The Nexus between Fire and Soil Bacterial Diversity in the African Miombo Woodlands of Niassa Special Reserve, Mozambique
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Sampling
2.3. DNA Extraction, Amplification, and Sequencing of 16S rRNA Genes
2.4. Assembly of Reads and Taxonomical Assignment
2.5. Diversity Analysis
2.6. Functional Prediction
2.7. Isolation and Characterization of Root Nodule Bacteria Using a Trap Host Legume
2.8. Isolation of Rhizosphere Bacteria and Characterization of In Vitro Plant Growth Promoting Activities
3. Results and Discussion
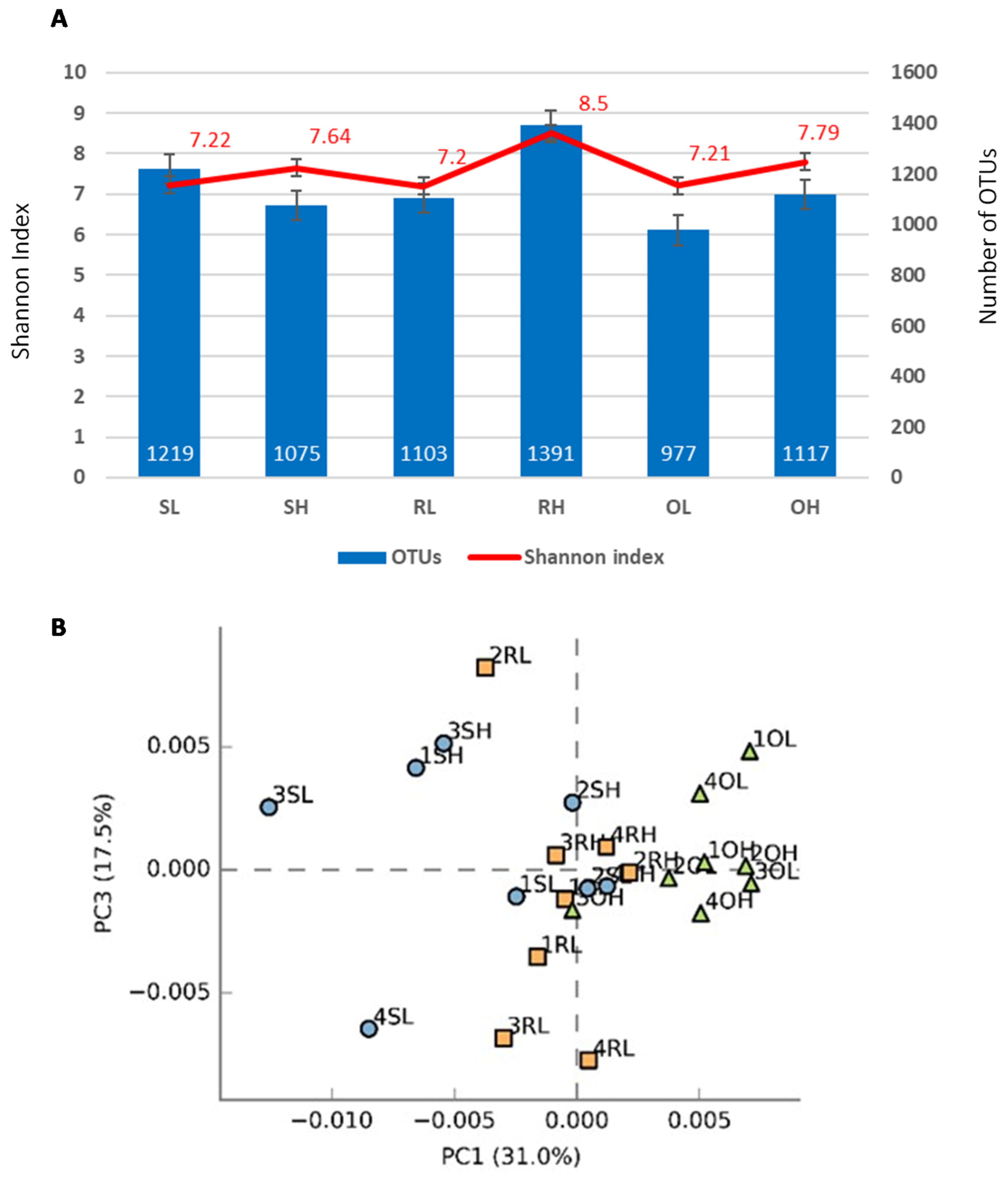
3.1. Sequencing and Distribution of OTUs
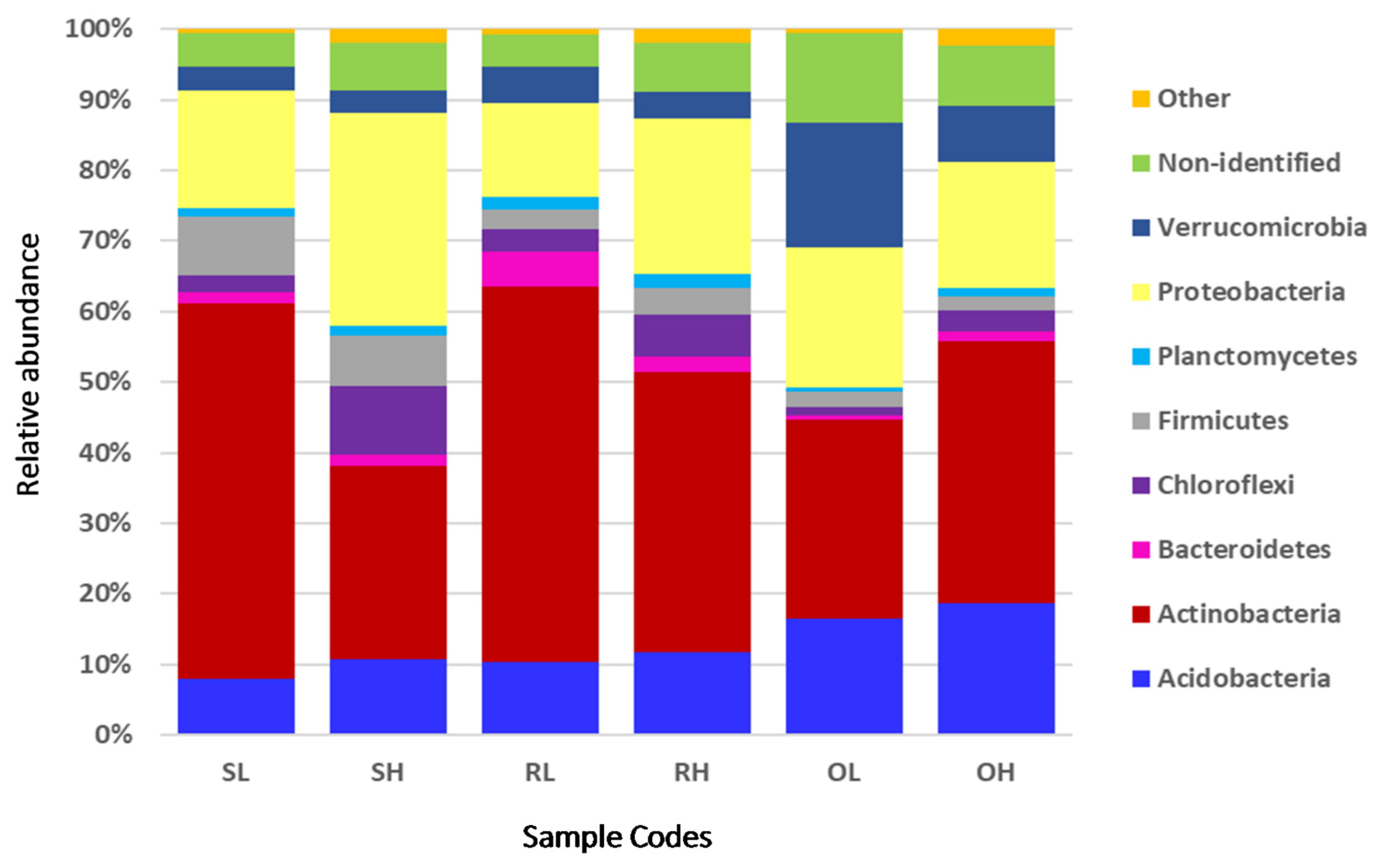
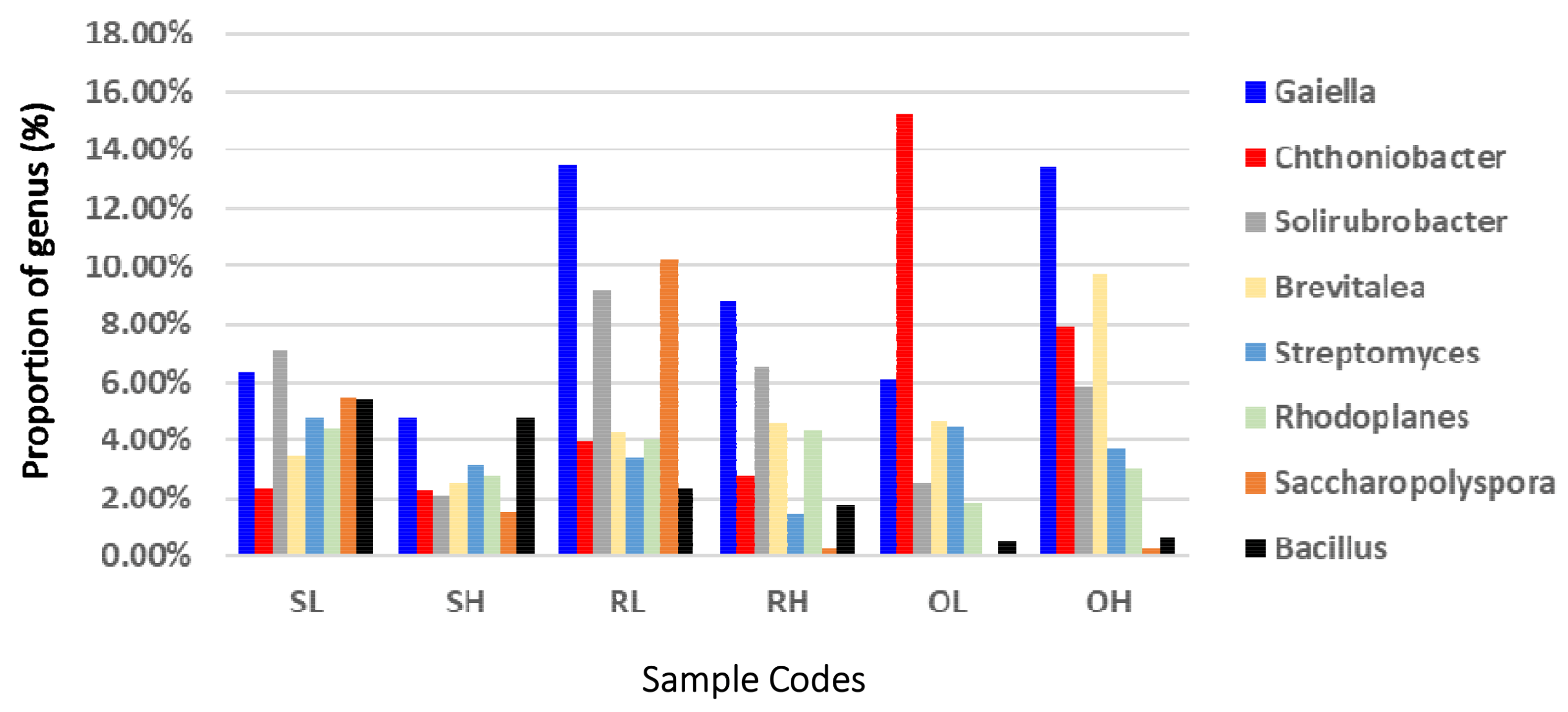
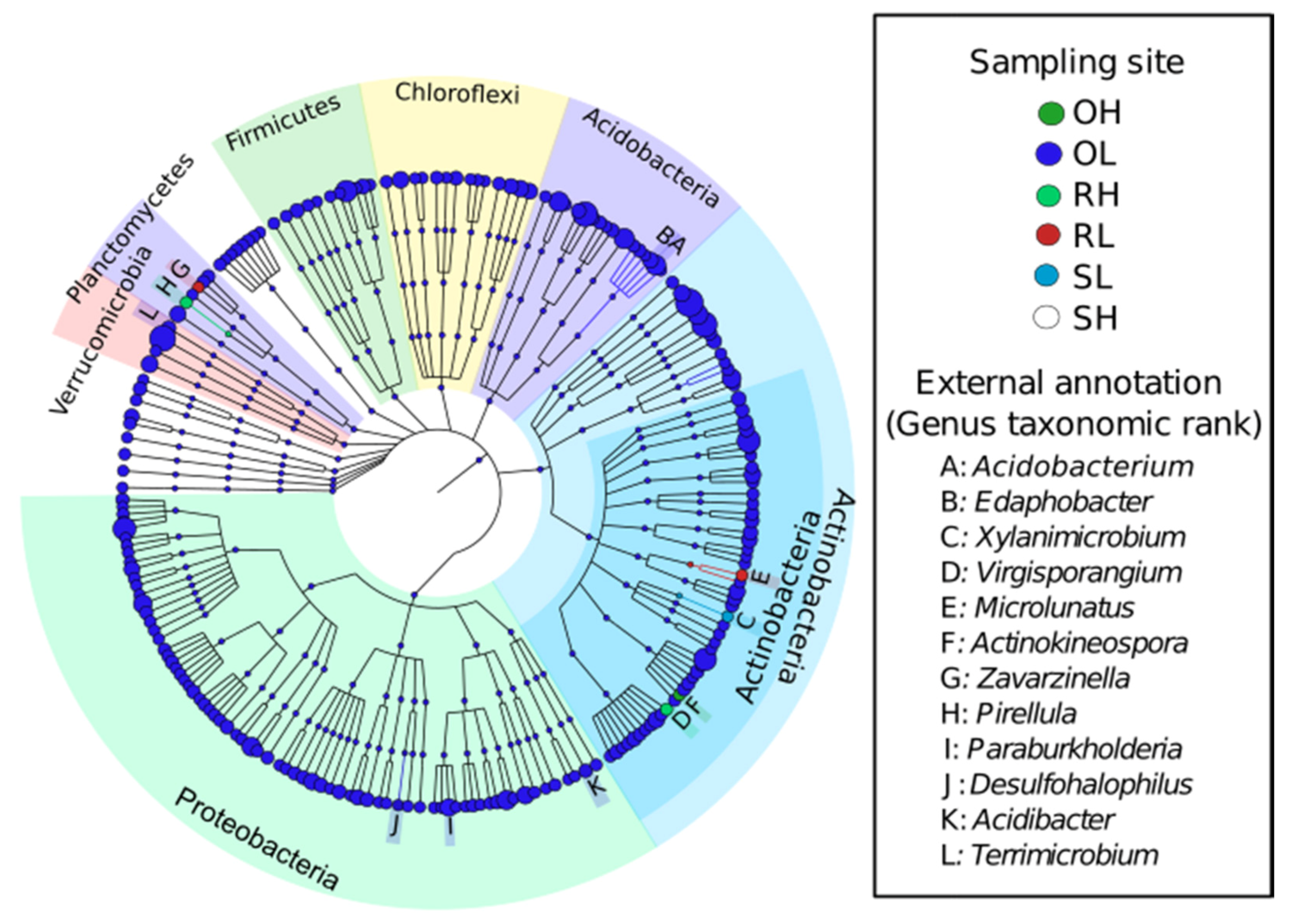
3.2. Taxonomic Composition of the Microbial Communities
3.3. Vigna unguiculata as a Trap for Rhizobia Bacteria
3.4. Characterization of In Vitro Plant Growth Promoting Activities and Taxonomic Analysis of Bacteria Isolated from Soils
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardenas, E.; Kranabetter, J.M.; Hope, G.; Maas, K.R.; Hallam, S.; Mohn, W.W. Forest harvesting reduces the soil metagenomic potential for biomass decomposition. ISME J. 2015, 9, 2465–2476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyle, J.R.; Powers, R.F. Forest soils. In Reference Module in Earth Systems and Environmental Sciences; Elias, S.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 1–11. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neary, D.G.; Klopatek, C.C.; DeBano, L.F.; Ffolliott, P.F. Fire effects on belowground sustainability: A review and synthesis. For. Ecol. Manag. 1999, 122, 51–71. [Google Scholar] [CrossRef]
- Kaur, R.; Rajesh, C.; Sharma, R.; Boparai, J.; Sharma, P. metagenomic investigation of bacterial diversity of hot spring soil from ecological genetics and genomics metagenomic investigation of bacterial diversity of hot spring soil from Manikaran, Himachal Pradesh, India. Ecol. Genet. Genom. 2018, 6, 16–21. [Google Scholar] [CrossRef]
