Colonization with Altered Schaedler Flora Impacts Leukocyte Adhesion in Mesenteric Ischemia-Reperfusion Injury
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Successful Transfer of Altered Schaedler Flora from C3H/HeN Mice on the C57BL/6J Background, with Mouse Strain-Specific Impact on the Abundance of ASF 500
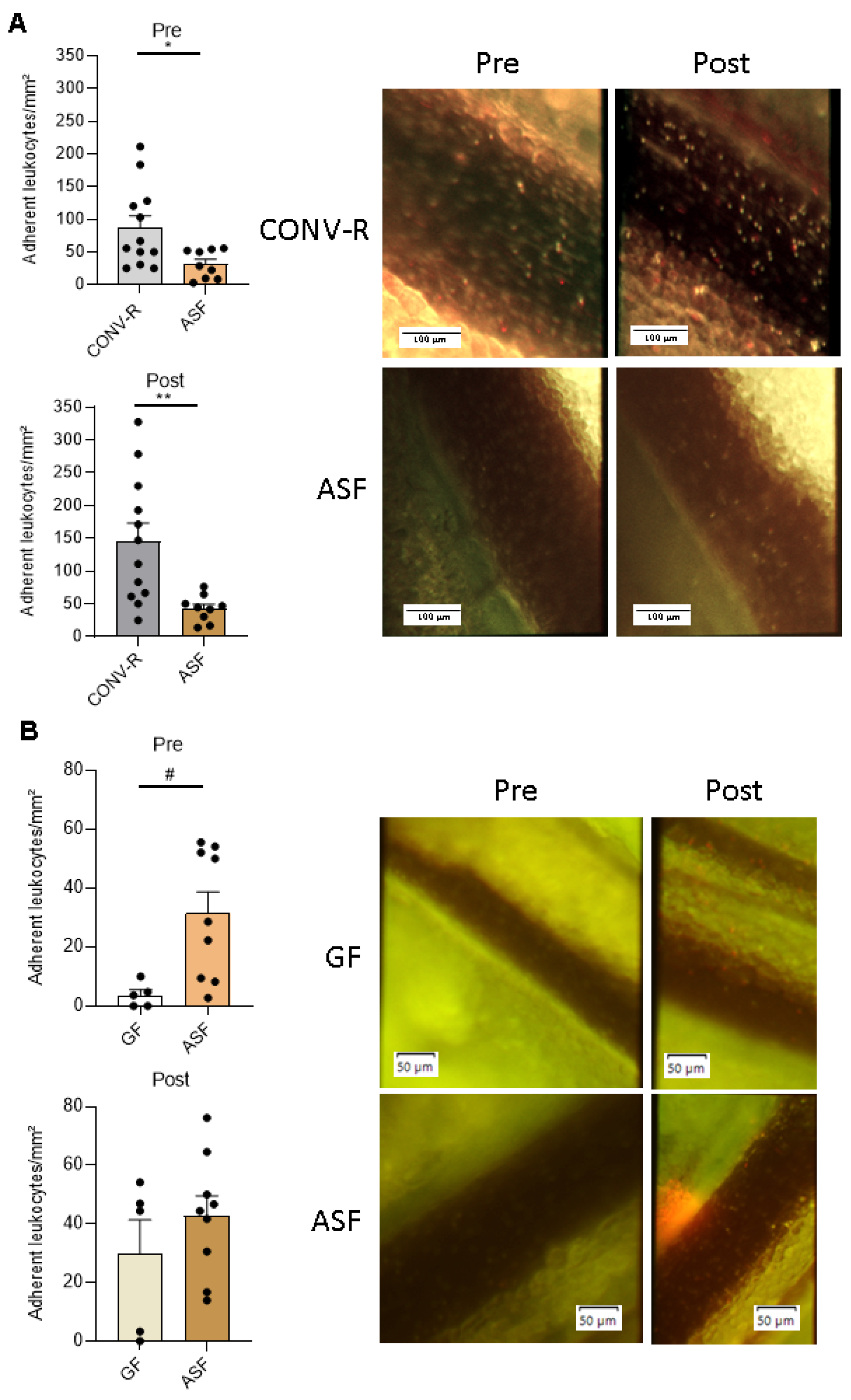
3.2. Altered Schaedler Flora-Colonization Protects from Leukocyte Adhesion to Mesenteric Venules, Prior to and after Ischemia-Reperfusion Injury
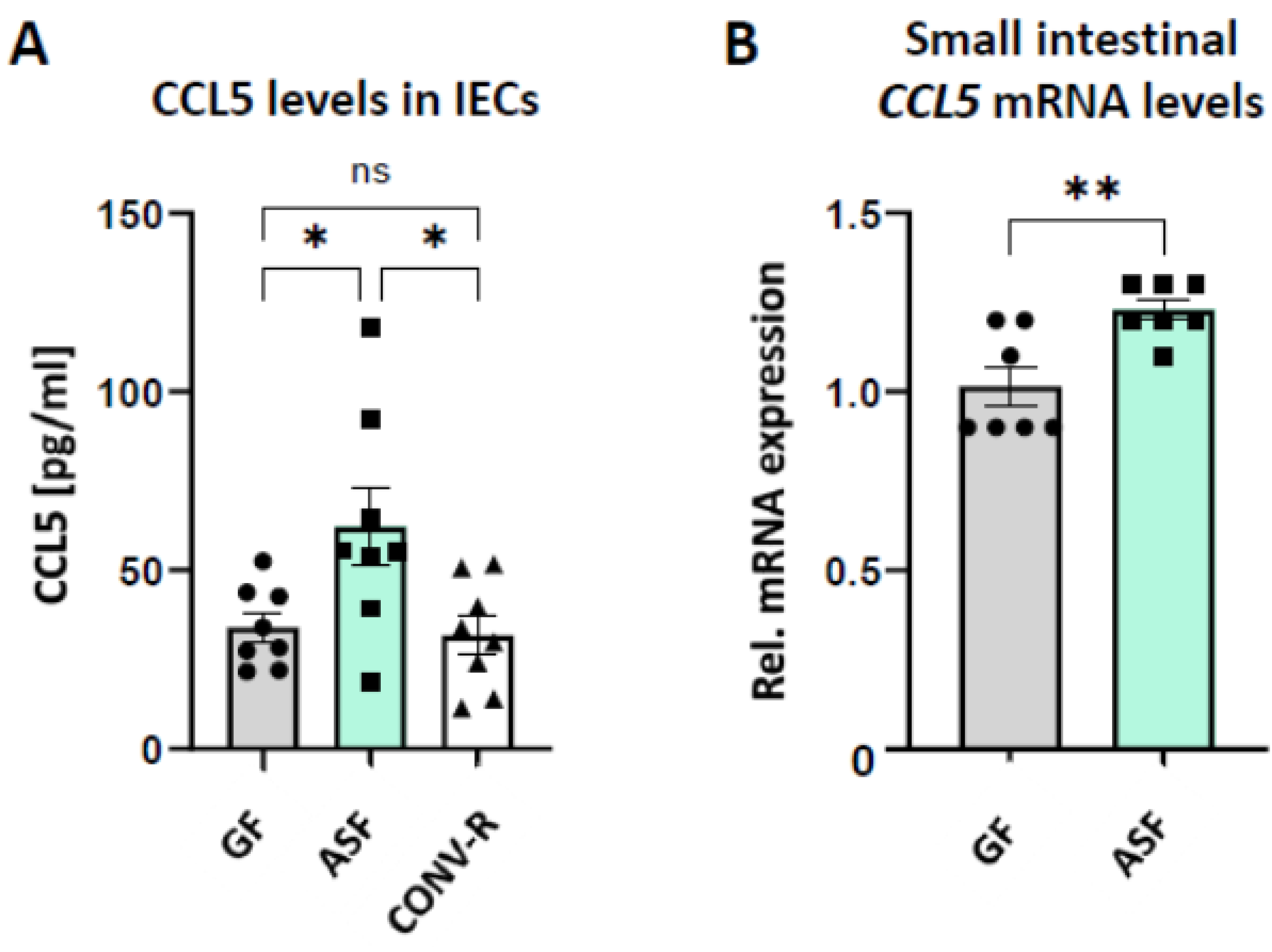
3.3. Altered Schaedler Flora Modulates the Expression of Epithelial-Derived Leukocyte-Attracktant Chemokine CCL5
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, C.; Reigstad, C.S.; Bäckhed, F. Intestinal microbiota during infancy and its implicatons for obesity. J. Pediatr. Gastroenterol. Nutr. 2009, 48, 249–256. [Google Scholar] [CrossRef]
- Milani, C.; Lugli, G.A.; Duranti, A.; Turroni, F.; Mancabelli, L.; Ferrario, C.; Mangifesta, M.; Hevia, A.; Viappiani, A.; Scholz, M.; et al. Bifidobacteria exhibit social behaviour through carbohydrate resource sharing in the gut. Sci. Rep. 2015, 5, 15782. [Google Scholar] [CrossRef] [PubMed]
- Bry, L.; Falk, P.G.; Midtvedt, T.; Gordon, J.I. A model of host-microbial interactions in an open mammalian ecosystem. Science 1996, 273, 1380–1383. [Google Scholar] [CrossRef]
- Bayer, F.; Ascher, S.; Pontarollo, G.; Reinhardt, C. Antibiotic treatment protocols and germ-free mouse models in vascular research. Front. Immunol. 2019, 10, 2174. [Google Scholar] [CrossRef]
- Clavel, T.; Lagkouvardos, I.; Stecher, B. From complex gut communities to minimal microbiomes via cultivation. Curr. Opin. Microbiol. 2017, 38, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Lagkouvardos, I.; Pukall, R.; Abt, B.; Foesel, B.U.; Meier-Kolthoff, J.P.; Kumar, N.; Bresciani, A.; Martinez, I.; Just, S.; Ziegler, C.; et al. The mouse intestinal bacterial collection (miBC) provides host-specific insights into cultured diversity and functional potential of the gut microbiota. Nat. Microbiol. 2016, 1, 16131. [Google Scholar] [PubMed]
