Genetic and Phenotypic Diversities in Experimental Populations of Diploid Inter-Lineage Hybrids in the Human Pathogenic Cryptococcus
Abstract
1. Introduction
2. Materials and Methods
2.1. Ancestral Clones
2.2. Mutation Accumulation (MA) Experiments
2.3. Ploidy Analysis of D120 Cultures
2.4. Detecting the Loss of Heterozygosity (LOH)
2.5. Growth Studies in Nine Conditions
2.6. Susceptibility to Fluconazole
2.7. Statistical Analyses
3. Results
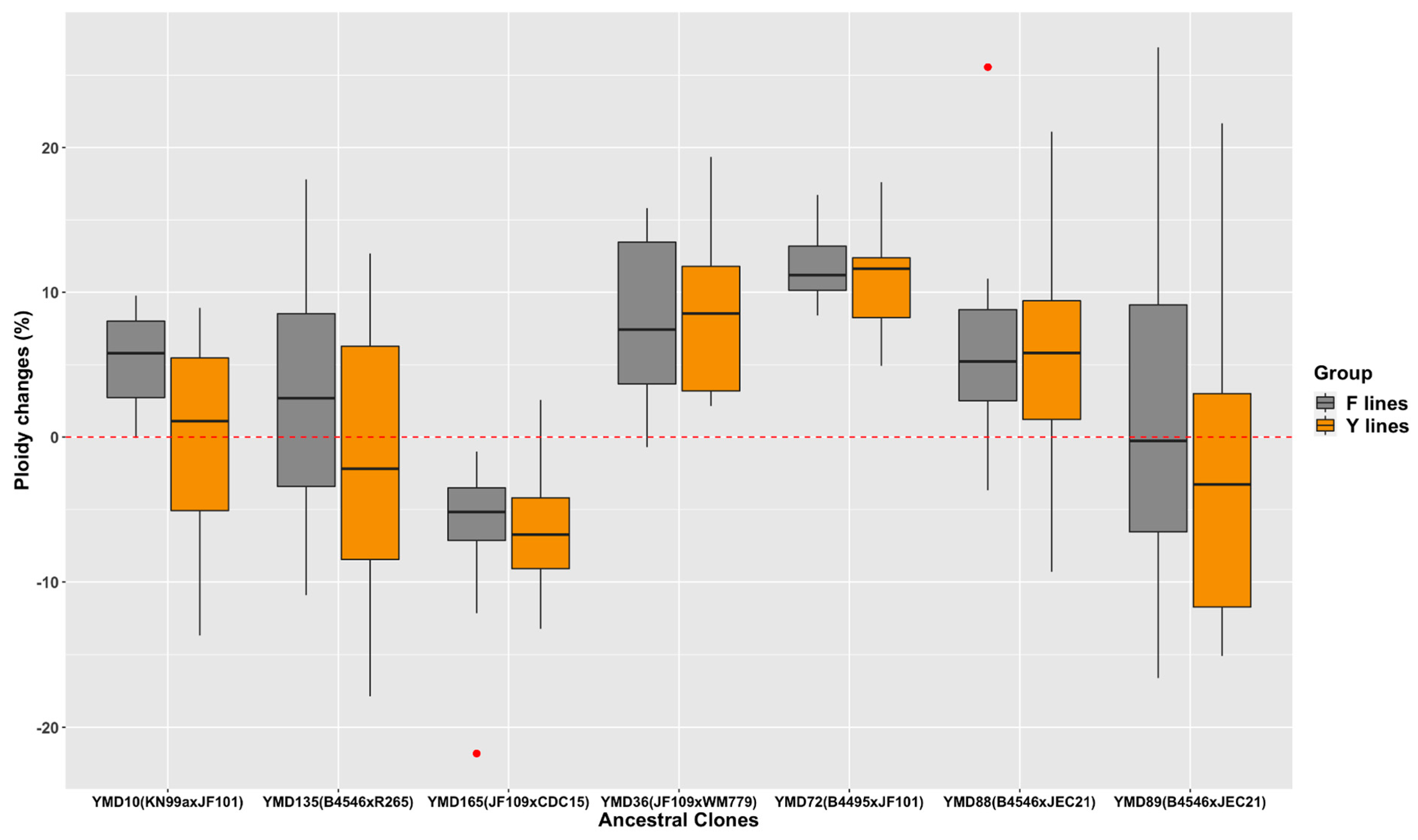
3.1. Ploidy Changes among D120 Cultures of 140 MA Lines
3.2. Genotypic Diversity among D120 Cultures of 140 MA Lines
3.3. LOH among 140 MA Lines
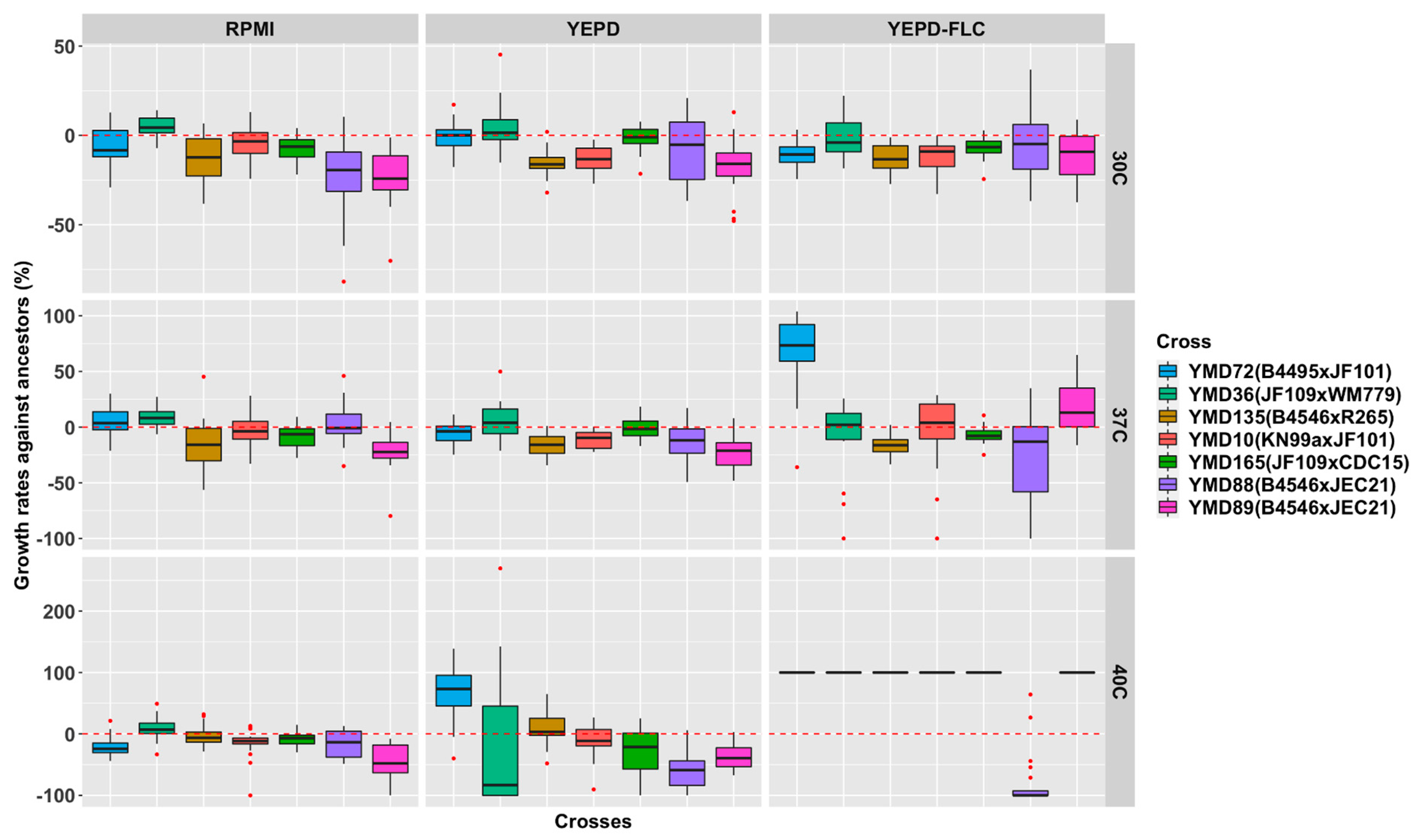
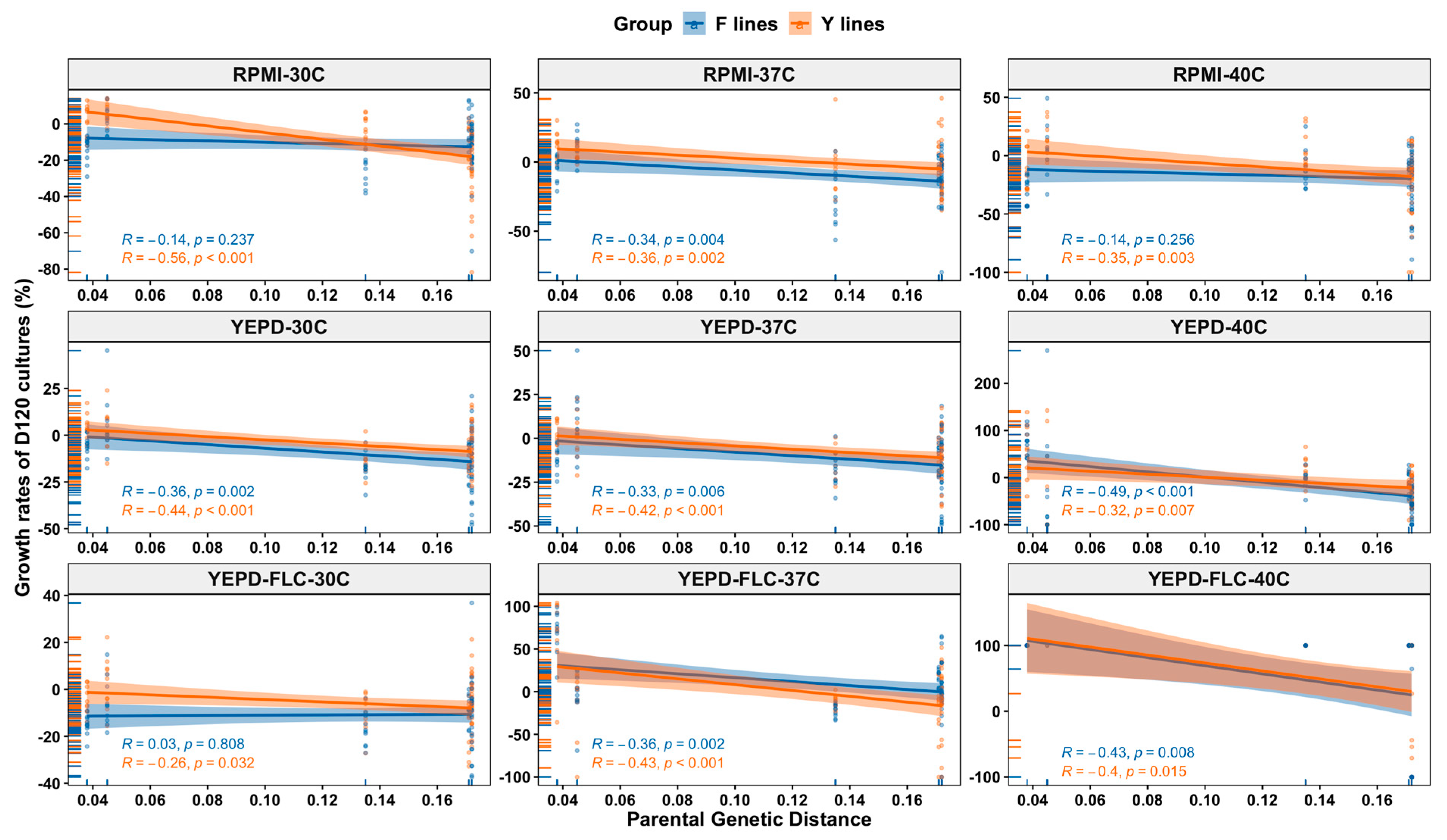
3.4. Growth Rates of D120 Cultures under Nine Environmental Conditions
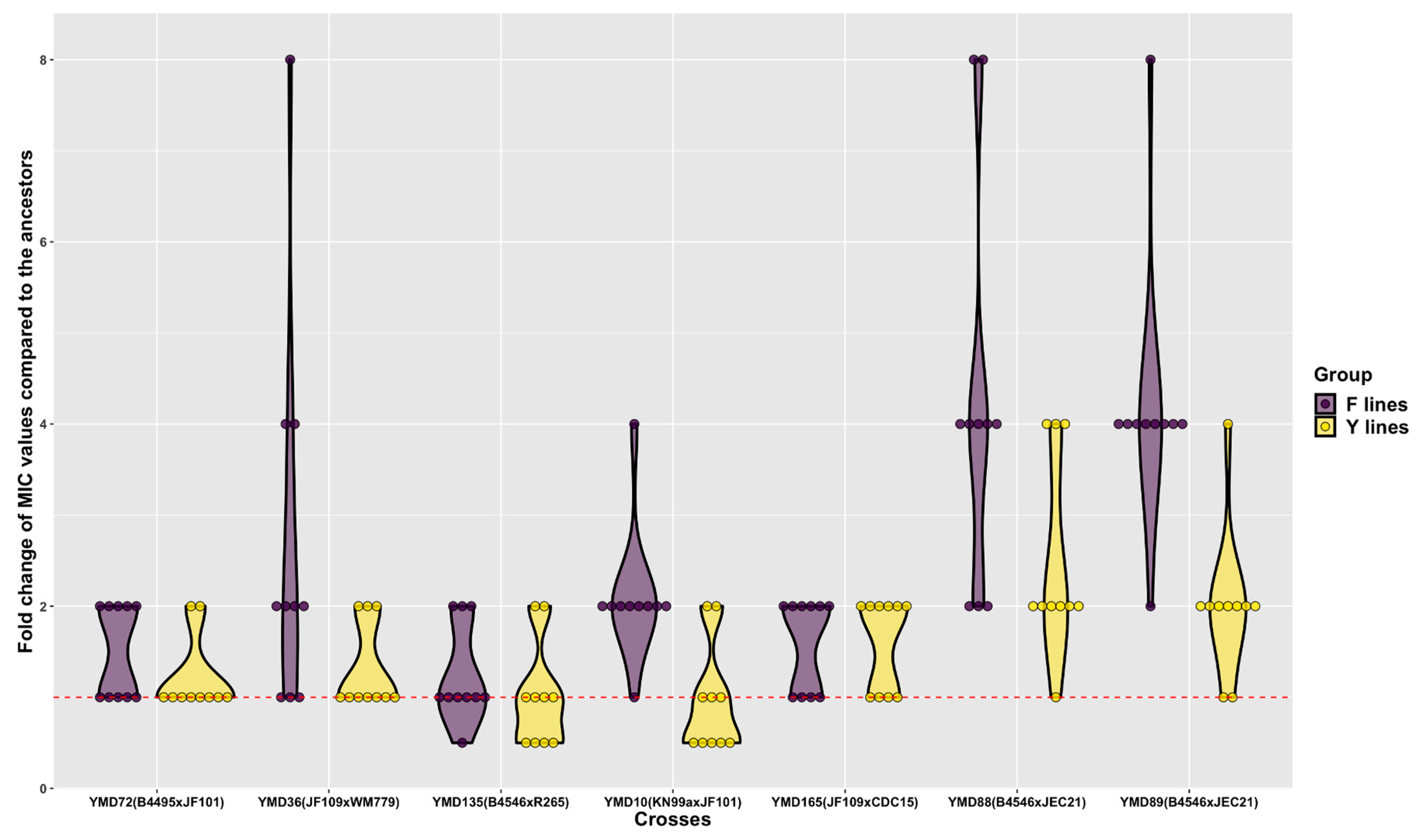
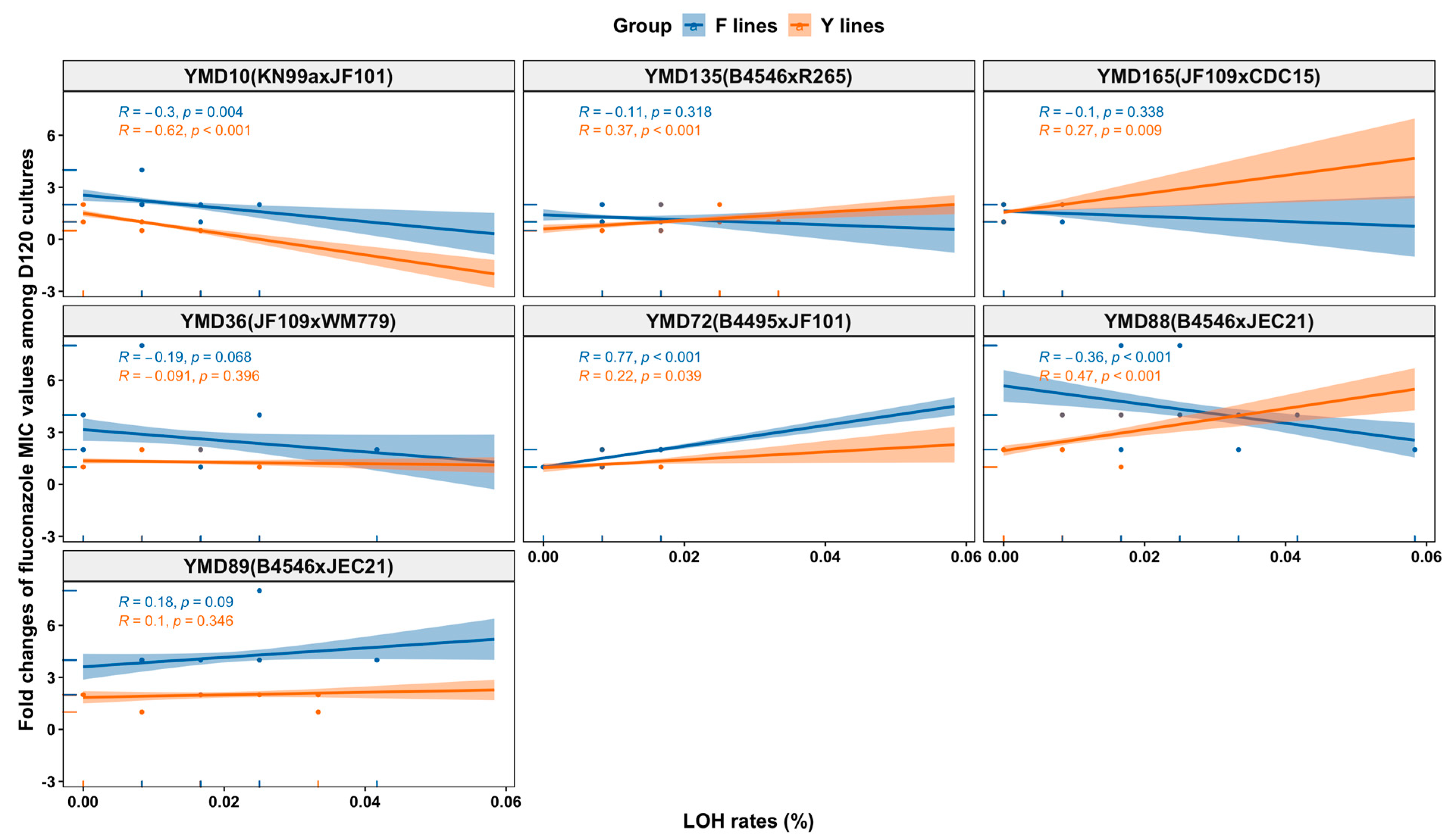
3.5. Fluconazole MIC Changes of D120 Cultures
3.6. Relationship between Parental Genetic Divergence and the Genetic and Phenotypic Changes among MA Lines
3.7. Impacts of Fluconazole Stress on Genetic and Phenotypic Changes among MA Lines
3.8. Relationships between the Observed Genetic Changes and Phenotypic Changes among MA Lines
4. Discussion
4.1. Contributions of Parental Divergence to Genetic Changes
4.2. Contributions of Fluconazole Stress to Genetic Changes
4.3. Interaction Effects of Factors on the Observed Changes among MA Lines
4.4. Relationships between Ploidy Changes and Phenotypic Changes
4.5. Relationships between LOH and Phenotypic Changes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pâques, F.; Haber, J. Multiple Pathways of Recombination Induced by Double-Strand Breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999, 63, 349–404. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, R.P.; Kumar, A.; Tyagi, M.B.; Sinha, R.P. Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair. J. Nucl. Acids 2010, 2010, 592980. [Google Scholar] [CrossRef] [PubMed]