- Doran, J.W.; Zeiss, M.R. Soil Health and Sustainability: Managing the biotic component of soil quality. Appl. Soil Ecol. 2000, 15, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Waldrop, M.P.; Balser, T.C.; Firestone, M.K. Linking microbial community composition to function in a tropical soil. Soil Biol. Biochem. 2000, 32, 1837–1846. [Google Scholar] [CrossRef]
- Yao, H.; He, Z.; Wilson, M.J.; Campbell, C.D. Microbial biomass and community structure in a sequence of soils with increasing fertility and changing land use. Microb. Ecol. 2000, 40, 223–237. [Google Scholar] [CrossRef]
- Maquia, I.S.; Fareleira, P.; Castro, I.V.E.; Brito, D.R.A.; Soares, R.; Chaúque, A.; Ferreira-Pinto, M.M.; Lumini, E.; Berruti, A.; Ribeiro, N.S.; et al. Mining the microbiome of key species from african savanna woodlands: Potential for soil health improvement and plant growth promotion. Microorganisms 2020, 8, 1291. [Google Scholar] [CrossRef]
- Macrae, A.; Coelho, R.R.R.; Peixoto, R.; Rosado, A.S. Tropical soil microbial communities. In The Prokaryotes: Prokaryotic Communities and Ecophysiology; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 85–95. [Google Scholar] [CrossRef]
- Mittermeier, R.A.; Mittermeier, C.G.; Brooks, T.M.; Pilgrim, J.D.; Konstant, W.R.; da Fonseca, G.A.B.; Kormos, C. Wilderness and biodiversity conservation. Proc. Nat. Acad. Sci. USA 2003, 100, 10309–10313. [Google Scholar] [CrossRef] [Green Version]
- Mittermeier, C.G.; Gil, P.R.; Mittermeier, R.; Fonseca, G.; Pilgrim, J. Wilderness: Earth’s Last Wild Places, 1st ed.; Conservation International: Washington, DC, USA, 2002; p. 576. [Google Scholar]
- Dziba, L.; Ramoelo, A.; Ryan, C.; Harrison, S.; Pritchard, R.; Tripathi, H.; Sitas, N.; Selomane, O.; Engelbrecht, F.; Pereira, L.; et al. Scenarios for Just and Sustainable Futures in the Miombo Woodlands. In Miombo Woodlands in a Changing Environment: Securing the Resilience and Sustainability of People and Woodlands; Ribeiro, N.S., Katerere, Y., Chirwa, P.W., Grundy, I.M., Eds.; Springer: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Frost, P. The Ecology of Miombo Woodlands. In The Miombo in Transition: Woodlands and Welfare in Africa; Campbell, B.M., Ed.; Center for International Forestry Research (CIFOR): Bogor, Indonesia, 1996; pp. 11–57. [Google Scholar]