- Schaedler, R.W.; Dubs, R.; Costello, R. Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 1965, 122, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chien, C.C.; Paster, B.J.; Ericson, R.L.; Orcutt, R.P.; Schauer, D.B.; Fox, J.G. Phylogeny of the defined murine microbiota: Altered Schaedler flora. Appl. Environ. Microbiol. 1999, 65, 3287–3289. [Google Scholar] [CrossRef]
- Wannemuehler, M.J.; Overstreet, A.-M.; Ward, D.V.; Phillips, G.J. Draft genome sequences of the altered Schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc. 2014, 2, e00287. [Google Scholar] [CrossRef]
- Becker, N.; Kunath, J.; Loh, G.; Blaut, M. Human intestinal microbiota: Characterization of a simplified and stable gnotobiotic rat model. Gut Microbes 2011, 2, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Macfarlane, G.T. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003, 62, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Hörmann, N.; Brandão, I.; Jäckel, S.; Ens, N.; Lillich, M.; Walter, U.; Reinhardt, C. Gut microbial colonization orchestrates TLR2 expression, signaling and epithelial proliferation in the small intestinal mucosa. PLoS ONE. 2014, 9, e113080. [Google Scholar] [CrossRef]
- Schaupp, L.; Muth, S.; Rogell, L.; Kofoed-Branzk, M.; Melchior, F.; Lienenklaus, S.; Ganal-Vonarburg, S.C.; Klein, M.; Guendel, F.; Hain, T.; et al. Microbiota-induced type I interferons instruct a poised basal state of dendritic cells. Cell 2020, 181, 1080–1096. [Google Scholar] [CrossRef] [PubMed]
- Siemer, S.; Hahlbrock, A.; Vallet, C.; CcClements, D.J.; Balszuweit, J.; Voskuhl, J.; Docter, D.; Wessler, S.; Kneuer, S.K.; Westmeier, D.; et al. Nanosized food additives impact beneficial and pathogenic bacteria in the human gut: A simulated gastrointestinal study. NPJ Sci. Food. 2018, 2, 22. [Google Scholar] [CrossRef]
- Stauber, R.H.; Siemer, S.; Becker, S.; Ding, G.B.; Strieth, S.; Knauer, S.K. Small meets smaller: Effects of nanomaterials on microbial biology, pathology, and ecology. ACS Nano. 2018, 12, 6351–6359. [Google Scholar] [CrossRef]
- Westmeier, D.; Hahlbrock, A.; Reinhardt, C.; Fröhlich-Nowoisky, J.; Wessler, S.; Vallet, C.; Poschl, U.; Knauer, S.K.; Stauber, R.H. Nanomaterial-microbe cross-talk: Physicochemical principles and (patho)biological consequences. Chem. Soc. Rev. 2018, 47, 5312–5337. [Google Scholar] [CrossRef]