- Garm, C.; Moreno-Villanueva, M.; Bürkle, A.; Larsen, L.A.; Bohr, V.A.; Christensen, K.; Stevnsner, T. Genetic and environmental influence on DNA strand break repair: A twin study. Environ. Mol. Mutagen. 2013, 54, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, A.; Kuzmin, A.; Otto, S.; Gerstein, A.; Kuzmin, A.; Otto, S. Loss-of-heterozygosity facilitates passage through Haldane’s sieve for Saccharomyces cerevisiae undergoing adaptation. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Dunkel, N.; Blass, J.; Rogers, P.D.; Morschhäuser, J. Mutations in the multi-drug resistance regulatorMRR1, followed by loss of heterozygosity, are the main cause ofMDR1overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 2008, 69, 827–840. [Google Scholar] [CrossRef]
- Hiraoka, M.; Watanabe, K.-I.; Umezu, K.; Maki, H. Spontaneous Loss of Heterozygosity in Diploid Saccharomyces cerevisiae Cells. Genetics 2000, 156, 1531–1548. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Wu, B.; Long, H.; Zhang, N.; Patterson, C.; Simpson, S.; Morris, K.; Thomas, W.K.; Lynch, M.; Hao, W. Variable Spontaneous Mutation and Loss of Heterozygosity among Heterozygous Genomes in Yeast. Mol. Biol. Evol. 2020, 37, 3118–3130. [Google Scholar] [CrossRef]
- Ciudad, T.; Hickman, M.; Bellido, A.; Berman, J.; Larriba, G. Phenotypic Consequences of a Spontaneous Loss of Heterozygosity in a Common Laboratory Strain of Candida albicans. Genetics 2016, 203, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Forche, A.; Abbey, D.; Pisithkul, T.; Weinzierl, M.A.; Ringstrom, T.; Bruck, D.; Petersen, K.; Berman, J. Stress Alters Rates and Types of Loss of Heterozygosity in Candida albicans. mBio 2011, 2. [Google Scholar] [CrossRef]
- Li, W.; Averette, A.F.; Desnos-Ollivier, M.; Ni, M.; Dromer, F.; Heitman, J. Genetic Diversity and Genomic Plasticity of Cryptococcus neoformans AD Hybrid Strains. G3 Genes Genomes Genetics 2012, 2, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Nielsen, K.; Patel, S.M.; Heitman, J. Impact of Mating Type, Serotype, and Ploidy on the Virulence of Cryptococcus neoformans. Infect. Immun. 2008, 76, 2923–2938. [Google Scholar] [CrossRef]