- Ribeiro, N.S.; Katerere, Y.; Chirwa, P.W.; Grundy, I.M. (Eds.) Miombo Woodlands in a Changing Environment: Securing the Resilience and Sustainability of People and Woodlands; Springer: Cham, Switzerland, 2020; pp. 1–245. [Google Scholar] [CrossRef]
- IPBES (Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services). Summary for Policymakers of the Regional Assessment Report on Biodiversity and Ecosystem Services for Africa of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services; IPBES Secretariat: Bonn, Germany, 2018; p. 49. Available online: https://ipbes.net/assessment-reports/africa (accessed on 12 July 2021).
- IPBES. Global Assessment Report on Biodiversity and Ecosystem Service; IPBES Secretariat: Bonn, Germany, 2019; p. 1148. [Google Scholar] [CrossRef]
- Gonçalves, F.M.P.; Revermann, R.; Gomes, A.L.; Aidar, M.P.M.; Finckh, M.; Juergens, N. Tree species diversity and composition of miombo woodlands in South-Central Angola: A chronosequence of forest recovery after shifting cultivation. Int. J. For. Res. 2017, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Dewees, P.A.; Campbell, B.M.; Katerere, Y.; Sitoe, A.; Cunningham, A.B.; Angelsen, A.; Wunder, S. Managing the miombo woodlands of Southern Africa: Policies, incentives and options for the rural poor. J. Nat. Resour. Policy Res. 2010, 2, 57–73. [Google Scholar] [CrossRef] [Green Version]
- Malmer, A.; van Noordwijk, M.; Brujinzeel, L.A. Effects of shifting cultivation and forest fire. In Forest Water People in the Humid Tropics: Past Present and Future Hydrological Research for Integrated Land and Water Management; Bonell, M., Brujinzeel, L.A., Eds.; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Bruijnzeel, L.A. Hydrology of moist tropical forests and effects of conversion: A state of knowledge review. J. Hydrol. 1991, 129, 397–399. [Google Scholar]
- Higgins, S.I.; Bond, W.; February, E.C.; Bronn, A.; Euston-Brown, D.I.W.; Enslin, B.; Govender, N.; Rademan, L.; O’Regan, S.; Potgieter, A.L.F.; et al. Effects of four decades of fire manipulation on woody vegetation structure in savanna. Ecology 2007, 88, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Higgins, S.I.; Bond, W.J.; Trollope, W.S.W. Fire, resprouting and variability: A recipe for grass-tree coexistence in savanna. J. Ecol. 2000, 88, 213–229. [Google Scholar] [CrossRef]
