 0.3 a | 8.1 ± 0.4 a | 6.6 ± 0.6 a | 6.7 ± 0.3 a | 5.2 ± 0.3 a | 5.8 ± 0.3 a | 5.7 ± 0.7 a | 5.5 ± 0.5 ab | 5.8 ± 0.2 b | 7.5 ± 0.3 a |
| LN12 | 5.6 ± 0.6 b | 8.1 ± 0.3 a | 5.1 ± 0.4 b | 5.8 ± 0.2 b | 4.7 ± 0.3 a | 5.5 ± 0.1 ab | 5.3 ± 0.2 ab | 5.8 ± 0.3 a | 6.7 ± 0.4 a | 6.4 ± 0.2 b |
| LN19 | 5.3 ± 0.5 b | 8.0 ± 0.3 a | 4.8 ± 0.2 bc | 5.2 ± 0.3 b | 4.8 ± 0.3 a | 4. ± 0.2 bcd | 5.7 ± 0.4 a | 4.8 ± 0.2 bc | 3.1 ± 0.2 d | 4.9 ± 0.1 c |
| INO-10 | 5.5 ± 0.2 b | 6.7 ± 0.2 b | 3.9 ± 0.4 cd | 3.60 ± 0.2 c | 3.5 ± 0.2 b | 5. ± 0.8 abc | 4.2 ± 0.2 bc | 3.8 ± 0.2 de | 4.0 ± 0.3 c | 0 e |
| INO-17 | 3.6 ± 0.2 c | 5.5 ± 0.3 c | 3.9 ± 0.4 cd | 5.2 ± 0.2 b | 3.50 ± 0.7 b | 4.1 ± 0.3 cd | 0 d | 3.2 ± 0.3 e | 3.1 ± 0.2 d | 3.1 ± 0.1 d |
| INO-31 | 3.8 ± 0.5 c | 5.3 ± 0.4 c | 3.1 ± 0.3 d | 3.8 ± 0.6 c | 3.77 ± 0.1 b | 3.3 ± 0.3 d | 0 d | 3.3 ± 0.2 e | 3.6 ± 0.3 cd | 4.6 ± 0.3 c |
| INO-11 | 4.6 ± 0.2 bc | 6.8 ± 0.4 b | 3.8 ± 0.2 d | 0 d | 3.3 ± 0.2 b | 3.5 ± 0.5 d | 3.6 ± 0.6 c | 4.4 ± 0.2 cd | 3.5 ± 0.3 cd | 3.2 ± 0.2 d |
| mMRS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, F.; Yang, H. Effects of Lactobacillus salivarius LN12 in Combination with Amoxicillin and Clarithromycin on Helicobacter pylori Biofilm In Vitro. Microorganisms 2021, 9, 1611. https://doi.org/10.3390/microorganisms9081611
Jin F, Yang H. Effects of Lactobacillus salivarius LN12 in Combination with Amoxicillin and Clarithromycin on Helicobacter pylori Biofilm In Vitro. Microorganisms. 2021; 9(8):1611. https://doi.org/10.3390/microorganisms9081611
Chicago/Turabian StyleJin, Fang, and Hong Yang. 2021. "Effects of Lactobacillus salivarius LN12 in Combination with Amoxicillin and Clarithromycin on Helicobacter pylori Biofilm In Vitro" Microorganisms 9, no. 8: 1611. https://doi.org/10.3390/microorganisms9081611
APA StyleJin, F., & Yang, H. (2021). Effects of Lactobacillus salivarius LN12 in Combination with Amoxicillin and Clarithromycin on Helicobacter pylori Biofilm In Vitro. Microorganisms, 9(8), 1611. https://doi.org/10.3390/microorganisms9081611