- Westmeier, D.; Siemer, S.; Vallet, C.; Steinmann, J.; Docter, D.; Buer, J.; Knauer, S.K.; Stauber, R.H. Boosting nanotoxicity to combat multidrug-resistant bacteria in pathophysiological environments. Nanoscale Adv. 2020, 2, 5428–5440. [Google Scholar] [CrossRef]
- Westmeier, D.; Posselt, G.; Hahlbrock, A.; Bartfeld, S.; Vallet, C.; Abfalter, C.; Docter, D.; Knauer, S.K.; Wessler, S.; Stauber, R.H. Nanoparticle binding attenuates the pathobiology of gastric cancer-associated Helicobacter pylori. Nanoscale 2018, 10, 1453–1463. [Google Scholar] [CrossRef]
- Busch, C.J.-L.; Hendrikx, T.; Weismann, D.; Jäckel, S.; Walenbergh, S.M.A.; Rendeiro, A.F.; Weißer, J.; Puhm, F.; Hladik, A.; Göderle, L.; et al. Malondialdehyde epitopes are sterile mediators of hepatic inflammation in hypercholesterolemic mice. Hepatology 2017, 65, 1181–1195. [Google Scholar] [CrossRef]
- Stepankova, R.; Tonar, Z.; Bartova, J.; Nedorost, L.; Rossman, P.; Poledne, R.; Schwarzer, M.; Tlaskalova-Hogenova, H. Absence of microbiota (germ-free conditions) accelerates the atherosclerosis in ApoE-deficient mice fed standard low cholesterol diet. J. Atheroscler. Thromb. 2010, 17, 796–804. [Google Scholar] [CrossRef]
- Ascher, S.; Reinhardt, C. The gut microbiota: An emerging risk factor for cardiovascular and cerebrovascular disease. Eur. J. Immunol. 2018, 48, 564–575. [Google Scholar] [CrossRef]
- Van Esch, B.C.A.M.; Porbahaie, M.; Abbring, S.; Garssen, J.; Potaczek, D.P.; Savelkoul, H.F.J.; van Neerven, R.J.J. The impact of milk and its components on epigenetic protramming of immune function in early life and beyond: Implications for allergy and asthma. Front. Immunol. 2020, 11, 2141. [Google Scholar] [CrossRef]
- Acevedo, N.; Alhamwe, B.A.; Caraballo, L.; Ding, M.; Ferrante, A.; Garn, H.; Garssen, J.; Hii, C.S.; Irvine, J.; Llinás-Caballero, K.; et al. Perinatal and early-life nutrition, epigenetics, and allergy. Nutrients 2021, 13, 724. [Google Scholar] [CrossRef] [PubMed]