- Ribeiro, N.S.; Cangela, A.; Chauque, A.; Bandeira, R.R.; Ribeiro-Barros, A.I. Characterisation of spatial and temporal distribution of the fire regime in Niassa National Reserve, northern Mozambique. Int. J. Wildland Fire 2017, 26, 1021–1029. [Google Scholar] [CrossRef]
- Saket, M. Tendencies of Forest Fires in Mozambique. Annex 4 to Proposal of a Model of Integrated Forest Management Plan for the Timber Concession of Maciambose Cheringoma, North of Sofala; República de Moçambique, Ministério da Agricultura e Pesca, Direcção Nacional de Florestas e Fauna Bravia ADB/ETC UK LTD: Beira, Mozambique, 1999.
- Chidumayo, E.N. Species structure in zambian miombo woodland. J. Trop. Ecol. 1987, 3, 109–118. [Google Scholar] [CrossRef]
- Mataix-Solera, J.; Guerrero, C.; García-Orenes, F.; Bárcenas, G.M.; Torres, M.P. Forest fire effects on soil microbiology. In Fire Effects on Soils and Restoration Strategies; Cerda, A., Robichaud, P., Eds.; Science Publisher: Oxford, UK, 2009; pp. 133–175. [Google Scholar]
- Certini, G. Effects of fire on properties of forest soils: A review. Oecologia 2005, 143, 1–10. [Google Scholar] [CrossRef]
- Frost, P.G.H.; Robertson, F. The Ecological Effects of Fire in Savannas. In Determinants of Tropical Savannas; Walker, B.H., Ed.; IRL Press: Oxford, UK; Harare, Zimbabwe, 1985; pp. 93–140. [Google Scholar]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Somers, E.; Vanderleyden, J.; Srinivasan, M. Rhizosphere bacterial signalling: A love parade beneath our feet. Crit. Rev. Microbiol. 2004, 30, 205–240. [Google Scholar] [CrossRef]
- Ganzin, N.; Poilecot, P.; Prin, T. Vegetation Survey of Niassa National Reserve Oriented for Vegetation Mapping and Range Resources Assessment Using Satellite Imagery. 2010. Available online: https://www.biofund.org.mz/wp-content/uploads/2019/01/1548766114-F0821.Mission_report_Niassa_Nganzin_Ppoilecot_june2010_final.pdf (accessed on 12 July 2021).