- Dong, K.; You, M.; Xu, J. Genetic Changes in Experimental Populations of a Hybrid in the Cryptococcus neoformans Species Complex. Pathogens 2019, 9, 3. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J.; Bennett, J.E. Distribution of α and α mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 1978, 108, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Lengeler, K.B.; Cox, G.M.; Heitman, J. Serotype AD Strains of Cryptococcus neoformans Are Diploid or Aneuploid and Are Heterozygous at the Mating-Type Locus. Infect. Immun. 2001, 69, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Bovers, M.; Hagen, F.; Kuramae, E.; Hoogveld, H.L.; Dromer, F.; St-Germain, G.; Boekhout, T. AIDS Patient Death Caused by Novel Cryptococcus neoformans × C.gattii Hybrid. Emerg. Infect. Dis. 2008, 14, 1105–1108. [Google Scholar] [CrossRef]
- Bovers, M.; Hagen, F.; Kuramae, E.; Diaz, M.R.; Spanjaard, L.; Dromer, F.; Hoogveld, H.L.; Boekhout, T. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006, 6, 599–607. [Google Scholar] [CrossRef]
- Lin, X.; Litvintseva, A.P.; Nielsen, K.; Patel, S.M.; Floyd, A.; Mitchell, T.G.; Heitman, J. αADα Hybrids of Cryptococcus neoformans: Evidence of Same-Sex Mating in Nature and Hybrid Fitness. PLoS Genet. 2007, 3, e186. [Google Scholar] [CrossRef]
- Ni, M.; Feretzaki, M.; Li, W.; Floyd-Averette, A.; Mieczkowski, P.; Dietrich, F.S.; Heitman, J. Unisexual and Heterosexual Meiotic Reproduction Generate Aneuploidy and Phenotypic Diversity De Novo in the Yeast Cryptococcus neoformans. PLoS Biol. 2013, 11, e1001653. [Google Scholar] [CrossRef]
- Samarasinghe, H.; Xu, J. Hybrids and hybridization in the Cryptococcus neoformans and Cryptococcus gattii species complexes. Infect. Genet. Evol. 2018, 66, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Cogliati, M.; Esposto, M.C.; Tortorano, A.M.; Viviani, M.A. Cryptococcus neoformans population includes hybrid strains homozygous at mating-type locus. FEMS Yeast Res. 2006, 6, 608–613. [Google Scholar] [CrossRef]
- Vogan, A.A.; Khankhet, J.; Xu, J.; Vogan, A.A.; Khankhet, J.; Xu, J. Evidence for Mitotic Recombination within the Basidia of a Hybrid Cross of Cryptococcus neoformans. PLoS ONE 2013, 8, e62790. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Xu, J. What Are the Best Parents for Hybrid Progeny? An Investigation into the Human Pathogenic Fungus Cryptococcus. J. Fungi 2021, 7, 299. [Google Scholar] [CrossRef]
