Oligosaccharide Metabolism and Lipoteichoic Acid Production in Lactobacillus gasseri and Lactobacillus paragasseri
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Oligosaccharide Metabolic Properties
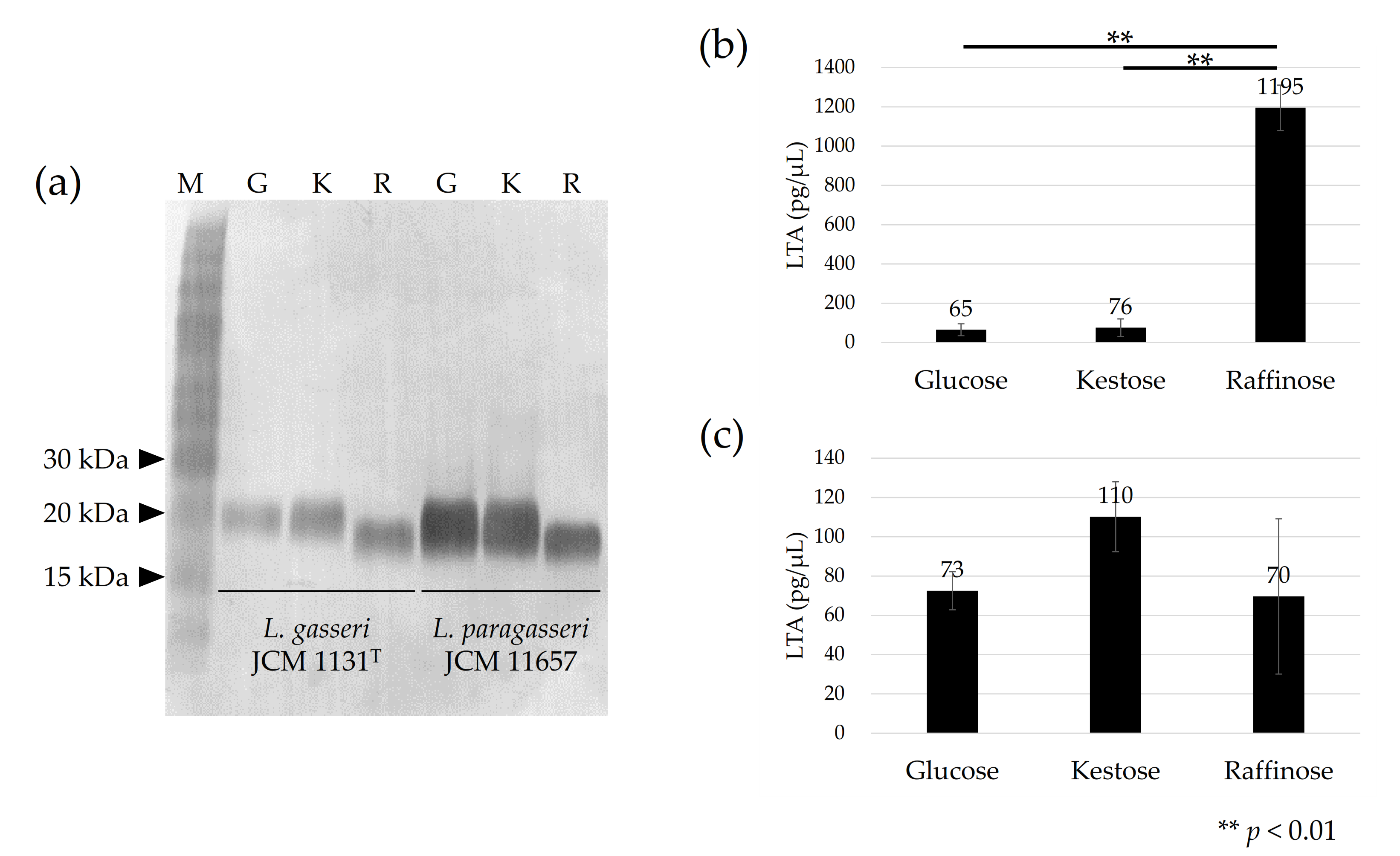
2.3. Immunoblotting
2.4. Quantification of LTA
2.5. Draft Genome Sequencing and Acquisition of Genomic Data
2.6. Genome Analysis
2.7. RNA Sequencing of L. gasseri and L. paragasseri
3. Results
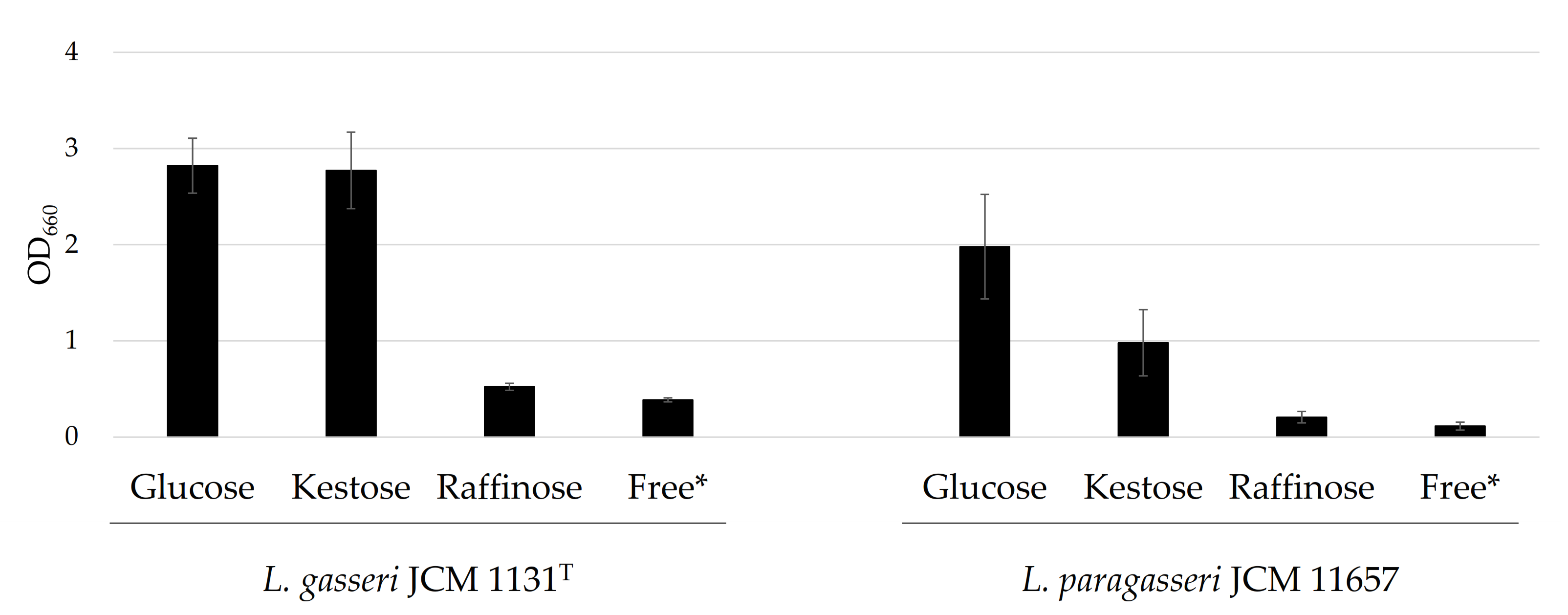
3.1. Growth in the Presence of Different Carbohydrates
3.2. Detection of LTA in Cells Cultured with Different Carbohydrates
3.3. Genomic Characteristics
3.3.1. Genes Involved in Kestose Metabolism
3.3.2. Genes Involved in LTA Synthesis
3.4. Transcriptomic Analysis
3.4.1. Transcriptomic Analysis of Genes Involved in Carbohydrate Metabolism
3.4.2. Transcriptomic Analysis of Genes Involved in LTA Synthesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Ose, R.; Hirano, K.; Maeno, S.; Nakagawa, J.; Salminen, S.; Tochio, T.; Endo, A. The ability of human intestinal anaerobes to metabolize different oligosaccharides: Novel means for microbiota modulation? Anaerobe 2018, 51, 110–119. [Google Scholar] [CrossRef]
- Tanno, H.; Fujii, T.; Hirano, K.; Maeno, S.; Tonozuka, T.; Sakamoto, M.; Ohkuma, M.; Tochio, T.; Endo, A. Characterization of fructooligosaccharide metabolism and fructooligosaccharide-degrading enzymes in human commensal butyrate producers. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef]
- Lawley, B.; Sims, I.M.; Tannock, G.W. Whole-transcriptome shotgun sequencing (RNA-seq) screen reveals upregulation of cellobiose and motility operons of Lactobacillus ruminis L5 during growth on tetrasaccharides derived from barley β-glucan. Appl. Environ. Microbiol. 2013, 79, 5661–5669. [Google Scholar] [CrossRef]
- Nishiyama, K.; Yamamoto, Y.; Sugiyama, M.; Takaki, T.; Urashima, T.; Fukiya, S.; Yokota, A.; Okada, N.; Mukai, T. Bifidobacterium Bifidum Extracellular Sialidase Enhances Adhesion to the Mucosal Surface and Supports Carbohydrate Assimilation. mBio 2017, 8, e00928-17. [Google Scholar] [CrossRef] [PubMed]
- Zabel, B.; Yde, C.C.; Roos, P.; Marcussen, J.; Jensen, H.M.; Salli, K.; Hirvonen, J.; Ouwehand, A.C.; Morovic, W. Novel Genes and Metabolite Trends in Bifidobacterium longum subsp. infantis Bi-26 Metabolism of Human Milk Oligosaccharide 2′-fucosyllactose. Sci. Rep. 2019, 9, 7983. [Google Scholar] [CrossRef]