- Ribeiro, N.S.; Matos, C.N.; Moura, I.R.; Washington-Allen, R.A.; Ribeiro, A.I. Monitoring vegetation dynamics and carbon stock density in miombo woodlands. Carbon Balance Manag. 2013, 8, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, N.S.; Shugart, H.H.; Washington-Allen, R. The effects of fire and elephants on species composition and structure of the Niassa Reserve, northern Mozambique. For. Ecol. Manag. 2008, 255, 1626–1636. [Google Scholar] [CrossRef]
- Craig, G.C. Aerial Survey of Wildlife in the Niassa Reserve and Adjacent Areas; Technical Report; Sociedade de Gestão e Desenvolvimento da Reserva do Niassa (SGDRN): Maputo, Mozambique, 2009. [Google Scholar]
- Riley, D.; Barber, S.A. Salt accumulation at the soybean (Glycine max. (L.) Merr.) root-soil interface. Soil. Sci. Soc. Am. J. 1970, 34, 154–155. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- López-García, A.; Pineda-Quiroga, C.; Atxaerandio, R.; Pérez, A.; Hernández, I.; García-Rodríguez, A.; González-Recio, O. Comparison of Mothur and QIIME for the analysis of rumen microbiota composition based on 16S rRNA amplicon sequences. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. uchime improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.A.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [Green Version]
- Statistica (Data Analysis Software System); Release 13; StataCorp LP: College Station, TX, USA, 2013.
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Asnicar, F.; Weingart, G.; Tickle, T.L.; Huttenhower, C.; Segata, N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 2015, 3, e1029. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Svetlana, Y.N.; Brown, J.R.; Taylor, C.M.; Hutternhower, C.; Langille, M.G.I. PICRUSt2: An improved and customizable approach for metagenome inference. BioRxiv Prepr. 2020. [Google Scholar] [CrossRef] [Green Version]
- Louca, S.; Doebeli, M. Efficient comparative phylogenetics on large trees. Bioinformatics 2018, 34, 1053–1055. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Doak, T.G. A Parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput. Biol. 2009, 5, e1000465. [Google Scholar] [CrossRef] [Green Version]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2019, 48, D445–D453. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical Analysis of Taxonomic and Functional Profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bluman, A. Elementary Statistics: A Step by Step Approach; McGraw-Hill Companies: New York, NY, USA, 2007. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Gomes, A.M.F.; Draper, D.; Talhinhas, P.; Santos, P.B.; Simões, F.; Nhantumbo, N.; Massinga, R.; Ramalho, J.C.; Marques, I.; Ribeiro-Barros, A.I. Genetic diversity among cowpea (Vigna unguiculata (L.) Walp.) landraces suggests central Mozambique as an important hotspot of variation. Agronomy 2020, 10, 1893. [Google Scholar] [CrossRef]
- Gomes, A.M.F.; Draper, D.; Nhantumbo, N.; Massinga, R.; Ramalho, J.C.; Marques, I.; Ribeiro-Barros, A.I. Diversity of cowpea Vigna unguiculata (L.) Walp. landraces in Mozambique: New opportunities for crop improvement and future breeding programs. Agronomy 2021, 11, 991. [Google Scholar] [CrossRef]
- Jensen, H.L. Nitrogen fixation in leguminous plants. I. General characters of root-nodule bacteria isolated from species of Medicago and Trifolium in Australia. Proc. Linn. Soc. NSW 1942, 66, 98–108. Available online: http://biostor.org/reference/68107 (accessed on 12 July 2021).
- Ferreira, E.M.; Marques, J.F. Selection of portuguese Rhizobium leguminosarum bv. trifolii strains for production of legume inoculants. Plant Soil 1992, 147, 151–158. [Google Scholar] [CrossRef]
- Vincent, J.M. A Manual for the Practical Study of Root-Nodule Bacteria, I.B.P. Handbook; Blackwell Scientific Publisher: Oxford, UK, 1970; Volume 8, pp. 8–9. [Google Scholar]
- Beringer, J.E. R Factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burk, D.; Lineweaver, H. The influence of fixed nitrogen on azotobacter. J. Bacteriol. 1930, 19, 389–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castanheira, N.; Dourado, A.C.; Alves, P.I.; Cortés-Pallero, A.M.; Delgado-Rodríguez, A.I.; Prazeres, A.; Borges, N.; Sánchez, C.; Crespo, M.T.B.; Fareleira, P. Annual ryegrass-associated bacteria with potential for plant growth promotion. Microbiol. Res. 2014, 169, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Asghar, H.N.; Zahir, Z.A.; Arshad, M.; Khaliq, A. Relationship between in vitro production of auxins by rhizobacteria and their growth-promoting activities in Brassica juncea L. Biol. Fertil. Soils 2002, 35, 231–237. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.S.; Fernández, F.J. O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 2007, 70, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Peix, A.; Rivas-Boyero, A.A.; Mateos, P.F.; Rodriguez-Barrueco, C.; Martínez-Molina, E.; Velazquez, E. Growth promotion of chickpea and barley by a phosphate solubilizing strain of mesorhizobium mediterraneum under growth chamber conditions. Soil. Biol. Biochem. 2001, 33, 103–110. [Google Scholar] [CrossRef]
- Verma, S.C.; Ladha, J.K.; Tripathi, A.K. Evaluation of plant growth promoting and colonization ability of endophytic diazotrophs from deep water rice. J. Biotechnol. 2001, 91, 127–141. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T.; Claeyssens, M.; Montagu, M.V. Cloning, expression in Escherichia coli, and characterization of cellulolytic enzymes of Azoarcus sp., a root-invading diazotroph. J. Bacteriol. Res. 1993, 175, 7056–7065. [Google Scholar] [CrossRef] [Green Version]