- Ravichandran, M.C.; Fink, S.; Clarke, M.N.; Hofer, F.C.; Campbell, C.S. Genetic interactions between specific chromosome copy number alterations dictate complex aneuploidy patterns. Genes Dev. 2018, 32, 1485–1498. [Google Scholar] [CrossRef]
- Birchler, J.A. Aneuploidy in plants and flies: The origin of studies of genomic imbalance. Semin. Cell Dev. Biol. 2013, 24, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.; Stelkens, R. Aneuploidy in yeast: Segregation error or adaptation mechanism? Yeast 2019, 36, 525–539. [Google Scholar] [CrossRef]
- Todd, R.T.; Forche, A.; Selmecki, A. Ploidy Variation in Fungi: Polyploidy, Aneuploidy, and Genome Evolution. Microbiol. Spectr. 2017, 5, 599–618. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.J.; Forche, A.; Berman, J.; Bennett, R.J.; Forche, A.; Berman, J. Rapid Mechanisms for Generating Genome Diversity: Whole Ploidy Shifts, Aneuploidy, and Loss of Heterozygosity. Cold Spring Harb. Perspect. Med. 2014, 4, a019604. [Google Scholar] [CrossRef] [PubMed]
- Weil, T.; Santamaría, R.; Lee, W.; Rung, J.; Tocci, N.; Abbey, D.; Bezerra, A.; Carreto, L.; Moura, G.R.; Bayés, M.; et al. Adaptive Mistranslation Accelerates the Evolution of Fluconazole Resistance and Induces Major Genomic and Gene Expression Alterations in Candida albicans. mSphere 2017, 2. [Google Scholar] [CrossRef]
- Huang, M.; McClellan, M.; Berman, J.; Kao, K.C. Evolutionary Dynamics of Candida albicans during In Vitro Evolution. Eukaryot. Cell 2011, 10, 1413–1421. [Google Scholar] [CrossRef]
- Rodero, L.; Mellado, E.; Rodriguez, A.C.; Salve, A.; Guelfand, L.; Cahn, P.; Cuenca-Estrella, M.; Davel, G.; Rodriguez-Tudela, J.L. G484S Amino Acid Substitution in Lanosterol 14-α Demethylase (ERG11) Is Related to Fluconazole Resistance in a Recurrent Cryptococcus neoformans Clinical Isolate. Antimicrob. Agents Chemother. 2003, 47, 3653–3656. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Sanguinetti, M.; Sanglard, D.; La Sorda, M.; Boccia, S.; Romano, L.; Morace, G.; Fadda, G. Identification and characterization of a Cryptococcus neoformans ATP binding cassette (ABC) transporter-encoding gene, CnAFR1, involved in the resistance to fluconazole. Mol. Microbiol. 2003, 47, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Modrich, P.; Lahue, R. Mismatch Repair in Replication Fidelity, Genetic Recombination, and Cancer Biology. Annu. Rev. Biochem. 1996, 65, 101–133. [Google Scholar] [CrossRef]
