Glucosidase Inhibitors Screening in Microalgae and Cyanobacteria Isolated from the Amazon and Proteomic Analysis of Inhibitor Producing Synechococcus sp. GFB01
Abstract
1. Introduction
2. Materials and Methods
2.1. Organisms and Culture Conditions
2.2. Extraction and Fraction of Intracellular Inhibitors
2.3. Screening in Agar Plates
2.4. Inhibitory Potential Assay for α and β-Glucosidase in Microplate
2.5. Search for Biosynthetic Enzymes of Glucosidase Inhibitors
2.6. Protein Extraction and Peptide Digestion
2.7. D-UPLC- Mass Spectrometric Analysis
2.8. Data Analysis and Statistics
3. Results
3.1. Screening of β-Glucosidase Inhibitory Activity
3.2. Growth Curve, Chlorophyll a and Protein Content
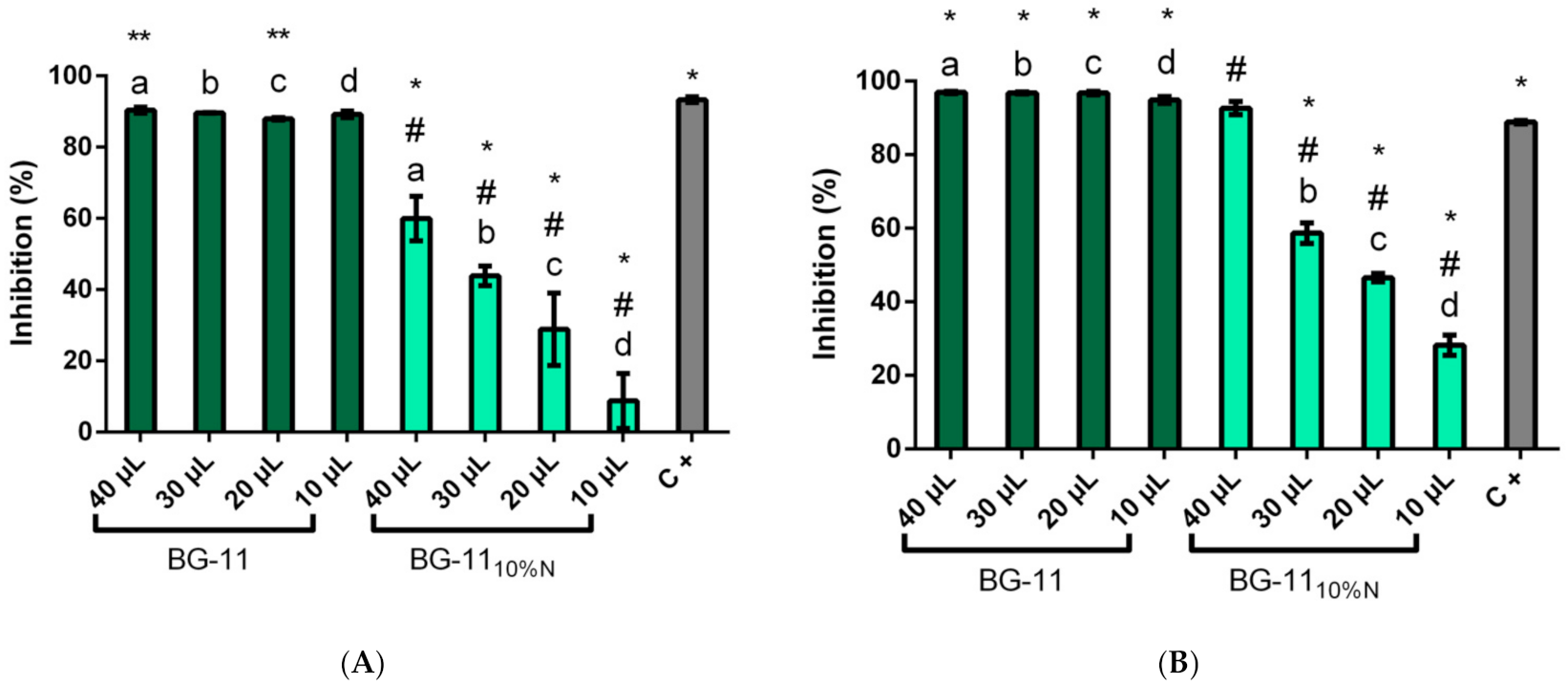
3.3. Colorimetric Inhibition Assay for α and β-Glucosidase
3.4. Potential Biosynthetic Enzymes
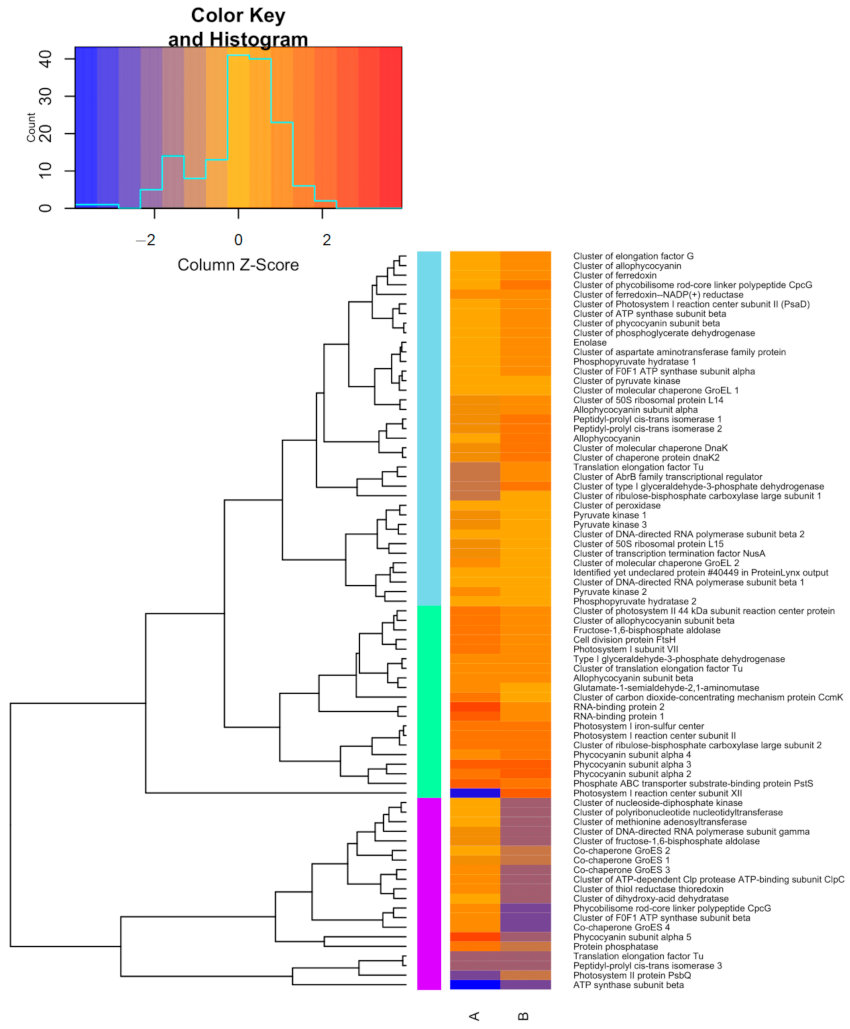
3.5. Protein Profile of Synechococcus sp. GFB01 Subjected to Nutritional Stress
3.6. Differentially Expressed Proteins
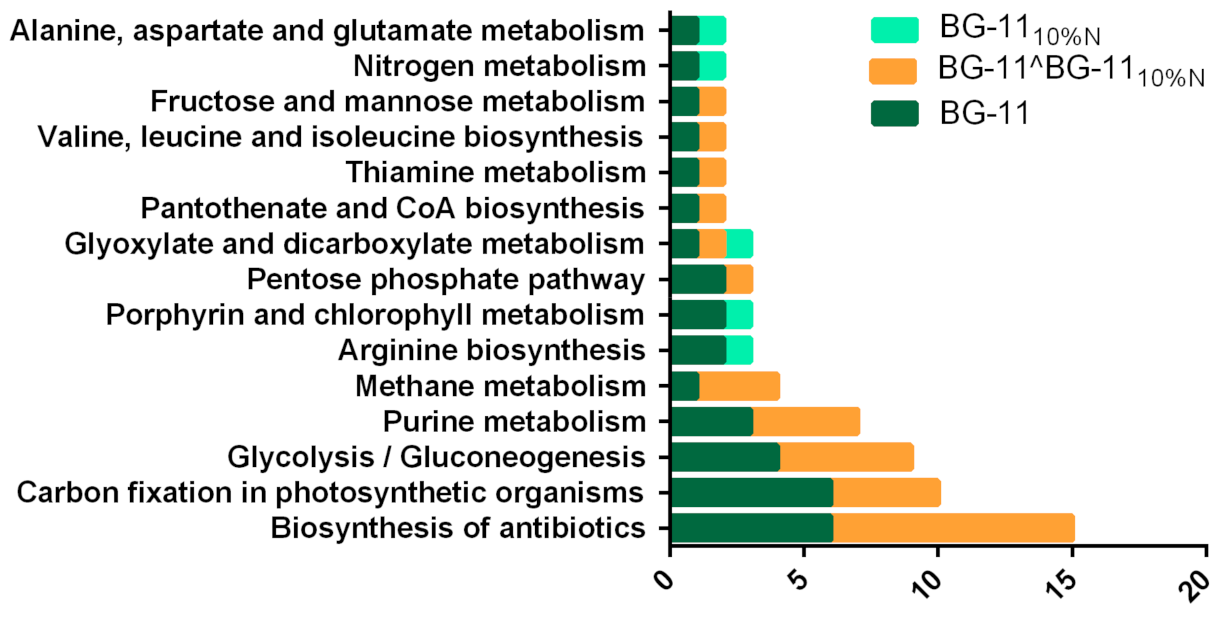
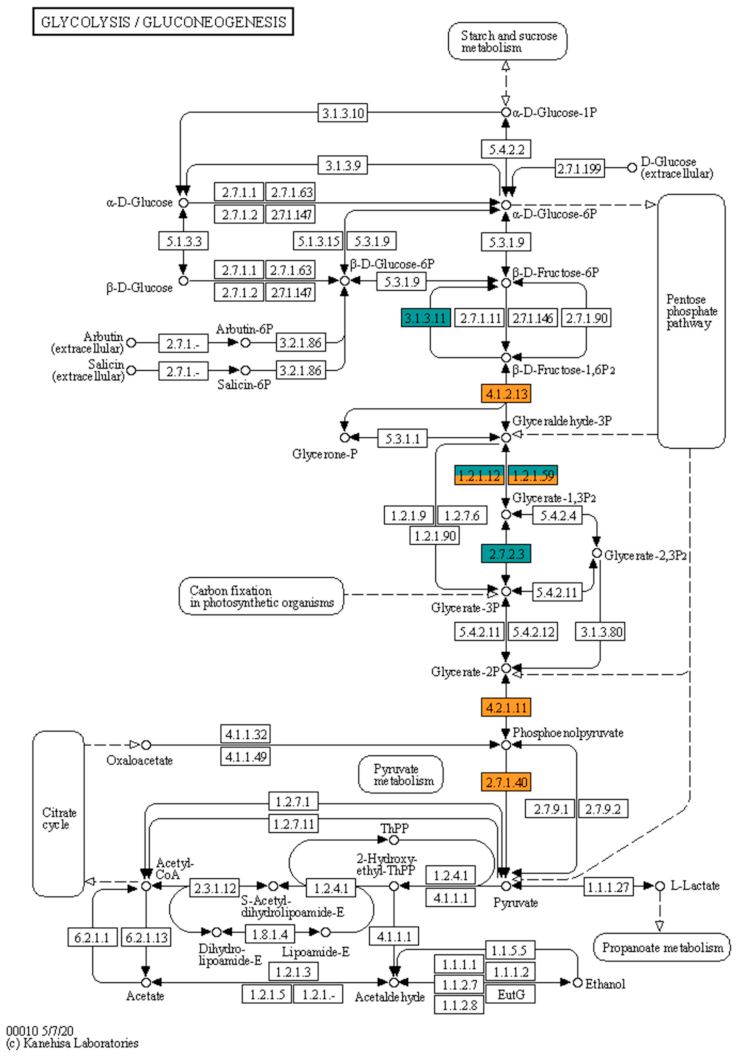
3.7. Proteins in Metabolic Pathways
4. Discussion
4.1. Growth Curve: Influence of Nitrate Concentration
4.2. Inhibitory Potential of the Methanolic Extract of Synechococcus sp. GFB01
4.3. Differential Proteome of Synechococcus sp. GFB01: Stress Response
4.4. Changes in Metabolic Pathways in Response to Nutritional Limitation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asano, N. Glycosidase Inhibitors: Update and Perspectives on Practical Use. Glycobiology 2003, 13, 93R–104R. [Google Scholar] [CrossRef] [PubMed]
- Stefano, E.D.; Oliviero, T.; Udenigwe, C. Functional Significance and Structure–Activity Relationship of Food-Derived α-Glucosidase Inhibitors. Curr. Opin. Food Sci. 2018, 20, 7–12. [Google Scholar] [CrossRef]
- Sharon, N.; Lis, H. Carbohydrates in Cell Recognition. Sci. Am. 1993, 268, 82–89. [Google Scholar] [CrossRef]
- Butters, T.D.; Dwek, R.A.; Platt, F.M. Imino Sugar Inhibitors for Treating the Lysosomal Glycosphingolipidoses. Glycobiology 2005, 15, 43R–52R. [Google Scholar] [CrossRef]