- Mateos, P.F.; Jimenez-Zurdo, J.I.; Chen, J.; Squartini, A.S.; Haack, S.K.; Martinez- Molina, E.; Hubbell, D.H.; Dazzo, F.B. Cell-associated pectinolytic and cellulolytic enzymes in Rhizobium leguminosarum biovar trifolii. Appl. Environ. Microbiol. 1992, 58, 1816–1822. [Google Scholar] [CrossRef] [Green Version]
- Villadas, P.J.; Díaz-Díaz, S.; Rodríguez-Rodríguez, A.; del Arco-Aguilar, M.; Fernández-González, A.J.; Pérez-Yépez, J.; Arbelo, C.; González-Mancebo, J.M.; Fernández-López, M.; León-Barrios, M. The soil microbiome of the laurel forest in Garajonay National Park (La Gomera, Canary Islands): Comparing unburned and burned habitats after a wildfire. Forests 2019, 10, 1051. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.; González-Pérez, J.A.; Turmero, A.; Hernández, M.; Ball, A.S.; González-Vila, F.J.; Arias, M.E. Physico-chemical and microbial perturbations of andalusian pine forest soils following a wildfire. Sci. Total Environ. 2018, 634, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Hamman, S.T.; Burke, I.C.; Stromberger, M.E. Relationships between microbial community structure and soil environmental conditions in a recently burned system. Soil. Biol. Biochem. 2007, 39, 1703–1711. [Google Scholar] [CrossRef]
- Frost, P.G.H. The responses and survival of organisms in fire-prone environments. In Ecological Effects of Fire in South African Ecosystems; Booysen, P.V., Tainton, N.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1984; pp. 273–309. [Google Scholar]
- Maquia, I.; Ribeiro, N.S.; Silva, V.; Bessa, F.; Goulao, L.F.; Ribeiro, A.I. Genetic diversity of Brachystegia boehmii Taub. and Burkea africana Hook. f. across a fire gradient in Niassa National Reserve, northern Mozambique. Biochem. Syst. Ecol. 2013, 48, 238–247. [Google Scholar] [CrossRef] [Green Version]
- Parr, C.L.; Andersen, A.N. Patch mosaic burning for biodiversity conservation: A critique of the pyrodiversity paradigm. Conserv. Biol. 2006, 20, 1610–1619. [Google Scholar] [CrossRef]
- Kara, O.; Bolat, I. Short-term effects of wildfire on microbial biomass and abundance in black pine plantation soils in Turkey. Ecol. Indic. 2009, 9, 1151–1155. [Google Scholar] [CrossRef]
- Janzen, C.; Tobin-Janzen, T. Microbial communities in fire-affected soils. In Microbiology of Extreme Soils; Dion, P., Nautiyal, C.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 299–316. [Google Scholar] [CrossRef]
- Liang, C.; Ding, Y.; Yue, Y.; Zhang, X.Y.; Song, M.H.; Gao, J.Q.; Yu, F.H. Litter affects CO2 emission from Alpine wetland soils experiencing drying-rewetting cycles with different intensities and frequencies. Catena 2020. [Google Scholar] [CrossRef]
- Fernández-González, A.J.; Martínez-Hidalgo, P.; Cobo-Díaz, J.F.; Villadas, P.J.; Martínez-Molina, E.; Toro, N.; Tringe, S.G.; Fernández-López, M. The rhizosphere microbiome of burned holm-oak: Potential role of the genus arthrobacter in the recovery of burned soils. Sci. Rep. 2017, 7, 6008. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The role of soil microorganisms in plant mineral nutrition—Current knowledge and future directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.; Ver, L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [Green Version]
- Severino, R.; Froufe, H.J.C.; Barroso, C.; Albuquerque, L.; Lobo-da-Cunha, A.; da Costa, M.S.; Egas, C. High-quality draft genome sequence of Gaiella occulta isolated from a 150 m deep mineral water borehole and comparison with the genome sequences of other deep-branching lineages of the phylum Actinobacteria. Microbiologyopen 2019, e840. [Google Scholar] [CrossRef] [Green Version]
- Egas, C.; Barroso, C.; Froufe, H.J.C.; Pacheco, J.; Albuquerque, L.; Costa, M.S. Complete genome sequence of the radiation-resistant bacterium Rubrobacter radiotolerans RSPS-4. Stand. Genom. Sci. 2014, 9, 1062–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mun, C.L.; Ling, C.M.W.V. Effects of elevated temperature on the tropical soil bacterial diversity. Sains Malays. 2020, 49, 2335–2344. [Google Scholar] [CrossRef] [Green Version]
- Parente, C.E.T.; Brito, E.M.S.; Carreta, C.A.; Cervantes-Rodríguez, E.A.; Fábila-Canto, A.P.; Vollú, R.E.; Seldin, L.; Malm, O. Bacterial diversity changes in agricultural soils influenced by poultry litter fertilization. Braz. J. Microbiol. 2021, 52, 675–686. [Google Scholar] [CrossRef] [PubMed]
- León, A.; Del-Ángel, M.; Ávila, J.L.; Delgado, G. Phthalides: Distribution in nature, chemical reactivity, synthesis, and biological activity. Prog. Chem. Org. Nat. Prod. 2017, 104. [Google Scholar] [CrossRef]
- Pertile, M.; Sousa, R.M.S.; Mendes, L.W.; Antunes, J.E.L.; Oliveira, L.M.S.; Araujo, F.F.; Melo, V.M.M.; Araujo, A.S.F. Response of soil bacterial communities to the application of the herbicides imazethapyr and flumyzin. Eur. J. Soil Biol. 2021, 102, 103252. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, X.; Tai, X.; Sun, L.; Wu, M.; Zhang, W.; Chen, X.; Zhang, G.; Chen, T.; Liu, G.; et al. Variation in actinobacterial community composition and potential function in different soil ecosystems belonging to the arid heihe river basin of northwest China. Front. Microbiol. 2019, 10, 2209. [Google Scholar] [CrossRef]