- Selle, K.; Klaenhammer, T.R. Genomic and phenotypic evidence for probiotic influences of Lactobacillus gasseri on human health. FEMS Microbiol. Rev. 2013, 37, 915–935. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Tada, I.; Kobayashi, H.; Endo, A.; Maeno, S.; Toyoda, A.; Arita, M.; Nakamura, Y.; Sakamoto, M.; Ohkuma, M.; et al. Lactobacillus paragasseri sp. nov., a sister taxon of Lactobacillus gasseri, based on whole-genome sequence analyses. Int. J. Syst. Evol. Microbiol. 2018, 68, 3512–3517. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yang, B.; Stanton, C.; Ross, R.P.; Zhao, J.; Zhang, H.; Chen, W. Comparative analysis of Lactobacillus gasseri from Chinese subjects reveals a new species-level taxa. BMC Genom. 2020, 21, 119. [Google Scholar] [CrossRef] [PubMed]
- Paveljšek, D.; Ivičak-Kocjan, K.; Treven, P.; Benčina, M.; Jerala, R.; Rogelj, I. Distinctive probiotic features share common TLR2-dependent signalling in intestinal epithelial cells. Cell Microbiol. 2021, 23, e13264. [Google Scholar] [CrossRef]
- Endo, A.; Nakamura, S.; Konishi, K.; Nakagawa, J.; Tochio, T. Variations in prebiotic oligosaccharide fermentation by intestinal lactic acid bacteria. Int. J. Food Sci. Nutr. 2016, 67, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Yokota, S.; Morita, N.; Fukiya, S.; Tomita, S.; Tanaka, N.; Okada, S.; Yokota, A. Characterization of a Lactobacillus gasseri JCM 1131T lipoteichoic acid with a novel glycolipid anchor structure. Appl. Environ. Microbiol. 2013, 79, 3315–3318. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Yokota, S.; Fukiya, S.; Yokota, A. Structural diversity and biological significance of lipoteichoic acid in Gram-positive bacteria: Focusing on beneficial probiotic lactic acid bacteria. Biosci. Microbiota Food Health 2016, 35, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Grangette, C.; Nutten, S.; Palumbo, E.; Morath, S.; Hermann, C.; Dewulf, J.; Pot, B.; Hartung, T.; Hols, P.; Mercenier, A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc. Natl. Acad. Sci. USA 2005, 102, 10321–10326. [Google Scholar] [CrossRef]
- Noh, S.Y.; Kang, S.S.; Yun, C.H.; Han, S.H. Lipoteichoic acid from Lactobacillus plantarum inhibits Pam2CSK4-induced IL-8 production in human intestinal epithelial cells. Mol. Immunol. 2015, 64, 183–189. [Google Scholar] [CrossRef]
- Palomino, M.M.; Allievi, M.C.; Gründling, A.; Sanchez-Rivas, C.; Ruzal, S.M. Osmotic stress adaptation in Lactobacillus casei BL23 leads to structural changes in the cell wall polymer lipoteichoic acid. Microbiology 2013, 159, 2416–2426. [Google Scholar] [CrossRef]
- Li, M.; Lai, Y.; Villaruz, A.E.; Cha, D.J.; Sturdevant, D.E.; Otto, M. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. USA 2007, 104, 9469–9474. [Google Scholar] [CrossRef]