- Costa, J. Glycoconjugates from Extracellular Vesicles: Structures, Functions and Emerging Potential as Cancer Biomarkers. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2017, 1868, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Santos, C.J.; Kuehn, H.S.; Rosenzweig, S.D. N-Glycan Modification in Covid-19 Pathophysiology: In Vitro Structural Changes with Limited Functional Effects. J. Clin. Immunol. 2021, 41, 335–344. [Google Scholar] [CrossRef]
- Junge, B.; Matzke, M.; Stoltefuss, J. Chemistry and Structure-Activity Relationships of Glucosidase Inhibitors. In Oral Antidiabetics; Kuhlmann, J., Puls, W., Eds.; Springer: Berlin/Heidelberg, Germany, 1996; Volume 119, pp. 411–482. ISBN 9783540624561. [Google Scholar]
- Gao, X.; Cai, X.; Yang, W.; Chen, Y.; Han, X.; Ji, L. Meta-Analysis and Critical Review on the Efficacy and Safety of Alpha-Glucosidase Inhibitors in Asian and Non-Asian Populations. J. Diabetes Investig. 2018, 9, 321–331. [Google Scholar] [CrossRef]
- Lamine, J.B.; Boujbiha, M.A.; Dahane, S.; Cherifa, A.B.; Khlifi, A.; Chahdoura, H.; Yakoubi, M.T.; Ferchichi, S.; El Ayeb, N.; Achour, L. α-Amylase and α-Glucosidase Inhibitor Effects and Pancreatic Response to Diabetes Mellitus on Wistar Rats of Ephedra Alata Areal Part Decoction with Immunohistochemical Analyses. Environ. Sci. Pollut. Res. 2019, 26, 9739–9754. [Google Scholar] [CrossRef]
- Asano, N.; Nash, R.J.; Molyneux, R.J.; Fleet, G.W.J. Sugar-Mimic Glycosidase Inhibitors: Natural Occurrence, Biological Activity and Prospects for Therapeutic Application. Tetrahedron Asymmetry 2000, 11, 1645–1680. [Google Scholar] [CrossRef]
- De Melo, E.B.; Gomes, A.D.S.; Carvalho, I. α- and β-Glucosidase Inhibitors: Chemical Structure and Biological Activity. Tetrahedron 2006, 62, 10277–10302. [Google Scholar] [CrossRef]
- WHO Global Diabetes Compact. Available online: https://cdn.who.int/media/docs/default-source/world-diabetes-day/global-diabetes-compact-final.pdf?sfvrsn=63bd1e20_2 (accessed on 9 April 2021).