- Kant, R.; Van Passel, M.W.J.; Palva, A.; Lucas, S.; Lapidus, A.; Del Rio, T.G.; Dalin, E.; Tice, H.; Bruce, D.; Goodwin, L.; et al. Genome sequence of Chthoniobacter flavus ellin428, an aerobic heterotrophic soil bacterium. J. Bacteriol. Res. 2011, 193, 2902–2903. [Google Scholar] [CrossRef] [Green Version]
- Sangwan, P.; Chen, X.; Hugenholtz, P.; Janssen, P.H. Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl. Environ. Microbiol. 2004, 70, 5857–5881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herlemann, D.P.R.; Lundin, D.; Labrenz, M.; Jürgens, K.; Zheng, Z.; Aspeborg, H.; Andersson, A.F. Metagenomic De Novo assembly of an aquatic representative of the verrucomicrobial class Spartobacteria. MBio 2013, 4, e0569-12. [Google Scholar] [CrossRef] [Green Version]
- Wüst, P.K.; Foesel, B.U.; Geppert, A.; Huber, K.J.; Luckner, M.; Wanner, G.; Overmann, J. Brevitalea aridisoli, B. deliciosa and Arenimicrobium luteum, three novel species of Acidobacteria subdivision 4 (class Blastocatellia) isolated from savanna soil and description of the novel family PyrinomonadaceaeInt. J. Syst. Evol. Microbiol. 2016, 66, 3355–3366. [Google Scholar] [CrossRef]
- Jia, D.; Dong, X.; Li, Y. Effect of strip harvesting on bacterial diversity of forest soils in the Daxing’an mountains. Soil Sci. Soc. Am. J. 2020, 84, 512–521. [Google Scholar] [CrossRef]
- Armbruster, M.; Goodall, T.; Hirsch, P.R.; Ostle, N.; Puissant, J.; Fagan, K.C.; Pywell, R.F.; Griffiths, R.I. Bacterial and archaeal taxa are reliable indicators of soil restoration across distributed calcareous grasslands. Eur. J. Soil Sci. 2020, 1–15. [Google Scholar] [CrossRef]
- Aguiar, L.M.; Souza, M.F.; Laia, M.L.; Melo, J.O.; Costa, M.R.; Gonçalves, J.F.; Silva, D.V.; Santos, J.B. Metagenomic analysis reveals mechanisms of atrazine biodegradation promoted by tree species. Environ. Pollut. 2020, 267, 115636. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Niu, X.; Tian, Y.; Xiao, Y. Assessment of PAH degradation potential of native species from a coking plant through identifying of the beneficial bacterial community within the rhizosphere soil. Chemosphere 2021, 264, 128513. [Google Scholar] [CrossRef]
- Zhang, C.; Tayyab, M.; Abubakar, A.Y.; Yang, Z.; Pang, Z.; Islam, W.; Lin, Z.; Li, S.; Luo, J.; Fan, X.; et al. Bacteria with different assemblages in the soil profile drive the diverse nutrient cycles in the sugarcane straw retention ecosystem. Diversity 2019, 11, 194. [Google Scholar] [CrossRef] [Green Version]
- Too, C.C.; Keller, A.; Sickel, W.; Lee, S.M.; Yule, C.M. microbial community structure in a Malaysian tropical peat swamp forest: The influence of tree species and depth. Front. Microbiol. 2018, 9, 2859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatar, D. Isolation, phylogenetic analysis and antimicrobial activity of halophilic actinomycetes from different saline environments located near Çorum Province. Biologia 2021. [Google Scholar] [CrossRef]
- Messaoudi, O.; Wink, J.; Bendahou, M. Diversity of actinobacteria isolated from date palms rhizosphere and saline environments: Isolation, identification and biological activity evaluation. Microorganisms 2020, 8, 1853. [Google Scholar] [CrossRef]
- Falagán, C.; Johnson, D.B. Acidibacter ferrireducens gen. nov., sp. nov.: An acidophilic ferric iron-reducing Gammaproteobacterium. Extremophiles 2014, 18, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- García-Fraile, P.; Benada, O.; Cajthaml, T.; Baldrian, P.; Lladó, S. Terracidiphilus gabretensis gen. nov., sp. nov., an abundant and active forest soil acidobacterium important in organic matter transformation. Appl. Environ. Microbiol. 2016, 82, 560–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; He, X.; Sun, J.; Ma, Y. A degeneration gradient of poplar trees contributes to the taxonomic, functional, and resistome diversity of bacterial communities in rhizosphere soils. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [Green Version]
- Blum, J.S.; Kulp, T.R.; Han, S.; Lanoil, B.; Saltikov, C.W.; Stolz, J.F.; Miller, L.G.; Oremland, R.S. Desulfohalophilus alkaliarsenatis gen. nov., sp. nov., an extremely halophilic sulfate- and arsenate-respiring bacterium from Searles Lake, California. Extremophiles 2012, 16, 727–742. [Google Scholar] [CrossRef] [Green Version]
- Hemmat-Jou, M.H.; Safari-Sinegani, A.A.; Mirzaie-Asl, A.; Tahmourespour, A. Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach. Ecotoxicology 2018, 27, 1281–1291. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Qiu, W. Linking microbial mechanism with bioelectricity production in sludge matrix-fed microbial fuel cells: Freezing/thawing liquid versus fermentation liquor. Sci. Total Environ. 2021, 752, 141907. [Google Scholar] [CrossRef]
- Dias, M.A.M.; Bomfim, C.S.G.; Rodrigues, D.R.; Silva, A.F.; Santos, J.C.S.; Nascimento, T.R.; Martins, L.M.V. Paraburkholderia spp. are the main rhizobial microsymbionts of Mimosa tenuiflora (Willd.) Poir. in soils of the Brazilian tropical dry forests (Caatinga biome). Syst. Appl. Microbiol. 2021, 44, 126208. [Google Scholar] [CrossRef] [PubMed]
- Alhadrami, H.A.; Thissera, B.; Hassan, M.H.A.; Behery, F.A.; Ngwa, C.J.; Hassan, H.M.; Pradel, G.; Abdelmohsen, U.R.; Rateb, M.E. Bio-guided isolation of antimalarial metabolites from the coculture of two red sea sponge-derived actinokineospora and Rhodococcus spp. Mar. Drugs 2021, 19, 109. [Google Scholar] [CrossRef]