- De Storme, N.; Mason, A. Plant speciation through chromosome instability and ploidy change: Cellular mechanisms, molecular factors and evolutionary relevance. Curr. Plant Biol. 2014, 1, 10–33. [Google Scholar] [CrossRef]
- Xu, J. Estimating the Spontaneous Mutation Rate of Loss of Sex in the Human Pathogenic Fungus Cryptococcus neoformans. Genetics 2002, 162, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Sia, R.A.; Lengeler, K.B.; Heitman, J.; Sia, R.A.; Lengeler, K.B.; Heitman, J. Diploid Strains of the Pathogenic Basidiomycete Cryptococcus neoformans Are Thermally Dimorphic. Fungal Genet. Biol. 2000, 29, 153–163. [Google Scholar] [CrossRef]
- Clinical Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard, 3rd ed.; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Vogan, A.A.; Khankhet, J.; Samarasinghe, H.; Xu, J. Identification of QTLs Associated with Virulence Related Traits and Drug Resistance in Cryptococcus neoformans. G3 Genes Genomes Genetics 2016, 6, 2745–2759. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.; Grünwald, N. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Farrer, R.A.; Chang, M.; Davis, M.J.; van Dorp, L.; Yang, D.-H.; Shea, T.; Sewell, T.R.; Meyer, W.; Balloux, F.; Edwards, H.M.; et al. A New Lineage of Cryptococcus gattii (VGV) Discovered in the Central Zambezian Miombo Woodlands. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Hagen, F.; Khayhan, K.; Theelen, B.; Kolecka, A.; Polacheck, I.; Sionov, E.; Falk, R.; Parnmen, S.; Lumbsch, T.; Boekhout, T. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 2015, 78, 16–48. [Google Scholar] [CrossRef]
- Kidd, S.E.; Guo, H.; Bartlett, K.H.; Xu, J.; Kronstad, J. Comparative Gene Genealogies Indicate that Two Clonal Lineages of Cryptococcus gattii in British Columbia Resemble Strains from Other Geographical Areas. Eukaryot. Cell 2005, 4, 1629–1638. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Vilgalys, R.; Mitchell, T.G. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 2000, 9, 1471–1481. [Google Scholar] [CrossRef] [PubMed]