- Lang, W.K.; Glassey, K.; Archibald, A.R. Influence of phosphate supply on teichoic acid and teichuronic acid content of Bacillus subtilis cell walls. J. Bacteriol. 1982, 151, 367–375. [Google Scholar] [CrossRef]
- Shiraishi, T.; Yokota, S.; Sato, Y.; Ito, T.; Fukiya, S.; Yamamoto, S.; Sato, T.; Yokota, A. Lipoteichoic acids are embedded in cell walls during logarithmic phase, but exposed on membrane vesicles in Lactobacillus gasseri JCM 1131T. Benef. Microbes 2018, 9, 653–662. [Google Scholar] [CrossRef]
- Kajitani, R.; Yoshimura, D.; Ogura, Y.; Gotoh, Y.; Hayashi, T.; Itoh, T. Platanus_B: An accurate de novo assembler for bacterial genomes using an iterative error-removal process. DNA Res. 2020, 27, dsaa014. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Fujisawa, T.; Kaminuma, E.; Nakamura, Y.; Arita, M. DFAST and DAGA: Web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 2016, 35, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Maeno, S.; Tanizawa, Y.; Kanesaki, Y.; Kubota, E.; Kumar, H.; Dicks, L.; Salminen, S.; Nakagawa, J.; Arita, M.; Endo, A. Genomic characterization of a fructophilic bee symbiont Lactobacillus kunkeei reveals its niche-specific adaptation. Syst. Appl. Microbiol. 2016, 39, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Su, W.; Sun, J.; Shimizu, K.; Kadota, K. TCC-GUI: A Shiny-based application for differential expression analysis of RNA-Seq count data. BMC Res. Notes 2019, 12, 133. [Google Scholar] [CrossRef] [PubMed]
- Trollope, K.M.; van Wyk, N.; Kotjomela, M.A.; Volschenk, H. Sequence and structure-based prediction of fructosyltransferase activity for functional subclassification of fungal GH32 enzymes. FEBS J. 2015, 282, 4782–4796. [Google Scholar] [CrossRef]
- Anwar, M.A.; Kralj, S.; Piqué, A.V.; Leemhuis, H.; van der Maarel, M.J.E.C.; Dijkhuizen, L. Inulin and levan synthesis by probiotic Lactobacillus gasseri strains: Characterization of three novel fructansucrase enzymes and their fructan products. Microbiology 2010, 156, 1264–1274. [Google Scholar] [CrossRef] [PubMed]
- Mohamadzadeh, M.; Pfeiler, E.A.; Brown, J.B.; Zadeh, M.; Gramarossa, M.; Managlia, E.; Bere, P.; Sarraj, B.; Khan, M.W.; Pakanati, K.C.; et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. USA 2011, 108, 4623–4630. [Google Scholar] [CrossRef] [PubMed]
- Perea Vélez, M.; Verhoeven, T.L.; Draing, C.; Von Aulock, S.; Pfitzenmaier, M.; Geyer, A.; Lambrichts, I.; Grangette, C.; Pot, B.; Vanderleyden, J.; et al. Functional analysis of D-alanylation of lipoteichoic acid in the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2007, 73, 3595–3604. [Google Scholar] [CrossRef]