- Dwek, R.A.; Butters, T.D.; Platt, F.M.; Zitzmann, N. Targeting Glycosylation as a Therapeutic Approach. Nat. Rev. Drug Discov. 2002, 1, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Block, T.M.; Lu, X.; Platt, F.M.; Foster, G.R.; Gerlich, W.H.; Blumberg, B.S.; Dwek, R.A. Secretion of Human Hepatitis B Virus Is Inhibited by the Imino Sugar N-Butyldeoxynojirimycin. Proc. Natl. Acad. Sci. USA 1994, 91, 2235–2239. [Google Scholar] [CrossRef] [PubMed]
- Zitzmann, N.; Mehta, A.S.; Carrouee, S.; Butters, T.D.; Platt, F.M.; McCauley, J.; Blumberg, B.S.; Dwek, R.A.; Block, T.M. Imino Sugars Inhibit the Formation and Secretion of Bovine Viral Diarrhea Virus, a Pestivirus Model of Hepatitis C Virus: Implications for the Development of Broad Spectrum Anti-Hepatitis Virus Agents. Proc. Natl. Acad. Sci. USA 1999, 96, 11878–11882. [Google Scholar] [CrossRef] [PubMed]
- Gruters, R.A.; Neefjes, J.J.; Tersmette, M.; De Goede, R.E.Y.; Tulp, A.; Huisman, H.G.; Miedema, F.; Ploegh, H.L. Interference with HIV-Induced Syncytium Formation and Viral Infectivity by Inhibitors of Trimming Glucosidase. Nature 1987, 330, 74–77. [Google Scholar] [CrossRef]
- Ratner, L. Glucosidase Inhibitors for Treatment of HIV-1 Infection. AIDS Res. Hum. Retrovir. 1992, 8, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Zitzmann, N.; Rudd, P.M.; Block, T.M.; Dwek, R.A. α-Glucosidase Inhibitors as Potential Broad Based Anti-Viral Agents. FEBS Lett. 1998, 430, 17–22. [Google Scholar] [CrossRef]
- Choudhary, S.K.; Margolis, D.M. Curing HIV: Pharmacologic Approaches to Target HIV-1 Latency. Annu. Rev. Pharmacol. Toxicol. 2011, 51, 397–418. [Google Scholar] [CrossRef][Green Version]
- Williams, S.J.; Goddard-Borger, E.D. α-Glucosidase Inhibitors as Host-Directed Antiviral Agents with Potential for the Treatment of COVID-19. Biochem. Soc. Trans. 2020, 48, 1287–1295. [Google Scholar] [CrossRef]
- Dadashpour, S.; Emami, S. Indole in the Target-Based Design of Anticancer Agents: A Versatile Scaffold with Diverse Mechanisms. Eur. J. Med. Chem. 2018, 150, 9–29. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, Z.; Yang, H.; Jiang, Y.; Wei, W.; Li, Q.; Mo, Q.; Liu, J. β-Glucosidase Inhibition Sensitizes Breast Cancer to Chemotherapy. Biomed. Pharmacother. 2017, 91, 504–509. [Google Scholar] [CrossRef]
- Li, Z.; Xu, D.; Tong, X.; Shan, C. Inhibition of β-Glucosidase Overcomes Gastric Cancer Chemoresistance through Inducing Lysosomal Dysfunction. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101456. [Google Scholar] [CrossRef] [PubMed]
- Kusano, G.; Shibano, M.; Tsukamoto, D. Polyhydroxylated Alkaloids with Lipophilic Moieties as Glycosidase Inhibitors from Higher Plants. Heterocycles 2002, 57, 1539. [Google Scholar] [CrossRef]
- Paramitha, V.; Lipton, A.; Thangaraj, M.M. Evaluation of α- and β-Glucosidase Inhibitory Properties of Macro-Algae Using Intestinal Extracts of Marine Snail, Thais Rudolphi (Lamarck, 1822). Indian J. Biotechnol. 2008, 7, 61–65. [Google Scholar]
- Kumar, S.; Narwal, S.; Kumar, V.; Prakashukne, O.; Viswanathan, V.; Phadatare, A.G. Alpha-Glucosidase Inhibitors from Plants: A Natural Approach to Treat Diabetes. Pharmacogn. Rev. 2011, 5, 19–29. [Google Scholar] [CrossRef]
- Kim, J.-S.; Kwon, Y.-S.; Sa, Y.-J.; Kim, M.-J. Isolation and Identification of Sea Buckthorn (Hippophae Rhamnoides) Phenolics with Antioxidant Activity and α-Glucosidase Inhibitory Effect. J. Agric. Food Chem. 2011, 59, 138–144. [Google Scholar] [CrossRef]
- Heo, S.-J.; Hwang, J.-Y.; Choi, J.-I.; Han, J.-S.; Kim, H.-J.; Jeon, Y.-J. Diphlorethohydroxycarmalol Isolated from Ishige Okamurae, a Brown Algae, a Potent α-Glucosidase and α-Amylase Inhibitor, Alleviates Postprandial Hyperglycemia in Diabetic Mice. Eur. J. Pharmacol. 2009, 615, 252–256. [Google Scholar] [CrossRef]
- Zhao, C.; Yang, C.; Liu, B.; Lin, L.; Sarker, S.D.; Nahar, L.; Yu, H.; Cao, H.; Xiao, J. Bioactive Compounds from Marine Macroalgae and Their Hypoglycemic Benefits. Trends Food Sci. Technol. 2018, 72, 1–12. [Google Scholar] [CrossRef]
- Pradhan, B.; Patra, S.; Behera, C.; Nayak, R.; Jit, B.P.; Ragusa, A.; Jena, M. Preliminary Investigation of the Antioxidant, Anti-Diabetic, and Anti-Inflammatory Activity of Enteromorpha Intestinalis Extracts. Molecules 2021, 26, 1171. [Google Scholar] [CrossRef]
- Cannell, R.J.P.; Kellam, S.J.; Owsianka, A.M.; Walker, J.M. Microalgae and Cyanobacteria as a Source of Glycosidase Inhibitors. Microbiology 1987, 133, 1701–1705. [Google Scholar] [CrossRef] [PubMed]
- Jüttner, F.; Wu, J.-T. Evidence of Allelochemical Activity in Subtropical Cyanobacterial Biofilms of Taiwan. Arch. Hydrobiol. 2000, 147, 505–517. [Google Scholar] [CrossRef]
- Jüttner, F. Liberation of 5,8,11,14,17-Eicosapentaenoic Acid and Other Polyunsaturated Fatty Acids from Lipids as a Grazer Defense Reaction in Epilithic Diatom Biofilms. J. Phycol. 2001, 37, 744–755. [Google Scholar] [CrossRef]
- Jüttner, F.; Wessel, H.P. Isolation of Di(Hydroxymethyl)Dihydroxypyrrolidine from The Cyanobacterial Genus Cylindrospermum That Effectively Inhibits Digestive Glucosidases of Aquatic Insects and Crustacean Grazers. J. Phycol. 2003, 39, 26–32. [Google Scholar] [CrossRef]
- Thammana, S.; Suzuki, H.; Lobkovsky, E.; Clardy, J.; Shimizu, Y. Isolation and Structure Assignment of an Iminotetrasaccharide from a Cultured Filamentous Cyanobacterium Anabaena Sp. J. Nat. Prod. 2006, 69, 365–368. [Google Scholar] [CrossRef]