- Ngamskulrungroj, P.; Gilgado, F.; Faganello, J.; Litvintseva, A.P.; Leal, A.L.; Tsui, C.; Mitchell, T.G.; Vainstein, M.; Meyer, W. Genetic Diversity of the Cryptococcus Species Complex Suggests that Cryptococcus gattii Deserves to Have Varieties. PLoS ONE 2009, 4, e5862. [Google Scholar] [CrossRef]
- Casadevall, A.; Freij, J.B.; Hann-Soden, C.; Taylor, J.; Casadevall, A.; Freij, J.B.; Hann-Soden, C.; Taylor, J. Continental Drift and Speciation of the Cryptococcus neoformans and Cryptococcus gattii Species Complexes. mSphere 2017, 2, e00103-17. [Google Scholar] [CrossRef]
- Sharpton, T.J.; Neafsey, D.E.; Galagan, J.E.; Taylor, J.W. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol. 2008, 9, R24. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Liu, I.; Sham, A.; E Stajich, J.; Dietrich, F.S.; Kronstad, J.W. Comparative hybridization reveals extensive genome variation in the AIDS-associated pathogen Cryptococcus neoformans. Genome Biol. 2008, 9, R41. [Google Scholar] [CrossRef]
- Vogan, A.A.; Xu, J. Evidence for genetic incompatibilities associated with post-zygotic reproductive isolation in the human fungal pathogen Cryptococcus neoformans. Genome 2014, 57, 335–344. [Google Scholar] [CrossRef]
- Sun, S.; Xu, J. Chromosomal Rearrangements between Serotype A and D Strains in Cryptococcus neoformans. PLoS ONE 2009, 4, e5524. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, C.A.; Kronstad, J.W.; Taylor, G.; Warren, R.; Yuen, M.; Hu, G.; Jung, W.H.; Sham, A.; Kidd, S.E.; Tangen, K.; et al. Genome Variation in Cryptococcus gattii, an Emerging Pathogen of Immunocompetent Hosts. mBio 2011, 2. [Google Scholar] [CrossRef]
- Fujitani, Y.; Kobayashi, I. Effect of DNA Sequence Divergence on Homologous Recombination as Analyzed by a Random-Walk Model. Genetics 1999, 153, 1973–1988. [Google Scholar] [CrossRef] [PubMed]
- Opperman, R.; Emmanuel, E.; Levy, A.A. The Effect of Sequence Divergence on Recombination Between Direct Repeats in Arabidopsis. Genetics 2004, 168, 2207–2215. [Google Scholar] [CrossRef] [PubMed]