- Hesser, A.R.; Matano, L.M.; Vickery, C.R.; Wood, B.M.; Santiago, A.G.; Morris, H.G.; Do, T.; Losick, R.; Walker, S. The length of lipoteichoic acid polymers controls Staphylococcus aureus cell size and envelope integrity. J. Bacteriol. 2020, 202, e00149-20. [Google Scholar] [CrossRef] [PubMed]
- Menéndez, C.; Martínez, D.; Pérez, E.R.; Musacchio, A.; Ramírez, R.; López-Munguía, A.; Hernández, L. Engineered thermostable β-fructosidase from Thermotoga maritima with enhanced fructooligosaccharides synthesis. Enzym. Microb. Technol. 2019, 125, 53–62. [Google Scholar] [CrossRef]
- Mao, B.; Tang, H.; Gu, J.; Li, D.; Cui, S.; Zhao, J.; Zhang, H.; Chen, W. In vitro fermentation of raffinose by the human gut bacteria. Food Funct. 2018, 9, 5824–5831. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.; Møller, M.S.; Poulsen, J.N.; Pichler, M.J.; Svensson, B.; Lo Leggio, L.; Goh, Y.J.; Abou Hachem, M. An 1,4-α-Glucosyltransferase Defines a New Maltodextrin Catabolism Scheme in Lactobacillus acidophilus. Appl. Environ. Microbiol. 2020, 86, e00661-20. [Google Scholar] [CrossRef]
- Hardy, L.; Jacques, N.A.; Forester, H.; Campbell, L.K.; Knox, K.W.; Wicken, A.J. Effect of fructose and other carbohydrates on the surface properties, lipoteichoic acid production, and extracellular proteins of Streptococcus mutans Ingbritt grown in continuous culture. Infect. Immun. 1981, 31, 78–87. [Google Scholar] [CrossRef]
- Jacques, N.A.; Hardy, L.; Campbell, L.K.; Knox, K.W.; Evans, J.D.; Wicken, A.J. Effect of carbohydrate source and growth conditions on the production of lipoteichoic acid by Streptococcus mutans Ingbritt. Infect. Immun. 1979, 26, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Wicken, A.J.; Broady, K.W.; Ayres, A.; Knox, K.W. Production of lipoteichoic acid by lactobacilli and streptococci grown in different environments. Infect. Immun. 1982, 36, 864–869. [Google Scholar] [CrossRef]
- MacArthur, A.E.; Archibald, A.R. Effect of culture pH on the D-alanine ester content of lipoteichoic acid in Staphylococcus aureus. J. Bacteriol. 1984, 160, 792–793. [Google Scholar] [CrossRef]
- Hurst, A.; Hughes, A.; Duckworth, M.; Baddiley, J. Loss of D-alanine during sublethal heating of Staphylococcus aureus S6 and magnesium binding during repair. J. Gen. Microbiol. 1975, 89, 277–284. [Google Scholar] [CrossRef][Green Version]
- Koprivnjak, T.; Mlakar, V.; Swanson, L.; Fournier, B.; Peschel, A.; Weiss, J.P. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 2006, 188, 3622–3630. [Google Scholar] [CrossRef]