- Leflaive, J.; Ten-Hage, L. Algal and Cyanobacterial Secondary Metabolites in Freshwaters: A Comparison of Allelopathic Compounds and Toxins. Freshw. Biol. 2007, 52, 199–214. [Google Scholar] [CrossRef]
- Priatni, S.; Budiwati, T.A.; Ratnaningrum, D.; Kosasih, W.; Andryani, R.; Susanti, H.; Susilaningsih, D. Antidiabetic Screening of Some Indonesian Marine Cyanobacteria Collection. Biodivers. J. Biol. Divers. 2016, 17. [Google Scholar] [CrossRef]
- Żak, A.; Kosakowska, A. Cyanobacterial and Microalgal Bioactive Compounds-the Role of Secondary Metabolites in Allelopathic Interactions. Oceanol. Hydrobiol. Stud. 2016, 45, 131–143. [Google Scholar] [CrossRef]
- Ghosh, T.; Bhayani, K.; Paliwal, C.; Maurya, R.; Chokshi, K.; Pancha, I.; Mishra, S. Cyanobacterial Pigments as Natural Anti-Hyperglycemic Agents: An In Vitro Study. Front. Mar. Sci. 2016, 3, 146. [Google Scholar] [CrossRef]
- Borowitzka, M.A. High-Value Products from Microalgae—Their Development and Commercialisation. J. Appl. Phycol. 2013, 25, 743–756. [Google Scholar] [CrossRef]
- Gheda, S.F.; Ismail, G.A. Natural Products from Some Soil Cyanobacterial Extracts with Potent Antimicrobial, Antioxidant and Cytotoxic Activities. An. Acad. Bras. Ciênc. 2020, 92, e20190934. [Google Scholar] [CrossRef]
- Higuchi-Takeuchi, M.; Numata, K. Acetate-Inducing Metabolic States Enhance Polyhydroxyalkanoate Production in Marine Purple Non-Sulfur Bacteria Under Aerobic Conditions. Front. Bioeng. Biotechnol. 2019, 7, 118. [Google Scholar] [CrossRef] [PubMed]
- Gradíssimo, D.G.; Xavier, L.P.; Santos, A.V. Cyanobacterial Polyhydroxyalkanoates: A Sustainable Alternative in Circular Economy. Molecules 2020, 25, 4331. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Chen, F.; Wei, D.; Zhang, X.; Chen, G. Biodiesel Production by Microalgal Biotechnology. Appl. Energy 2010, 87, 38–46. [Google Scholar] [CrossRef]
- Aboim, J.B.; Oliveira, D.; Ferreira, J.E.; Siqueira, A.S.; Dall’Agnol, L.T.; Rocha Filho, G.N.; Gonçalves, E.C.; Nascimento, L.A. Determination of Biodiesel Properties Based on a Fatty Acid Profile of Eight Amazon Cyanobacterial Strains Grown in Two Different Culture Media. RSC Adv. 2016, 6, 109751–109758. [Google Scholar] [CrossRef]
- De Oliveira, D.T.; Vasconcelos, C.T.; Feitosa, A.M.T.; Aboim, J.B.; de Oliveira, A.D.N.; Xavier, L.P.; Santos, A.S.; Gonçalves, E.C.; da Rocha Filho, G.N.; do Nascimento, L.A.S. Lipid Profile Analysis of Three New Amazonian Cyanobacteria as Potential Sources of Biodiesel. Fuel 2018, 234, 785–788. [Google Scholar] [CrossRef]
- Gouda, K.G.M.; Kavitha, M.D.; Sarada, R. Antihyperglycemic, Antioxidant and Antimicrobial Activities of the Butanol Extract from Spirulina platensis: Antihyperglycemic Effect of Spirulina. J. Food Biochem. 2015, 39, 594–602. [Google Scholar] [CrossRef]
- Hu, S.; Fan, X.; Qi, P.; Zhang, X. Identification of Anti-Diabetes Peptides from Spirulina Platensis. J. Funct. Foods 2019, 56, 333–341. [Google Scholar] [CrossRef]
- Ramos-Romero, S.; Torrella, J.R.; Pagès, T.; Viscor, G.; Torres, J.L. Edible Microalgae and Their Bioactive Compounds in the Prevention and Treatment of Metabolic Alterations. Nutrients 2021, 13, 563. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Sang, Y.; Zhang, B.; Yu, X.; Xu, Q.; Xiu, Z.; Dong, Y. Synergistic Effects of Acarbose and an Oroxylum Indicum Seed Extract in Streptozotocin and High-Fat-Diet Induced Prediabetic Mice. Biomed. Pharmacother. 2017, 87, 160–170. [Google Scholar] [CrossRef]
- Mazur-Marzec, H.; Błaszczyk, A.; Felczykowska, A.; Hohlfeld, N.; Kobos, J.; Toruńska-Sitarz, A.; Devi, P.; Montalvão, S.; D’souza, L.; Tammela, P.; et al. Baltic Cyanobacteria–a Source of Biologically Active Compounds. Eur. J. Phycol. 2015, 50, 343–360. [Google Scholar] [CrossRef]
- Ahmed, B.E.; Badawi, M.H.; Mostafa, S.S.; Higazy, A.M. Human Anticancers and Antidiabetic Activities of the Cyanobacterium Fischerella Sp. BS1-EG Isolated from River Nile, Egypt. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 3473–3485. [Google Scholar] [CrossRef]
- Guimarães, P.I.; Leão, T.F.; De Melo, A.G.C.; Ramos, R.T.J.; Silva, A.; Fiore, M.F.; Schneider, M.P.C. Draft Genome Sequence of the Picocyanobacterium Synechococcus Sp. Strain GFB01, Isolated from a Freshwater Lagoon in the Brazilian Amazon. Genome Announc. 2015, 3, e00876-15. [Google Scholar] [CrossRef]
- Aryal, U.K.; Callister, S.J.; Mishra, S.; Zhang, X.; Shutthanandan, J.I.; Angel, T.E.; Shukla, A.K.; Monroe, M.E.; Moore, R.J.; Koppenaal, D.W.; et al. Proteome Analyses of Strains ATCC 51142 and PCC 7822 of the Diazotrophic Cyanobacterium Cyanothece Sp. under Culture Conditions Resulting in Enhanced H2 Production. Appl. Environ. Microbiol. 2013, 79, 1070–1077. [Google Scholar] [CrossRef]
- Meeks, J.C.; Castenholz, R.W. Growth and Photosynthesis in an Extreme Thermophile, Synechococcus Lividus (Cyanophyta). Archiv. Mikrobiol. 1971, 78, 25–41. [Google Scholar] [CrossRef]
- Fiore, M.D.F.; Neilan, B.A.; Copp, J.N.; Rodrigues, J.L.M.; Tsai, S.M.; Lee, H.; Trevors, J.T. Characterization of Nitrogen-Fixing Cyanobacteria in the Brazilian Amazon Floodplain. Water Res. 2005, 39, 5017–5026. [Google Scholar] [CrossRef]