- Altamirano, S.; Fang, D.; Simmons, C.; Sridhar, S.; Wu, P.; Sanyal, K.; Kozubowski, L. Fluconazole-Induced Ploidy Change in Cryptococcus neoformans Results from the Uncoupling of Cell Growth and Nuclear Division. mSphere 2017, 2, e00205-17. [Google Scholar] [CrossRef]
- Stone, N.R.; Rhodes, J.; Fisher, M.C.; Mfinanga, S.; Kivuyo, S.; Rugemalila, J.; Segal, E.S.; Needleman, L.; Molloy, S.F.; Kwon-Chung, J.; et al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Investig. 2019, 129, 999–1014. [Google Scholar] [CrossRef]
- Harrison, B.D.; Hashemi, J.; Bibi, M.; Pulver, R.; Bavli, D.; Nahmias, Y.; Wellington, M.; Sapiro, G.; Berman, J. A Tetraploid Intermediate Precedes Aneuploid Formation in Yeasts Exposed to Fluconazole. PLoS Biol. 2014, 12, e1001815. [Google Scholar] [CrossRef]
- Pavelka, N.; Rancati, G.; Zhu, J.; Bradford, W.D.; Saraf, A.; Florens, L.; Sanderson, B.W.; Hattem, G.; Li, R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nat. Cell Biol. 2010, 468, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Sionov, E.; Lee, H.; Chang, Y.C.; Kwon-Chung, K.J. Cryptococcus neoformans Overcomes Stress of Azole Drugs by Formation of Disomy in Specific Multiple Chromosomes. PLoS Pathog. 2010, 6, e1000848. [Google Scholar] [CrossRef] [PubMed]
- Semighini, C.P.; Averette, A.F.; Perfect, J.R.; Heitman, J.; Semighini, C.P.; Averette, A.F.; Perfect, J.R.; Heitman, J. Deletion of Cryptococcus neoformans AIF Ortholog Promotes Chromosome Aneuploidy and Fluconazole-Resistance in a Metacaspase-Independent Manner. PLoS Pathog. 2011, 7, e1002364. [Google Scholar] [CrossRef]
- Yang, F.; Kravets, A.; Bethlendy, G.; Welle, S.; Rustchenko, E. Chromosome 5 Monosomy of Candida albicans Controls Susceptibility to Various Toxic Agents, Including Major Antifungals. Antimicrob. Agents Chemother. 2013, 57, 5026–5036. [Google Scholar] [CrossRef] [PubMed]
- Selmecki, A.; Forche, A.; Berman, J. Aneuploidy and Isochromosome Formation in Drug-Resistant Candida albicans. Science 2006, 313, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Ngamskulrungroj, P.; Chang, Y.; Hansen, B.; Bugge, C.; Fischer, E.; Kwon-Chung, K.J. Characterization of the Chromosome 4 Genes That Affect Fluconazole-Induced Disomy Formation in Cryptococcus neoformans. PLoS ONE 2012, 7, e33022. [Google Scholar] [CrossRef] [PubMed]