- Hermann, C.; Spreitzer, I.; Schröder, N.W.; Morath, S.; Lehner, M.D.; Fischer, W.; Schütt, C.; Schumann, R.R.; Hartung, T. Cytokine induction by purified lipoteichoic acids from various bacterial species—Role of LBP, sCD14, CD14 and failure to induce IL-12 and subsequent IFN-γ release. Eur. J. Immunol. 2002, 32, 541–551. [Google Scholar] [CrossRef]
- Ryu, Y.H.; Baik, J.E.; Yang, J.S.; Kang, S.S.; Im, J.; Yun, C.H.; Kim, D.W.; Lee, K.; Chung, D.K.; Ju, H.R.; et al. Differential immunostimulatory effects of Gram-positive bacteria due to their lipoteichoic acids. Int. Immunopharmacol. 2009, 9, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Hatano, S.; Hirose, Y.; Yamamoto, Y.; Murosaki, S.; Yoshikai, Y. Scavenger receptor for lipoteichoic acid is involved in the potent ability of Lactobacillus plantarum strain L-137 to stimulate production of interleukin-12p40. Int. Immunopharmacol. 2015, 25, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Deininger, S.; Stadelmaier, A.; von Aulock, S.; Morath, S.; Schmidt, R.R.; Hartung, T. Definition of structural prerequisites for lipoteichoic acid-inducible cytokine induction by synthetic derivatives. J. Immunol. 2003, 170, 4134–4138. [Google Scholar] [CrossRef] [PubMed]
- Claes, I.J.; Segers, M.E.; Verhoeven, T.L.; Dusselier, M.; Sels, B.F.; De Keersmaecker, S.C.; Vanderleyden, J.; Lebeer, S. Lipoteichoic acid is an important microbe-associated molecular pattern of Lactobacillus rhamnosus GG. Microb. Cell Fact. 2012, 11, 161. [Google Scholar] [CrossRef]
- Smelt, M.J.; de Haan, B.J.; Bron, P.A.; van Swam, I.; Meijerink, M.; Wells, J.M.; Kleerebezem, M.; Faas, M.M.; de Vos, P. The impact of Lactobacillus plantarum WCFS1 teichoic acid D-alanylation on the generation of effector and regulatory T-cells in healthy mice. PLoS ONE 2013, 8, e63099. [Google Scholar] [CrossRef]
- Swanson, K.S.; Gibson, G.R.; Hutkins, R.; Reimer, R.A.; Reid, G.; Verbeke, K.; Scott, K.P.; Holscher, H.D.; Azad, M.B.; Delzenne, N.M.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 687–701. [Google Scholar] [CrossRef] [PubMed]


| Glc vs. Kes | Glc vs. Raf | Kes vs. Raf | ||||
|---|---|---|---|---|---|---|
| Glc ↑ a | Kes ↑ | Glc ↑ | Raf ↑ | Kes ↑ | Raf ↑ | |
| JCM 1131T | 38 | 31 | 650 | 404 | 679 | 405 |
| Sum | 69 | 1054 | 1084 | |||
| JCM 11657 | 231 | 237 | 348 | 379 | 286 | 343 |
| Sum | 468 | 727 | 629 | |||
| C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | X | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JCM 1131T | All CDSs a | 46 | 25 | 87 | 57 | 148 | 42 | 47 | 185 | 107 | 121 | 97 | 2 | 63 | 57 | 8 | 98 | 121 | 49 | 14 | 56 | 33 | |