- Pandey, S.; Sree, A.; Dash, S.S.; Sethi, D.P.; Chowdhury, L. Diversity of Marine Bacteria Producing Beta-Glucosidase Inhibitors. Microb. Cell Factories 2013, 12, 35. [Google Scholar] [CrossRef]
- Pandey, S.; Sree, A.; Dash, S.S.; Sethi, D.P. A Novel Method for Screening Beta-Glucosidase Inhibitors. BMC Microbiol. 2013, 13, 55. [Google Scholar] [CrossRef]
- Eberhart, B.; Cross, D.F.; Chase, L.R. β-glucosidase system of neurospora crassa I. β-glucosidase and cellulase activities of mutant and wild-type strains. J. Bacteriol. 1964, 87, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Shinde, J.; Taldone, T.; Barletta, M.; Kunaparaju, N.; Hu, B.; Kumar, S.; Placido, J.; Zito, S.W. α-Glucosidase Inhibitory Activity of Syzygium Cumini (Linn.) Skeels Seed Kernel in Vitro and in Goto–Kakizaki (GK) Rats. Carbohydr. Res. 2008, 343, 1278–1281. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Chadha, B.S.; Kumar, B.A.; Ghatora, S.K.; Saini, H.S. Purification and Characterization of SS-Glucosidase from Melanocarpus Sp. MTCC 3922. Electron. J. Biotechnol. 2007, 10, 260–270. [Google Scholar] [CrossRef]
- Grover, A.K.; MacMurchie, D.D.; Cushley, R.J. Studies on Almond Emulsin Βd-Glucosidase I. Isolation and Characterization of a Bifunctional Isozyme. Biochim. Biophys. Acta (BBA)-Enzym. 1977, 482, 98–108. [Google Scholar] [CrossRef]
- Clark, L.F.; Johnson, J.V.; Horenstein, N.A. Identification of a Gene Cluster That Initiates Azasugar Biosynthesis in Bacillus Amyloliquefaciens. ChemBioChem 2011, 12, 2147–2150. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.-D.; Cho, Y.S.; Song, J.H.; Park, Y.S.; Lee, J.Y.; Hwang, K.Y.; Rhee, S.K.; Chung, J.H.; Kwon, O.; Seong, S.-I. Identification of the Genes Involved in 1-Deoxynojirimycin Synthesis in Bacillus Subtilis MORI 3K-85. J. Microbiol. 2011, 49, 431–440. [Google Scholar] [CrossRef]
- Seo, M.-J.; Nam, Y.-D.; Lee, S.-Y.; Park, S.-L.; Yi, S.-H.; Lim, S.-I. Isolation of the Putative Biosynthetic Gene Cluster of 1-Deoxynojirimycin by Bacillus Amyloliquefaciens 140N, Its Production and Application to the Fermentation of Soybean Paste. Biosci. Biotechnol. Biochem. 2013, 77, 398–401. [Google Scholar] [CrossRef]
- Pearson, W.R. An Introduction to Sequence Similarity (“Homology”) Searching. Curr. Protoc. Bioinform. 2013, 42, 3.1.1–3.1.8. [Google Scholar] [CrossRef]
- Wang, W.; Vignani, R.; Scali, M.; Cresti, M. A Universal and Rapid Protocol for Protein Extraction from Recalcitrant Plant Tissues for Proteomic Analysis. Electrophoresis 2006, 27, 2782–2786. [Google Scholar] [CrossRef]
- Villén, J.; Gygi, S.P. The SCX/IMAC Enrichment Approach for Global Phosphorylation Analysis by Mass Spectrometry. Nat. Protoc. 2008, 3, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Wegener, K.M.; Singh, A.K.; Jacobs, J.M.; Elvitigala, T.; Welsh, E.A.; Keren, N.; Gritsenko, M.A.; Ghosh, B.K.; Camp, D.G.; Smith, R.D.; et al. Global Proteomics Reveal an Atypical Strategy for Carbon/Nitrogen Assimilation by a Cyanobacterium Under Diverse Environmental Perturbations. Mol. Cell. 2010, 9, 2678–2689. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento, S.V.D.; Magalhães, M.M.; Cunha, R.L.; Costa, P.H.D.O.; Alves, R.C.D.O.; Oliveira, G.C.D.; Valadares, R.B.D.S. Differential Accumulation of Proteins in Oil Palms Affected by Fatal Yellowing Disease. PLoS ONE 2018, 13, e0195538. [Google Scholar] [CrossRef]
- Trindade, F.C.; Ramos, S.J.; Gastauer, M.; Saraiva, A.M.M.; Caldeira, C.F.; Oliveira, G.; Valadares, R.B.D.S. Metaproteomes Reveal Increased Capacity for Stress Tolerance of Soil Microbes in Ferruginous Tropical Rocky Outcrops. Pedobiologia 2020, 81–82, 150664. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical Statistical Model to Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Moore, L.R.; Post, A.F.; Rocap, G.; Chisholm, S.W. Utilization of Different Nitrogen Sources by the Marine Cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 2002, 47, 989–996. [Google Scholar] [CrossRef]
- Bird, C.; Wyman, M. Nitrate/Nitrite Assimilation System of the Marine Picoplanktonic Cyanobacterium Synechococcus Sp. Strain WH 8103: Effect of Nitrogen Source and Availability on Gene Expression. AEM 2003, 69, 7009–7018. [Google Scholar] [CrossRef]
- Meenakshi, S.; Srisudha, S. Proteome Analysis of Nitrogen Starvation Responses in Cyanobacteria. Int. J. Adv. Agric. Sci. Technol. 2018, 5, 42–50. [Google Scholar]
- Lardon, L.; Hélias, A.; Sialve, B.; Steyer, J.-P.; Bernard, O. Life-Cycle Assessment of Biodiesel Production from Microalgae. Environ. Sci. Technol. 2009, 43, 6475–6481. [Google Scholar] [CrossRef]
- Ernst, A.; Deicher, M.; Herman, P.M.J.; Wollenzien, U.I.A. Nitrate and Phosphate Affect Cultivability of Cyanobacteria from Environments with Low Nutrient Levels. Appl. Environ. Microbiol. 2005, 71, 3379–3383. [Google Scholar] [CrossRef]
- Aboim, J.B.; De Oliveira, D.T.; De Mescouto, V.A.; Dos Reis, A.S.; Da Rocha Filho, G.N.; Santos, A.V.; Xavier, L.P.; Santos, A.S.; Gonçalves, E.C.; Nascimento, L.A.S.D. Optimization of Light Intensity and NaNO3 Concentration in Amazon Cyanobacteria Cultivation to Produce Biodiesel. Molecules 2019, 24, 2326. [Google Scholar] [CrossRef]
- Munawaroh, H.S.; Gumilar, G.G.; Nurjanah, F.; Yuliani, G.; Aisyah, S.; Kurnia, D.; Wulandari, A.P.; Kurniawan, I.; Ningrum, A.; Koyande, A.K.; et al. In-Vitro Molecular Docking Analysis of Microalgae Extracted Phycocyanin as an Anti-Diabetic Candidate. Biochem. Eng. J. 2020, 161, 107666. [Google Scholar] [CrossRef]