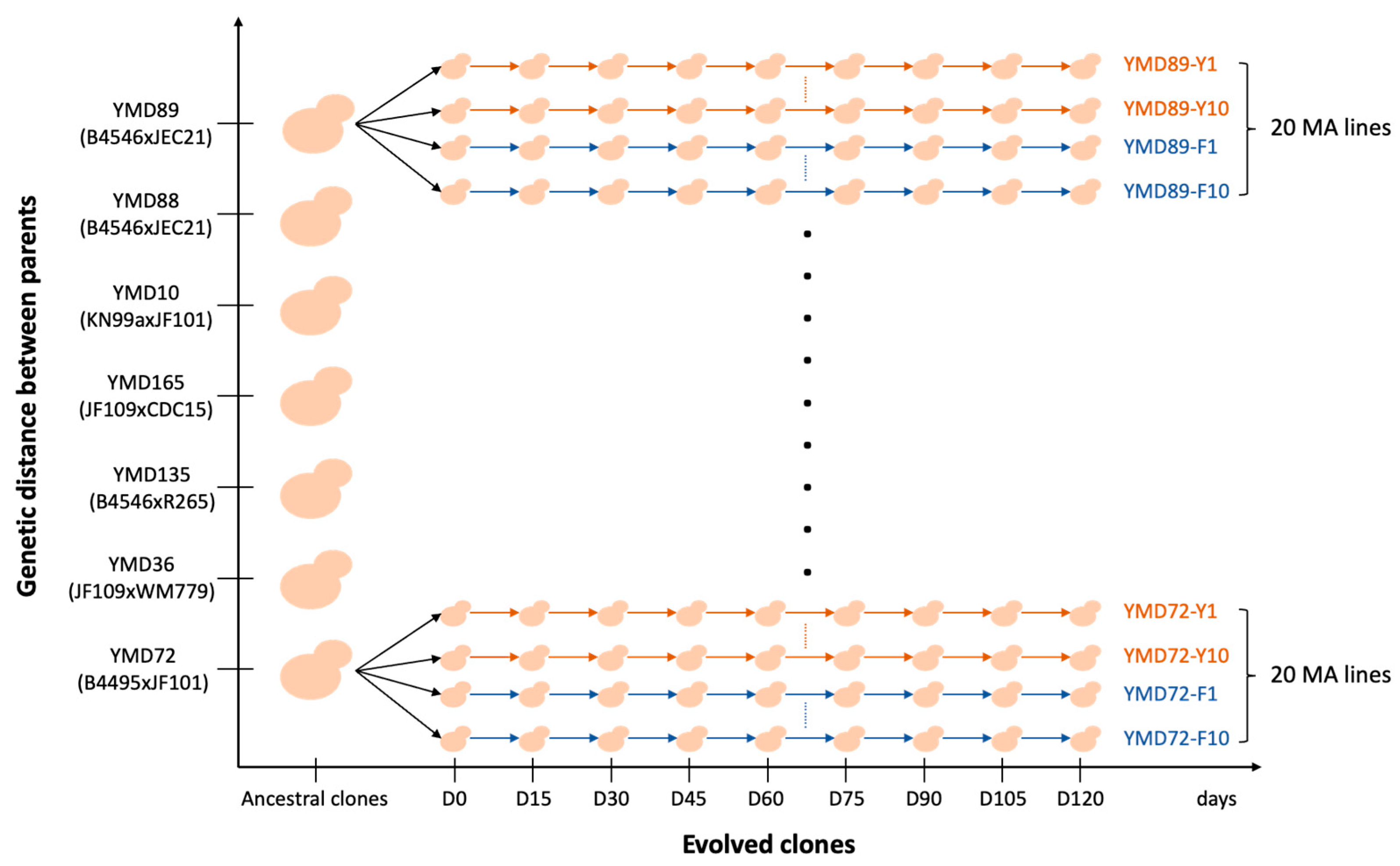


| Crosses | Ancestral Clones | MATa Parent (Lineage) | MATα Parent (Lineage) | Parental Haploid Genome Divergence | |
|---|---|---|---|---|---|
| Inter-lineage crosses within CGSC | VGI × VGIII | YMD72 | B4495 (VGI) | JF101 (VGIII) | 0.038 |
| VGIV × VGIII | YMD36 | JF109 (VGIII) | WM779 (VGIV) | 0.045 | |
| VGII × VGIII | YMD135 | B4546 (VGIII) | R265 (VGII) | 0.135 | |
| Inter-lineage crosses between CNSC and CGSC | VNI × VGIII | YMD165 | JF109 (VGIII) | CDC15 (VNI) | 0.171 |
| YMD10 | KN99a (VNI) | JF101 (VGIII) | 0.171 | ||
| VNIV × VGIII | YMD88 | B4546 (VGIII) | JEC21 (VNIV) | 0.172 | |
| YMD89 | B4546 (VGIII) | JEC21 (VNIV) | 0.172 | ||
| Ancestral Clones | Number of MA Lines | Number of Multi-Locus Genotypes (MLGs) | Shannon-Weiner Diversity Index (H) |
|---|---|---|---|
| YMD72 | 20 | 9 | 2.06 |
| YMD36 | 20 | 14 | 2.39 |
| YMD135 | 20 | 11 | 2.00 |
| YMD165 | 20 | 2 | 0.42 |
| YMD10 | 20 | 14 | 2.53 |
| YMD88 | 20 | 16 | 2.69 |
| YMD89 | 20 | 17 | 2.76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, M.; Lin, Y.M.; Dobrin, A.; Xu, J. Genetic and Phenotypic Diversities in Experimental Populations of Diploid Inter-Lineage Hybrids in the Human Pathogenic Cryptococcus. Microorganisms 2021, 9, 1579. https://doi.org/10.3390/microorganisms9081579
You M, Lin YM, Dobrin A, Xu J. Genetic and Phenotypic Diversities in Experimental Populations of Diploid Inter-Lineage Hybrids in the Human Pathogenic Cryptococcus. Microorganisms. 2021; 9(8):1579. https://doi.org/10.3390/microorganisms9081579
Chicago/Turabian StyleYou, Man, Yuxin Monica Lin, Annamaria Dobrin, and Jianping Xu. 2021. "Genetic and Phenotypic Diversities in Experimental Populations of Diploid Inter-Lineage Hybrids in the Human Pathogenic Cryptococcus" Microorganisms 9, no. 8: 1579. https://doi.org/10.3390/microorganisms9081579
APA StyleYou, M., Lin, Y. M., Dobrin, A., & Xu, J. (2021). Genetic and Phenotypic Diversities in Experimental Populations of Diploid Inter-Lineage Hybrids in the Human Pathogenic Cryptococcus. Microorganisms, 9(8), 1579. https://doi.org/10.3390/microorganisms9081579