- Stappenbeck, T.S.; Hooper, L.V.; Gordon, J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 2002, 99, 15451–15455. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, C.; Bergentall, M.; Greiner, T.U.; Schaffner, F.; Ostergren-Lundén, G.; Petersen, L.C.; Ruf, W.; Bäckhed, F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature 2012, 483, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.H.; Choe, K.; Hong, S.P.; Jeong, S.-H.; Mäkinen, T.; Kim, K.S.; Alitalo, K.; Surh, C.D.; Koh, G.Y.; Song, J.-H. Gut microbiota regulates lacteal integrity by inducing VEGF-C in intestinal villus macrophages. EMBO Rep. 2019, 20, e46927. [Google Scholar] [CrossRef]
- Komatsu, S.; Berg, R.D.; Russell, J.M.; Nimura, Y.; Granger, D.N. Enteric microflora contribute to constitutive ICAM-1 expression on vascular endothelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G186–G191. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, G.; Manwani, D.; Mortha, A.; Xu, C.; Faith, J.J.; Burk, R.D.; Kunisaki, Y.; Jang, J.-E.; Scheiermann, C.; et al. Neutrophil ageing is regulated by the microbiome. Nature 2015, 525, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Karbach, S.H.; Schönfelder, T.; Brandão, I.; Wilms, E.; Hörmann, N.; Jäckel, S.; Schüler, R.; Finger, S.; Knorr, M.; Lagrange, J.; et al. Gut microbiota promote angiotensin II-induced arterial hypertension and vascular dysfunction. J. Am. Heart Assoc. 2016, 5, e003698. [Google Scholar] [CrossRef]
- Lindskog Jonsson, A.; Caesar, R.; Akrami, R.; Reinhardt, C.; Fåk Hållenius, F.; Borén, J.; Bäckhed, F. Impact of gut microbiota and diet on the development of atherosclerosis in Apoe-/- mice. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2318–2326. [Google Scholar] [CrossRef]
- Kiouptsi, K.; Pontarollo, G.; Todorov, H.; Braun, J.; Jäckel, S.; Koeck, T.; Bayer, F.; Karwot, C.; Karpi, A.; Gerber, S.; et al. Germ-free housing conditions do not affect aortic root and aortic arch lesion size of late atherosclerotic low-density lipoprotein receptor-deficient mice. Gut Microbes 2020, 11, 1809–1823. [Google Scholar] [CrossRef] [PubMed]
- Pontarollo, G.; Kiouptsi, K.; Reinhardt, C. A holobiont view on thrombosis: Unravelling the microbiota’s incluence on arterial thrombus growth. Microb. Cell. 2020, 7, 28–31. [Google Scholar] [CrossRef]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef]