- Gross, E.M.; Wolk, C.P.; Juttner, F. Fischerellin, A New Allelochemical from the Freshwater Cyanobacterium Fischerella Muscicola1. J. Phycol. 1991, 27, 686–692. [Google Scholar] [CrossRef]
- Shibano, M.; Fujimoto, Y.; Kushino, K.; Kusano, G.; Baba, K. Biosynthesis of 1-Deoxynojirimycin in Commelina Communis: A Difference between the Microorganisms and Plants. Phytochemistry 2004, 65, 2661–2665. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-Y.; Asano, N.; Ikeda, K.; Wang, M.-X.; Butters, T.D.; Wormald, M.R.; Dwek, R.A.; Winters, A.L.; Nash, R.J.; Fleet, G.W.J. Looking Glass Inhibitors: L-DMDP, a More Potent and Specific Inhibitor of α-Glucosidases Than the Enantiomeric Natural Product DMDP. Chem. Commun. 2004, 1936–1937. [Google Scholar] [CrossRef]
- Anderson, D.C.; Campbell, E.L.; Meeks, J.C. A Soluble 3D LC/MS/MS Proteome of the Filamentous Cyanobacterium Nostoc p Unctiforme. J. Proteome Res. 2006, 5, 3096–3104. [Google Scholar] [CrossRef] [PubMed]
- Krasikov, V.; Aguirre Von Wobeser, E.; Dekker, H.L.; Huisman, J.; Matthijs, H.C.P. Time-Series Resolution of Gradual Nitrogen Starvation and Its Impact on Photosynthesis in the Cyanobacterium Synechocystis PCC 6803. Physiol. Plant. 2012, 145, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Mikkat, S.; Huang, F.; Huckauf, J.; Marin, K.; Norling, B.; Hagemann, M. Proteome Analysis of Salt Stress Response in the Cyanobacterium Synechocystis Sp. Strain PCC 6803. Proteomics 2006, 6, 2733–2745. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Nguyen, A.Y.; Dai, Z.; Su, D.; Gaffrey, M.J.; Moore, R.J.; Jacobs, J.M.; Monroe, M.E.; Smith, R.D.; Koppenaal, D.W.; et al. Proteome-Wide Light/Dark Modulation of Thiol Oxidation in Cyanobacteria Revealed by Quantitative Site-Specific Redox Proteomics. Mol. Cell. Proteom. 2014, 13, 3270–3285. [Google Scholar] [CrossRef] [PubMed]
- Selim, K.A.; Haffner, M. Heavy Metal Stress Alters the Response of the Unicellular Cyanobacterium Synechococcus Elongatus PCC 7942 to Nitrogen Starvation. Life 2020, 10, 275. [Google Scholar] [CrossRef]
- Verma, E.; Chakraborty, S.; Kharwar, S.; Tiwari, B.; Singh, S.S.; Mishra, A.K. Nitrogen Starvation–Induced Oxidative Stress Relieves PII-Mediated Inhibition of Acetyl-CoA Carboxylase (ACCase) Activity and Signals Enhanced Lipid Synthesis in Synechococcus PCC 7942. J. Appl. Phycol. 2021, 33, 313–329. [Google Scholar] [CrossRef]
- Pade, N.; Hagemann, M. Salt Acclimation of Cyanobacteria and Their Application in Biotechnology. Life 2014, 5, 25–49. [Google Scholar] [CrossRef]
- Ismaiel, M.M.S.; Piercey-Normore, M.D.; Rampitsch, C. Proteomic Analyses of the Cyanobacterium Arthrospira (Spirulina) Platensis under Iron and Salinity Stress. Environ. Exp. Bot. 2018, 147, 63–74. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Chen, X.-H.; Xue, C.; Zhang, H.; Sun, G.; Xie, Z.-X.; Lin, L.; Wang, D.-Z. Proteomic Response to Rising Temperature in the Marine Cyanobacterium Synechococcus Grown in Different Nitrogen Sources. Front. Microbiol. 2019, 10, 1976. [Google Scholar] [CrossRef]
- Tanaka, N.; Hiyama, T.; Nakamoto, H. Cloning, Characterization and Functional Analysis of GroESL Operon from Thermophilic Cyanobacterium Synechococcus Vulcanus. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzym. 1997, 1343, 335–348. [Google Scholar] [CrossRef]
- Asadulghani, M.; Nitta, K.; Kaneko, Y.; Kojima, K.; Fukuzawa, H.; Kosaka, H.; Nakamoto, H. Comparative Analysis of the HspA Mutant and Wild-Type Synechocystis Sp. Strain PCC 6803 under Salt Stress: Evaluation of the Role of HspA in Salt-Stress Management. Arch. Microbiol 2004, 182, 487–497. [Google Scholar] [CrossRef]
- Nitta, K.; Suzuki, N.; Honma, D.; Kaneko, Y.; Nakamoto, H. Ultrastructural Stability under High Temperature or Intensive Light Stress Conferred by a Small Heat Shock Protein in Cyanobacteria. FEBS Lett. 2005, 579, 1235–1242. [Google Scholar] [CrossRef]
- Singh, A.K.; Summerfield, T.C.; Li, H.; Sherman, L.A. The Heat Shock Response in the Cyanobacterium Synechocystis Sp. Strain PCC 6803 and Regulation of Gene Expression by HrcA and SigB. Arch. Microbiol. 2006, 186, 273–286. [Google Scholar] [CrossRef]
- Mehta, K.; Jaiswal, D.; Nayak, M.; Prasannan, C.B.; Wangikar, P.P.; Srivastava, S. Elevated Carbon Dioxide Levels Lead to Proteome-Wide Alterations for Optimal Growth of a Fast-Growing Cyanobacterium, Synechococcus Elongatus PCC 11801. Sci. Rep. 2019, 9, 6257. [Google Scholar] [CrossRef]
- Figge, R.M.; Schubert, M.; Brinkmann, H.; Cerff, R. Glyceraldehyde-3-Phosphate Dehydrogenase Gene Diversity in Eubacteria and Eukaryotes: Evidence for Intra- and Inter-Kingdom Gene Transfer. Mol. Biol. Evol. 1999, 16, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Koksharova, O.; Schubert, M.; Shestakov, S.; Cerff, R. [No Title Found]. Plant Mol. Biol. 1998, 36, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, T.; Gao, X.; Shi, M.; Wu, L.; Chen, L.; Zhang, W. Biosynthesis of Platform Chemical 3-Hydroxypropionic Acid (3-HP) Directly from CO2 in Cyanobacterium Synechocystis Sp. PCC 6803. Metab. Eng. 2016, 34, 60–70. [Google Scholar] [CrossRef]