| Glc vs. Kes | Glc | 2 | 0 | 2 | 1 | 10 | 0 | 1 | 3 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 3 | 2 | 2 | 0 | 0 | 0 | |
| Kes | 1 | 0 | 3 | 0 | 3 | 0 | 0 | 1 | 4 | 0 | 6 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 3 | 0 | ||
| Glc vs. Raf | Glc | 11 | 8 | 33 | 24 | 33 | 17 | 22 | 83 | 34 | 48 | 33 | 0 | 18 | 18 | 3 | 34 | 52 | 18 | 4 | 21 | 7 | |
| Raf | 8 | 4 | 14 | 7 | 52 | 5 | 6 | 19 | 27 | 24 | 19 | 0 | 12 | 15 | 1 | 25 | 19 | 9 | 2 | 10 | 14 | ||
| Kes vs. Raf | Kes | 16 | 8 | 38 | 21 | 45 | 13 | 17 | 85 | 29 | 52 | 37 | 1 | 21 | 20 | 2 | 40 | 49 | 19 | 7 | 21 | 8 | |
| Raf | 6 | 4 | 11 | 8 | 47 | 6 | 7 | 21 | 31 | 26 | 19 | 0 | 10 | 11 | 1 | 23 | 19 | 7 | 2 | 12 | 13 | ||
| JCM 11657 | All CDSs | 45 | 27 | 95 | 60 | 176 | 39 | 45 | 190 | 106 | 111 | 97 | 3 | 64 | 58 | 8 | 105 | 126 | 52 | 17 | 57 | 17 | |
| Glc vs. Kes | Glc | 3 | 1 | 12 | 2 | 36 | 6 | 2 | 31 | 10 | 25 | 5 | 2 | 5 | 11 | 1 | 17 | 14 | 6 | 1 | 4 | 0 | |
| Kes | 3 | 1 | 23 | 17 | 12 | 7 | 6 | 6 | 13 | 18 | 9 | 0 | 9 | 12 | 2 | 12 | 15 | 6 | 3 | 5 | 1 | ||
| Glc vs. Raf | Glc | 9 | 3 | 22 | 7 | 15 | 14 | 9 | 69 | 12 | 32 | 14 | 0 | 11 | 7 | 3 | 24 | 22 | 7 | 3 | 11 | 0 | |
| Raf | 15 | 3 | 8 | 13 | 60 | 1 | 6 | 13 | 39 | 15 | 27 | 0 | 8 | 13 | 0 | 25 | 16 | 10 | 5 | 12 | 4 | ||
| Kes vs. Raf | Kes | 12 | 0 | 29 | 14 | 16 | 12 | 12 | 29 | 11 | 17 | 14 | 0 | 12 | 11 | 1 | 10 | 15 | 7 | 3 | 9 | 0 | |
| Raf | 18 | 2 | 7 | 7 | 92 | 1 | 4 | 19 | 32 | 11 | 16 | 2 | 4 | 11 | 0 | 16 | 11 | 8 | 3 | 9 | 2 |
| Gene ID | Product | Glucose (G) vs. Kestose (K) | Glucose (G) vs. Raffinose (R) | Kestose (K) vs. Raffinose (R) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| log2 FC (G/K) | p | Significant Difference | log2 FC (G/R) | p | Significant Difference | log2 FC (K/R) | p | Significant Difference | ||
| 1268 | Levansucrase | −4.4 | <0.01 | K > G | −2.5 | <0.01 | R > G | 1.9 | <0.01 | K > R |
| 1269 | levansucrase (GH68) | −4.7 | <0.01 | K > G | −2.3 | <0.01 | R > G | 2.4 | <0.01 | K > R |
| 396 | PTS β-glucoside transporter subunit IIBCA | 0.7 | <0.01 | - a | −1.7 | <0.01 | R > G | −2.4 | <0.01 | R > K |
| 397 | sucrose-6-phosphate hydrolase (GH32) | 0.3 | 0.01 | - | −0.5 | <0.01 | - | −0.8 | <0.01 | - |
| 1783 | PTS β-glucoside transporter subunit IIBCA | −2.9 | <0.01 | K > G | −3.7 | <0.01 | R > G | −0.7 | <0.01 | - |
| 1784 | sucrose-6-phosphate hydrolase (GH32) | −1.1 | <0.01 | K > G | −2.2 | <0.01 | R > G | 1.0 | <0.01 | K > R |
| 0256 | α-galactosidase (GH36) | −0.2 | <0.01 | - | 0.1 | 0.03 | - | 0.3 | <0.01 | - |
| 1820 | D-alanyl transfer protein DltD | 0.2 | <0.01 | - | 0.6 | <0.01 | - | 0.4 | <0.01 | - |