- Jäckel, S.; Kiouptsi, K.; Lillich, M.; Hendrikx, T.; Khandagale, A.; Kollar, B.; Hörmann, N.; Reiss, C.; Subramaniam, S.; Wilms, E.; et al. Gut microbiota regulate hepatic von Willebrand factor synthesis and arterial thrombus formation via Toll-like receptor-2. Blood 2017, 130, 542–553. [Google Scholar] [CrossRef]
- Kiouptsi, K.; Jäckel, S.; Pontarollo, G.; Grill, A.; Kuijpers, M.J.E.; Wilms, E.; Weber, C.; Sommer, F.; Nagy, M.; Neideck, C.; et al. The microbiota promotes arterial thrombosis in low-density lipoprotein receptor-deficient mice. mBio 2019, 10, e02298. [Google Scholar] [CrossRef]
- Kiouptsi, K.; Finger, S.; Garlapati, V.S.; Knorr, M.; Brandt, M.; Walter, U.; Wenzel, P.; Reinhardt, C. Hypoxia evokes increased PDI and PDIA6 expression in the infarcted myocardium of ex-germ-free and conventionally raised mice. Biol. Open 2019, 8, bio038851. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.L.; Lee, D.M.; Li Puma, L.C.; Ecton, K.E.; Thomas, K.N.; Febvre, H.P.; Chicco, A.J.; Weir, T.L.; Gentile, C.L. Gut microbiota regulates cardiac ischemic tolerance and aortic stiffness in obesity. Am. J. Physiol. Heart Circ. Physiol. 2019, 17, H1210–H1220. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Gao, X.; Xia, G.; Chen, M.; Zeng, N.; Wang, S.; You, C.; Tian, X.; Di, H.; Tang, W.; et al. Rapid gut dysbiosis induced by stroke exacerbates brain infarction in turn. Gut 2021, 70, gutjnl-2020-323263. [Google Scholar] [CrossRef]
- Cicalese, L.; Billiar, T.R.; Rao, A.S.; Bauer, A.J. Interaction between ischemia/reperfusion-induced leukocyte emigration and translocating bacterial enterotoxins on enteric muscle function. Transplant. Proc. 1997, 29, 1815. [Google Scholar] [CrossRef]
- Thorburn, T.; Aali, M.; Lehmann, C. Immune response to systemic inflammation in the intestinal microcirculation. Front. Biosci. (Landmark. Ed.). 2018, 23, 782–795. [Google Scholar]
- Harward, T.R.; Brooks, D.L.; Flynn, T.C.; Seeger, J.M. Multiple organ dysfunction after mesenteric artery revascularization. J. Vasc. Surg. 1993, 18, 459–467. [Google Scholar] [CrossRef]