- Wang, X.; Xiong, X.; Sa, N.; Roje, S.; Chen, S. Metabolic Engineering of Enhanced Glycerol-3-Phosphate Synthesis to Increase Lipid Production in Synechocystis Sp. PCC 6803. Appl. Microbiol. Biotechnol. 2016, 100, 6091–6101. [Google Scholar] [CrossRef]
- Amaral, S.C.D.; Santos, A.V.; Da Cruz Schneider, M.P.; Da Silva, J.K.R.; Xavier, L.P. Determination of Volatile Organic Compounds and Antibacterial Activity of the Amazonian Cyanobacterium Synechococcus Sp. Strain GFB01. Molecules 2020, 25, 4744. [Google Scholar] [CrossRef] [PubMed]
- Marri, L.; Thieulin-Pardo, G.; Lebrun, R.; Puppo, R.; Zaffagnini, M.; Trost, P.; Gontero, B.; Sparla, F. CP12-Mediated Protection of Calvin–Benson Cycle Enzymes from Oxidative Stress. Biochimie 2014, 97, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Mata-Cabana, A.; Florencio, F.J.; Lindahl, M. Membrane Proteins from the Cyanobacterium Synechocystis Sp. PCC 6803 Interacting with Thioredoxin. Proteomics 2007, 7, 3953–3963. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, M.; Florencio, F.J. Systematic Screening of Reactive Cysteine Proteomes. Proteomics 2004, 4, 448–450. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chen, L.; Te, R.; Qiao, J.; Wang, J.; Zhang, W. Complementary ITRAQ Proteomics and RNA-Seq Transcriptomics Reveal Multiple Levels of Regulation in Response to Nitrogen Starvation in Synechocystis Sp. PCC 6803. Mol. BioSyst. 2013, 9, 2565. [Google Scholar] [CrossRef] [PubMed]
- Deschoenmaeker, F.; Facchini, R.; Leroy, B.; Badri, H.; Zhang, C.-C.; Wattiez, R. 2014 Proteomic and Cellular Views of Arthrospira Sp. PCC 8005 Adaptation to Nitrogen Depletion. Microbiology 2014, 160, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Depraetere, O.; Deschoenmaeker, F.; Badri, H.; Monsieurs, P.; Foubert, I.; Leys, N.; Wattiez, R.; Muylaert, K. Trade-Off between Growth and Carbohydrate Accumulation in Nutrient-Limited Arthrospira Sp. PCC 8005 Studied by Integrating Transcriptomic and Proteomic Approaches. PLoS ONE 2015, 10, e0132461. [Google Scholar] [CrossRef]
- Shimmori, Y.; Kanesaki, Y.; Nozawa, M.; Yoshikawa, H.; Ehira, S. Transcriptional Activation of Glycogen Catabolism and the Oxidative Pentose Phosphate Pathway by NrrA Facilitates Cell Survival Under Nitrogen Starvation in the Cyanobacterium Synechococcus Sp. Strain PCC 7002. Plant Cell Physiol. 2018, 59, 1225–1233. [Google Scholar] [CrossRef]
- Von Wobeser, E.; Ibelings, B.W.; Bok, J.; Krasikov, V.; Huisman, J.; Matthijs, H.C.P. Concerted Changes in Gene Expression and Cell Physiology of the Cyanobacterium Synechocystis Sp. Strain PCC 6803 during Transitions between Nitrogen and Light-Limited Growth. Plant Physiol. 2011, 155, 1445–1457. [Google Scholar] [CrossRef]
- Baier, K.; Nicklisch, S.; Grundner, C.; Reinecke, J.; Lockau, W. Expression of Two NblA -Homologous Genes Is Required for Phycobilisome Degradation in Nitrogen-Starved Synechocystis Sp. PCC6803. FEMS Microbiol. Lett. 2001, 195, 35–39. [Google Scholar] [CrossRef]
- Klotz, A.; Reinhold, E.; Doello, S.; Forchhammer, K. Nitrogen Starvation Acclimation in Synechococcus Elongatus: Redox-Control and the Role of Nitrate Reduction as an Electron Sink. Life 2015, 5, 888–904. [Google Scholar] [CrossRef]
- Karradt, A.; Sobanski, J.; Mattow, J.; Lockau, W.; Baier, K. NblA, a Key Protein of Phycobilisome Degradation, Interacts with ClpC, a HSP100 Chaperone Partner of a Cyanobacterial Clp Protease. J. Biol. Chem. 2008, 283, 32394–32403. [Google Scholar] [CrossRef]
- Esteves-Ferreira, A.A.; Inaba, M.; Fort, A.; Araújo, W.L.; Sulpice, R. Nitrogen Metabolism in Cyanobacteria: Metabolic and Molecular Control, Growth Consequences and Biotechnological Applications. Crit. Rev. Microbiol. 2018, 44, 541–560. [Google Scholar] [CrossRef] [PubMed]
- Tolonen, A.C.; Aach, J.; Lindell, D.; Johnson, Z.I.; Rector, T.; Steen, R.; Church, G.M.; Chisholm, S.W. Global Gene Expression of Prochlorococcus Ecotypes in Response to Changes in Nitrogen Availability. Mol. Syst. Biol. 2006, 2, 53. [Google Scholar] [CrossRef] [PubMed]
- Sauer, J.; Schreiber, U.; Schmid, R.; Völker, U.; Forchhammer, K. Nitrogen Starvation-Induced Chlorosis InSynechococcus PCC 7942. Low-Level Photosynthesis as a Mechanism of Long-Term Survival. Plant Physiol. 2001, 126, 233–243. [Google Scholar] [CrossRef]
- Ehira, S.; Ohmori, M. NrrA, a Nitrogen-Responsive Response Regulator Facilitates Heterocyst Development in the Cyanobacterium Anabaena Sp. Strain PCC 7120. Mol. Microbiol. 2006, 59, 1692–1703. [Google Scholar] [CrossRef] [PubMed]
- Harke, M.J.; Gobler, C.J. Global Transcriptional Responses of the Toxic Cyanobacterium, Microcystis Aeruginosa, to Nitrogen Stress, Phosphorus Stress, and Growth on Organic Matter. PLoS ONE 2013, 8, e69834. [Google Scholar] [CrossRef]
- González-Fernández, C.; Ballesteros, M. Linking Microalgae and Cyanobacteria Culture Conditions and Key-Enzymes for Carbohydrate Accumulation. Biotechnol. Adv. 2012, 30, 1655–1661. [Google Scholar] [CrossRef] [PubMed]
- Damrow, R.; Maldener, I.; Zilliges, Y. The Multiple Functions of Common Microbial Carbon Polymers, Glycogen and PHB, during Stress Responses in the Non-Diazotrophic Cyanobacterium Synechocystis Sp. PCC 6803. Front. Microbiol. 2016, 7, 996. [Google Scholar] [CrossRef] [PubMed]
- Osanai, T.; Imamura, S.; Asayama, M.; Shirai, M.; Suzuki, I.; Murata, N.; Tanaka, K. Nitrogen Induction of Sugar Catabolic Gene Expression in Synechocystis Sp. PCC 6803. DNA Res. 2006, 13, 185–195. [Google Scholar] [CrossRef]