| 1821 | D-alanyl carrier protein DltC | 0.2 | 0.01 | - | 0.7 | <0.01 | - | 0.5 | <0.01 | - |
| 1822 | D-alanyl transfer protein DltB | 0.2 | 0.01 | - | 1.1 | <0.01 | G > R | 0.9 | <0.01 | - |
| 1823 | D-alanine-activating enzyme DltA | 0.1 | 0.02 | - | 1.4 | <0.01 | G > R | 1.2 | <0.01 | K > R |
| 1824 | D-Ala-teichoic acid biosynthesis protein DltX | 0.1 | 0.24 | - | 1.5 | <0.01 | G > R | 1.4 | <0.01 | K > R |
| 1590 | phosphoglycerol transferase (LtaS) | 0.0 | 0.31 | - | 0.8 | <0.01 | - | 0.7 | <0.01 | - |
| 1591 | membrane protein | 0.2 | 0.18 | - | 0.9 | <0.01 | - | 0.7 | <0.01 | - |
| 1592 | glycosyl transferase (GT4) | 0.2 | 0.09 | - | 2.1 | <0.01 | G > R | 1.9 | <0.01 | K > R |
| 1593 | glycosyl transferase (GT4) | 0.2 | 0.05 | - | 2.6 | <0.01 | G > R | 2.4 | <0.01 | K > R |
| Gene ID | Product | Glucose (G) vs. Kestose (K) | Glucose (G) vs. Raffinose (R) | Kestose (K) vs. Raffinose (R) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| log2 FC (G/K) | p | Significant Difference | log2 FC (G/R) | p | Significant Difference | log2 FC (K/R) | p | Significant Difference | ||
| 5790 | Levansucrase | −4.2 | <0.01 | K > G | −2.7 | <0.01 | R > G | 1.4 | <0.01 | K > R |
| 5800 | levansucrase (GH68) | −4.9 | <0.01 | K > G | −2.6 | <0.01 | R > G | 2.2 | <0.01 | K > R |
| 12100 | sucrose-6-phosphate hydrolase (GH32) | −2.6 | <0.01 | K > G | −1.2 | <0.01 | R > G | 1.5 | <0.01 | K > R |
| 12110 | PTS system, sucrose-specific IIABC component | −3.9 | <0.01 | K > G | −3.7 | <0.01 | R > G | 0.2 | 0.01 | -a |
| 9630 | sucrose phosphorylase (GH13) | −3.1 | <0.01 | K > G | −9.8 | <0.01 | R > G | −6.7 | <0.01 | R > K |
| 9640 | beta-glucoside-specific PTS system IIABC component | −1.8 | <0.01 | K > G | −8.6 | <0.01 | R > G | −6.8 | <0.01 | R > K |
| 17400 | α-galactosidase (GH36) | 0.1 | 0.30 | - | 1.0 | <0.01 | G > R | 0.9 | <0.01 | - |
| 14610 | oligo-1,6-glucosidase (GH13) | 6.2 | <0.01 | G > K | 1.2 | <0.01 | G > R | −5.0 | <0.01 | R > K |
| 14620 | sugar ABC transporter permease protein | 6.4 | <0.01 | G > K | −0.1 | 0.33 | - | −6.5 | <0.01 | R > K |
| 14630 | sugar ABC transporter permease protein | 6.4 | <0.01 | G > K | −0.1 | 0.33 | - | −6.5 | <0.01 | R > K |
| 14640 | sugar ABC transporter substrate-binding protein | 6.6 | <0.01 | G > K | 0.2 | 0.07 | - | −6.4 | <0.01 | R > K |
| 14650 | glycerol-3-phosphate ABC transporter ATP-binding protein | 6.8 | <0.01 | G > K | 0.5 | <0.01 | - | −6.3 | <0.01 | R > K |
| 14660 | beta-phosphoglucomutase | 7.1 | <0.01 | G > K | 0.5 | <0.01 | - | −6.6 | <0.01 | R > K |