- Ascher, S.; Wilms, E.; Pontarollo, G.; Kiouptsi, K.; Malinarich, F.; Kittner, J.M.; Bosmann, M.; Jurk, K.; Reinhardt, C. Response by Ascher et al to Letter Regarding Article, “Gut Microbiota Restricts NETosis in Acute Mesenteric Ischemia-Reperfusion Injury”. Arterioscler. Thromb. Vasc. Biol. 2021, 41, e74–e75. [Google Scholar] [PubMed]
- Ascher, S.; Wilms, E.; Pontarollo, G.; Formes, H.; Bayer, F.; Müller, M.; Malinarich, F.; Grill, A.; Bosmann, M.; Saffarzadeh, M.; et al. Gut microbiota restricts NETosis in acute mesenteric ischemia-reperfusion injury. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2279–2292. [Google Scholar] [CrossRef]
- Perez-Chanona, E.; Mühlbauer, M.; Jobin, C. The microbiota protects against ischemia/reperfusion-induced intestinal injury through nucleotide-binding oligomerization domain-containing protein 2 (NOD2) signaling. Am. J. Pathol. 2014, 184, 2965–2975. [Google Scholar] [CrossRef][Green Version]
- Souza, D.G.; Senchenkova, E.Y.; Russell, J.; Granger, D.N. MyD88 mediates the protective effects of probiotics against teh arteriolar thrombosis and leukocyte recruitment associated with experimental colitis. Inflamm. Bowel Dis. 2015, 21, 888–900. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Souza, D.G.; Vieira, A.T.; Soares, A.C.; Pinho, V.; Nicoli, J.R.; Vieira, L.Q.; Teixeira, M.M. The essential role of the intestinal microbiota in facilitating acute inflammatory responses. J. Immunol. 2004, 173, 4137–4146. [Google Scholar] [CrossRef]
- Geuking, M.B.; Cahenzli, J.; Lawson, M.A.E.; Ng, D.C.K.; Slack, E.; Hapfelmeier, S.; McCoy, K.D.; Macpherson, A.J. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 2011, 34, 794–806. [Google Scholar] [CrossRef]
- Wymore, B.M.; Wannemuehler, M.J.; Phillips, G.J.; Proctor, A.; Overstreet, A.-M.; Jergens, A.E.; Orcutt, R.P.; Fox, J.G. The altered Schaedler flora: Continued applications of a defined murine microbial community. ILAR J. 2015, 56, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Neto, J.C.; Mantz, S.; Held, K.; Sinha, R.; Segura Munoz, R.R.; Schmaltz, R.; Benson, A.K.; Walter, J.; Ramer-Tait, A.E. A real-time PCR assay for accurate quantification of the individual members of the altered Schaedler flora microbiota in gnotobiotic mice. J. Microbiol. Methods 2017, 135, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Shin-Ya, M.; Kishida, T.; Urano, A.; Takada, R.; Sakagami, J.; Imanishi, J.; Kita, M.; Ueda, Y.; Iwakura, Y.; et al. Interferon-gamma is causatively involved in experimental inflammatory bowel disease. Clin. Exp. Immunol. 2006, 146, 330–338. [Google Scholar] [CrossRef]