- Tan, X.; Hou, S.; Song, K.; Georg, J.; Klähn, S.; Lu, X.; Hess, W.R. The Primary Transcriptome of the Fast-Growing Cyanobacterium Synechococcus Elongatus UTEX 2973. Biotechnol. Biofuels 2018, 11, 218. [Google Scholar] [CrossRef]
- Esteves-Ferreira, A.A.; Cavalcanti, J.H.F.; Vaz, M.G.M.V.; Alvarenga, L.V.; Nunes-Nesi, A.; Araújo, W.L.; Esteves-Ferreira, A.A.; Cavalcanti, J.H.F.; Vaz, M.G.M.V.; Alvarenga, L.V.; et al. Cyanobacterial Nitrogenases: Phylogenetic Diversity, Regulation and Functional Predictions. Genet. Mol. Biol. 2017, 40, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schreiber, K.; Appel, J.; Makowka, A.; Fähnrich, B.; Roettger, M.; Hajirezaei, M.R.; Sönnichsen, F.D.; Schönheit, P.; Martin, W.F.; et al. The Entner–Doudoroff Pathway Is an Overlooked Glycolytic Route in Cyanobacteria and Plants. Proc. Natl. Acad. Sci. USA 2016, 113, 5441–5446. [Google Scholar] [CrossRef]
- Maeda, S.; Omata, T. Nitrite Transport Activity of the ABC-Type Cyanate Transporter of the Cyanobacterium Synechococcus Elongatus. J. Bacteriol. 2009, 191, 3265–3272. [Google Scholar] [CrossRef] [PubMed]
- Amapá, Lei n° 0835, de 27 de maio de 2004. Dispõe Sobre a Ocupação Urbana e Periurbana, Reordenamento Territorial, uso Econômico e Gestão AMBIENTAL das áreas de Ressaca e Várzea Localizadas no Estado do Amapá e dá Outras Providencias. Available online: http://www.mpap.mp.br/images/PRODEMAC/legislacao/Lei_n%C2%BA_0835_de_27_de_maio_de_2004.pdf (accessed on 9 April 2021).
- Brasil, Lei n° 12.651, de 25 de maio de 2012. Dispõe sobre a Proteção da Vegetação Nativa; Altera as Leis n°s 6.938, de 31 de agosto de 1981, 9.393, de 19 de Dezembro de 1996, e 11.428, de 22 de Dezembro de 2006; Revoga as Leis n°s 4.771, de 15 de Setembro de 1965, e 7.754, de 14 de Abril de 1989, e a Medida Provisória n° 2.166-67, de 24 de agosto de 2001; e dá outras providências. Available online: http://www.planalto.gov.br/ccivil_03/_ato2011-2014/2012/lei/l12651.htm (accessed on 7 May 2021).
- Bastos, C.M.C.B. Ressaca e Comunidade Lagoa dos Índios Em Macapá/Ap: Entrelaçamento do Patrimônio Ambiental Cultural. Plan. Am. Rev. Int. Dir. Amb. Pol. Púb. 2020, 151, 151–161. [Google Scholar] [CrossRef]
- Takiyama, L.R.; Silva, A.Q.; Costa, W.J.P.; Nascimento, H.S. Qualidade das Águas das Ressacas das Bacias do Igarapé da Fortaleza e do Rio Curiaú. In Diagnóstico das Ressacas do Estado do Amapá: Bacias do Igarapé da Fortaleza e Rio Curiaú; Takiyama, L.R., Silva, A.Q., Eds.; CPAQ/IEPA and DGEO/SEMA IEPA: Macapá, Brazil, 2003; pp. 81–104. [Google Scholar]
- Furtado, M.F.D.M. Potencial Para Bioindicação e Bioprospecção em Microalgas e Cianobactérias na Lagoa dos Índios. Master′s Thesis, Federal University of Amapá/UNIFAP, Macapá, Brazil, 2018; p. 131. [Google Scholar]
- Mendes, L.B.; Vermelho, A. Allelopathy as a Potential Strategy to Improve Microalgae Cultivation. Biotechnol. Biofuels 2013, 6, 152. [Google Scholar] [CrossRef]


| Parameter (μg/mL) | BG-11 | BG-1110%N | ||
|---|---|---|---|---|
| Day 15 | Day 30 | Day 15 | Day 30 | |
| Chlorophyll α | 406,686 | 980,088 | 395,319 | 971,247 |
| Protein | 546 | 573 | 693 | 166 |
| ASNo. a | Organism | Identity (%) | E-Value | Bit Score | Log2 FC c |
|---|---|---|---|---|---|
| WP_038545752 b | S. sp. KORDI-100 | 27.612 | 2.76 × 10−38 | 139 | - |
| WP_041430506 | S. sp. GFB01 | 26.426 | 1.73 × 10−42 | 101 | BG-11 d |
| WP_071802316 | 27.711 | 1.49 × 10−25 | 100 | −0.97 | |
| WP_051847317 | 24.750 | 3.57 × 10−25 | 99.8 | 0.56 | |
| WP_015167166 | 25 | 1.03 × 10−24 | 98.6 | BG-11 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gradíssimo, D.G.; Oliveira da Silva, V.C.; Xavier, L.P.; do Nascimento, S.V.; Valadares, R.B.d.S.; Faustino, S.M.M.; Schneider, M.P.C.; Santos, A.V. Glucosidase Inhibitors Screening in Microalgae and Cyanobacteria Isolated from the Amazon and Proteomic Analysis of Inhibitor Producing Synechococcus sp. GFB01. Microorganisms 2021, 9, 1593. https://doi.org/10.3390/microorganisms9081593
Gradíssimo DG, Oliveira da Silva VC, Xavier LP, do Nascimento SV, Valadares RBdS, Faustino SMM, Schneider MPC, Santos AV. Glucosidase Inhibitors Screening in Microalgae and Cyanobacteria Isolated from the Amazon and Proteomic Analysis of Inhibitor Producing Synechococcus sp. GFB01. Microorganisms. 2021; 9(8):1593. https://doi.org/10.3390/microorganisms9081593
Chicago/Turabian StyleGradíssimo, Diana Gomes, Vivian Cássia Oliveira da Silva, Luciana Pereira Xavier, Sidney Vasconcelos do Nascimento, Rafael Borges da Silva Valadares, Silvia Maria Mathes Faustino, Maria Paula Cruz Schneider, and Agenor Valadares Santos. 2021. "Glucosidase Inhibitors Screening in Microalgae and Cyanobacteria Isolated from the Amazon and Proteomic Analysis of Inhibitor Producing Synechococcus sp. GFB01" Microorganisms 9, no. 8: 1593. https://doi.org/10.3390/microorganisms9081593
APA StyleGradíssimo, D. G., Oliveira da Silva, V. C., Xavier, L. P., do Nascimento, S. V., Valadares, R. B. d. S., Faustino, S. M. M., Schneider, M. P. C., & Santos, A. V. (2021). Glucosidase Inhibitors Screening in Microalgae and Cyanobacteria Isolated from the Amazon and Proteomic Analysis of Inhibitor Producing Synechococcus sp. GFB01. Microorganisms, 9(8), 1593. https://doi.org/10.3390/microorganisms9081593