| 14670 | maltose phosphorylase (GH65) | 6.8 | <0.01 | G > K | 0.3 | <0.01 | - | −6.6 | <0.01 | R > K |
| 14680 | alpha-amylase (GH13) | 3.8 | <0.01 | G > K | 1.3 | <0.01 | G > R | −2.5 | <0.01 | R > K |
| 14690 | alpha-glucosidase (GH13) | 3.6 | <0.01 | G > K | 0.9 | <0.01 | - | −2.7 | <0.01 | R > K |
| 10260 | D-Ala-teichoic acid biosynthesis protein DltX | −0.1 | 0.29 | - | 2.1 | <0.01 | G > R | 2.2 | <0.01 | K > R |
| 10270 | D-alanine-activating enzyme DltA | 0.5 | <0.01 | - | 2.0 | <0.01 | G > R | 1.6 | <0.01 | K > R |
| 10280 | D-alanyl transfer protein DltB | 1.2 | <0.01 | G > K | 2.4 | <0.01 | G > R | 1.2 | <0.01 | K > R |
| 10290 | D-alanyl carrier protein DltC | 1.2 | <0.01 | G > K | 2.1 | <0.01 | G > R | 0.9 | <0.01 | - |
| 10300 | D-alanyl transfer protein DltD | 1.4 | <0.01 | G > K | 2.0 | <0.01 | G > R | 0.6 | <0.01 | - |
| 18430 | phosphoglycerol transferase (LtaS) | 0.3 | <0.01 | - | 0.2 | <0.01 | - | −0.1 | 0.04 | - |
| 18440 | membrane protein | 0.6 | <0.01 | - | 0.7 | <0.01 | - | 0.1 | 0.19 | - |
| 18450 | glycosyl transferase (GT4) | 0.2 | 0.01 | - | 0.5 | <0.01 | - | 0.2 | <0.01 | - |
| 18460 | glycosyl transferase (GT4) | −0.1 | 0.04 | - | -0.0 | 0.57 | - | 0.1 | 0.14 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiraishi, T.; Maeno, S.; Kishi, S.; Fujii, T.; Tanno, H.; Hirano, K.; Tochio, T.; Tanizawa, Y.; Arita, M.; Yokota, S.-i.; et al. Oligosaccharide Metabolism and Lipoteichoic Acid Production in Lactobacillus gasseri and Lactobacillus paragasseri. Microorganisms 2021, 9, 1590. https://doi.org/10.3390/microorganisms9081590
Shiraishi T, Maeno S, Kishi S, Fujii T, Tanno H, Hirano K, Tochio T, Tanizawa Y, Arita M, Yokota S-i, et al. Oligosaccharide Metabolism and Lipoteichoic Acid Production in Lactobacillus gasseri and Lactobacillus paragasseri. Microorganisms. 2021; 9(8):1590. https://doi.org/10.3390/microorganisms9081590
Chicago/Turabian StyleShiraishi, Tsukasa, Shintaro Maeno, Sayoko Kishi, Tadashi Fujii, Hiroki Tanno, Katsuaki Hirano, Takumi Tochio, Yasuhiro Tanizawa, Masanori Arita, Shin-ichi Yokota, and et al. 2021. "Oligosaccharide Metabolism and Lipoteichoic Acid Production in Lactobacillus gasseri and Lactobacillus paragasseri" Microorganisms 9, no. 8: 1590. https://doi.org/10.3390/microorganisms9081590
APA StyleShiraishi, T., Maeno, S., Kishi, S., Fujii, T., Tanno, H., Hirano, K., Tochio, T., Tanizawa, Y., Arita, M., Yokota, S.-i., & Endo, A. (2021). Oligosaccharide Metabolism and Lipoteichoic Acid Production in Lactobacillus gasseri and Lactobacillus paragasseri. Microorganisms, 9(8), 1590. https://doi.org/10.3390/microorganisms9081590