- Stojanocvic, T.; Bedke, J.; Gröne, H.J.; Proudfoot, A.E.I.; Becker, H.; Markus, P.; Hecker, M. Met-RANTES inhibition of mucosal perfusion failure in acute intestinal transplant rejection–role of endothelial cell-leukocyte interaction. J. Vasc. Res. 2002, 39, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Biggs, M.B.; Medlock, G.L.; Moutinho, T.J.; Lees, H.J.; Swann, J.R.; Kolling, G.L.; Papin, J.A. Systems-level metabolism of the altered Schaedler flora, a complete gut microbiota. ISME J. 2017, 11, 426–438. [Google Scholar] [CrossRef]
- Moghadamrad, S.; Hassan, M.; McCoy, K.D.; Kirundi, J.; Kellmann, P.; De Gottardi, A. Attenuated fibrosis in specific pathogen-free microbiota in experimental cholestasis–and toxin-induced liver injury. FASEB J. 2019, 33, 12464–12476. [Google Scholar] [CrossRef]
- Tran, H.Q.; Bretin, A.; Adeshirlarijaney, A.; Yeoh, B.S.; Vijay-Kumar, M.; Zou, J.; Denning, T.L.; Chassaing, B.; Gewirtz, A.T. “Western diet”-induced adipose inflammation requires a complex gut microbiota. Cell Mol. Gastroenterol. Hepatol. 2020, 9, 313–333. [Google Scholar] [CrossRef] [PubMed]
- Massberg, S.; Grahl, L.; von Bruehl, M.-L.; Manukyan, D.; Pfeiler, S.; Goosmann, C.; Brinkmann, V.; Lorenz, M.; Bidzhekov, K.; Khandagale, A.B.; et al. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat. Med. 2010, 16, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Von Hundelshausen, P.; Koenen, R.R.; Sack, M.; Mause, S.R.; Adriaens, W.; Proudfoot, A.E.I.; Hackeng, T.M.; Weber, C. Heterophilic interactions of platelet factor 4 and RANTES promote monocyte arrest on endothelium. Blood 2005, 105, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Yoshiya, K.; Lapchak, P.H.; Thai, T.-H.; Kannan, L.; Rani, P.; Dalle Lucca, J.J.; Tsokos, G.C. Depletion of gut commensal bacteria attenuates intestinal ischemia/reperfusion injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G1020–G1030. [Google Scholar] [CrossRef] [PubMed]
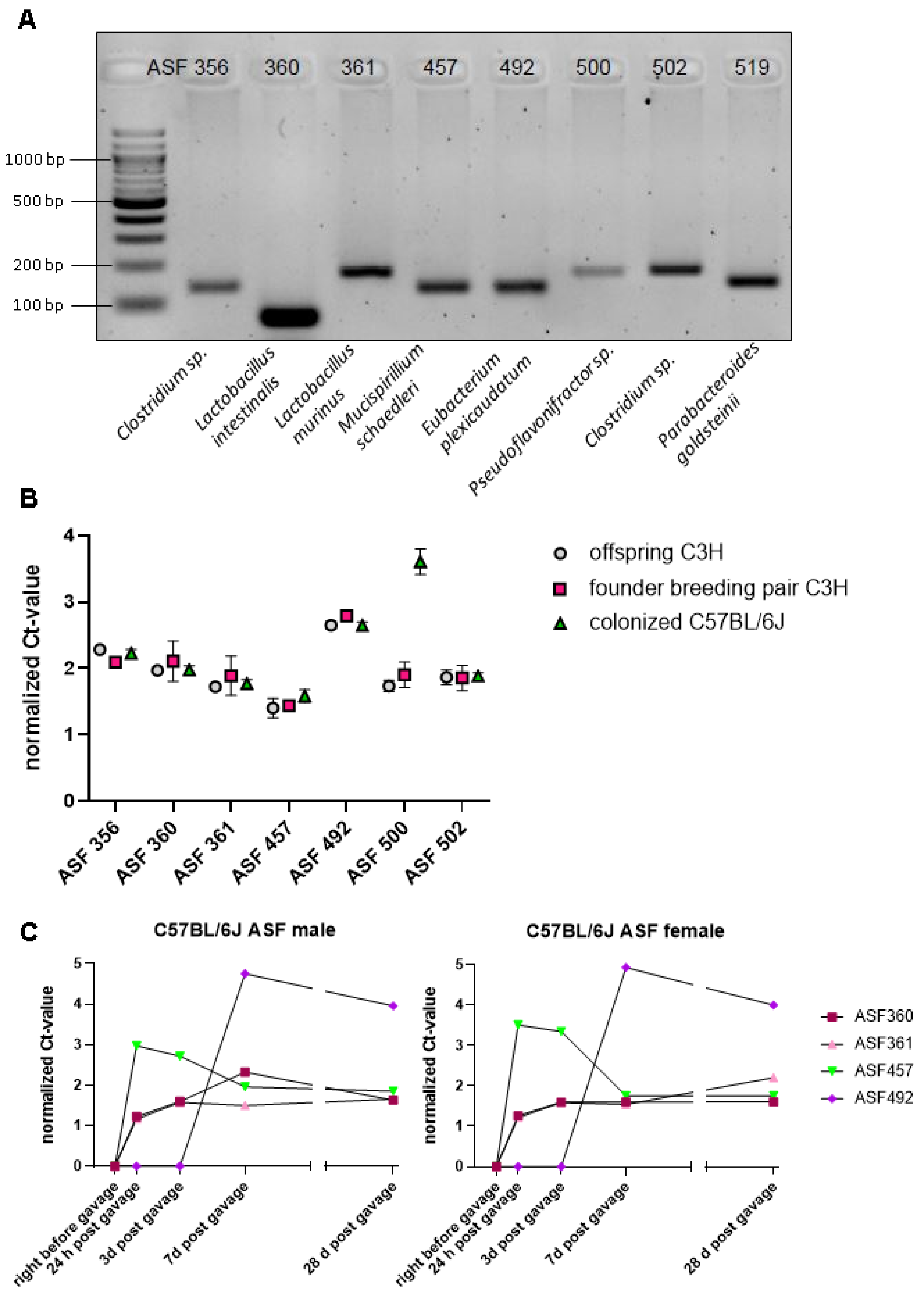
| Bacterial Species | Taxon ID | Primmer Sequences (5′ to 3′) | Amplicon Size (bp) |
|---|---|---|---|
| Clostridium sp. | 356 | Forward: AAAATAATTAGGAGCTTGCTTGCTTTTAA Reverse: TTAGAAGATGCCTCCTAAGAACC | 138 |
| Lactobacillus intestinalis | 360 | Forward:GGTGATGACGCTGGGAAC Reverse: AAGCAATAGCCATGCAGC | 130 |
| Lactobacillus murinus | 361 | Forward: GAACGAAACTTCTTTATCACC Reverse: TAGCATAGCCACCTTTTACA | 146 |
| Mucispirillum schaedleri | 457 | Forward: TCTCTTCGGGGATGATTAAAC Reverse: AACTTTTCCTATATAAACATGCAC | 135 |
| Eubacterium plexicaudatum | 492 | Forward:AATTCCTTCGGGGAGGAAGC Reverse: TAAAACCATGCGGTTTTAAAAAC | 137 |
| Pseudoflavonifractor sp. | 500 | Forward:ACGGAGGACCCCTGAAGG Reverse: AGCGATAAATCTTTGATGTCC | 172 |
| Clostridium sp. | 502 | Forward:GAGCGAAGCACTTTTTTAGAAC Reverse: TTACACCACCTCAGTTTTTACC | 177 |
| Parabacteroides goldsteinii | 519 | Forward:GCAGCACGATGTAGCAATACA Reverse: TTAACAAATATTTCCATGTGGAAC | 144 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayer, F.; Ascher, S.; Kiouptsi, K.; Kittner, J.M.; Stauber, R.H.; Reinhardt, C. Colonization with Altered Schaedler Flora Impacts Leukocyte Adhesion in Mesenteric Ischemia-Reperfusion Injury. Microorganisms 2021, 9, 1601. https://doi.org/10.3390/microorganisms9081601
Bayer F, Ascher S, Kiouptsi K, Kittner JM, Stauber RH, Reinhardt C. Colonization with Altered Schaedler Flora Impacts Leukocyte Adhesion in Mesenteric Ischemia-Reperfusion Injury. Microorganisms. 2021; 9(8):1601. https://doi.org/10.3390/microorganisms9081601
Chicago/Turabian StyleBayer, Franziska, Stefanie Ascher, Klytaimnistra Kiouptsi, Jens M. Kittner, Roland H. Stauber, and Christoph Reinhardt. 2021. "Colonization with Altered Schaedler Flora Impacts Leukocyte Adhesion in Mesenteric Ischemia-Reperfusion Injury" Microorganisms 9, no. 8: 1601. https://doi.org/10.3390/microorganisms9081601
APA StyleBayer, F., Ascher, S., Kiouptsi, K., Kittner, J. M., Stauber, R. H., & Reinhardt, C. (2021). Colonization with Altered Schaedler Flora Impacts Leukocyte Adhesion in Mesenteric Ischemia-Reperfusion Injury. Microorganisms, 9(8), 1601. https://doi.org/10.3390/microorganisms9081601