- Arn, F.; Frasson, D.; Kroslakova, I.; Rezzonico, F.; Pothie, J.F.; Riedl, R.; Sievers, M. Isolation and identification of actinomycetes strains from Switzerland and their biotechnological potential. Chimia (Aarau) 2020, 74, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Oberhofer, M.; Hess, J.; Leutgeb, M.; Gössnitzer, F.; Rattei, T.; Wawrosch, C.; Zotchev, S.B. Exploring Actinobacteria associated with rhizosphere and endosphere of the native alpine medicinal plant Leontopodium nivale subspecies alpinum. Front. Microbiol. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Heidari, B.; Mohammadipanah, F. Isolation and identification of two alkaloid structures with radical scavenging activity from Actinokineospora sp. UTMC 968, a new promising source of alkaloid compounds. Mol. Biol. Rep. 2018, 45, 2325–2332. [Google Scholar] [CrossRef]
- Ivanova, A.A.; Philippov, D.A.; Kulichevskaya, I.S.; Dedysh, S.N. Distinct diversity patterns of Planctomycetes associated with the freshwater macrophyte Nuphar lutea (L.) Smith. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2017, 111, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Jroundi, F.; Martinez-Ruiz, F.; Merroun, M.L.; Gonzalez-Muñoz, M.T. Exploring bacterial community composition in Mediterranean deep-sea sediments and their role in heavy metal accumulation. Sci. Total Environ. 2020, 712, 135660. [Google Scholar] [CrossRef]
- Fan, M.; Li, J.; Tang, Z.; Shangguan, Z. Soil bacterial community succession during desertification in a desert steppe ecosystem. L. Degrad. Dev. 2020, 31, 1662–1674. [Google Scholar] [CrossRef]
- Nagkirti, P.D.; Engineer, A.S.; Dhakephalkar, P.K. Xylanimonas oleitrophica sp. nov., a novel petroleum hydrocarbon degrading bacterium isolated from an Indian oil reservoir. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2021, 114, 129–136. [Google Scholar] [CrossRef]
- Moe, L.A. Amino acids in the rhizosphere: From plants to microbes. Am. J. Bot. 2013, 100, 1692–1705. [Google Scholar] [CrossRef] [PubMed]
- Auffret, M.D.; Karhu, K.; Khachane, A.; Dungait, J.; Fraser, F.; Hopkins, D.; Wookey, P.; Singh, B.; Freitag, T.E.; Hartley, I.P.; et al. The role of microbial community composition in controlling soil respiration responses to temperature. PLoS ONE 2016, 11, e0165448. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Liang, J.; Zhao, D.L.; Meng, C.; Xu, Z.C.; Xie, Z.H.; Zhang, C.S. The root nodule microbiome of cultivated and wild halophytic legumes showed similar diversity but distinct community structure in yellow river delta saline soils. Microorganisms 2020, 8, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, S.E.D.; Beuf, K.D.; Vekeman, B.; Willems, A. A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol. Biochem. 2015, 83, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Muresu, R.; Polone, E.; Sulas, L.; Baldan, B.; Tondello, A.; Delogu, G.; Cappuccinelli, P.; Alberghini, S.; Benhizia, Y.; Benhizia, H.; et al. Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol. Ecol. 2008, 63, 383–400. [Google Scholar] [CrossRef] [Green Version]
- Kan, F.L.; Chen, Z.Y.; Wang, E.T.; Tian, C.F.; Sui, X.H.; Chen, W.X. Characterization of symbiotic and endophytic bacteria isolated from root nodules of herbaceous legumes grown in Qinghai-Tibet plateau and in other zones of China. Arch. Microbiol. 2007, 188, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Messiha, N.A.S.; van Diepeningen, A.D.; Farag, N.S.; Abdallah, S.A.; Janse, J.D.; van Bruggen, A.H.C. Stenotrophomonas maltophilia: A new potential biocontrol agent of Ralstonia solanacearum, causal agent of potato brown rot. Eur. J. Plant Pathol. 2007, 118, 211–225. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, P.; Roy, P. Genomic potential of Stenotrophomonas maltophilia in bioremediation with an assessment of its multifaceted role in our environment. Front. Microbiol. 2016, 7, 967. [Google Scholar] [CrossRef]
- Berg, G.; Marten, P.; Ballin, G. Stenotrophomonas maltophilia in the rhizosphere of oilseed rape-occurrence, characterization and interaction with phytopathogenic fungi. Microbiol. Res. 1996, 151, 19–27. [Google Scholar] [CrossRef]
- Gopi, K.; Jinal, H.N.; Prittesh, P.; Kartik, V.P.; Amaresan, N. Effect of copper-resistant Stenotrophomonas maltophilia on Maize (Zea mays) growth, physiological properties, and copper accumulation: Potential for phytoremediation into biofortification. Int. J. Phytoremed. 2020. [Google Scholar] [CrossRef] [PubMed]
- An, S.Q.; Berg, G. Stenotrophomonas maltophilia. Trends Microbiol. 2018. [Google Scholar] [CrossRef] [Green Version]
- Naz, I.; Bano, A. Assessment of phytohormones producing capacity of Stenotrophomonas maltophilia ssa and its interaction with Zea Mays L. Pak. J. Bot. 2012, 44, 465–469. [Google Scholar]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.; Yu, M.; Lai, E.-M. Agrobacterium—Mediated plant transformation: Biology and applications. Arab. Book 2017, 15. [Google Scholar] [CrossRef] [Green Version]
- Krenek, P.; Samajova, O.; Luptovciak, I.; Doskocilova, A.; Komis, G.; Samaj, J. Transient plant transformation mediated by Agrobacterium tumefaciens: Principles, methods and applications. Biotechnol. Adv. 2015, 1, 1024–1042. [Google Scholar] [CrossRef]
- Veena, V.; Taylor, C.G. Agrobacterium rhizogenes: Recent developments and promising applications. Vitr. Cell. Dev. Biol. Plant 2007, 43, 83–403. [Google Scholar] [CrossRef]
- Rogel, M.A.; Hernández-Lucas, I.; Kuykendall, L.D.; Balkwill Martinez-Romero, E. Nitrogen-fixing nodules with Ensifer adhaerens harboring Rhizobium tropici symbiotic plasmids. Appl. Environ. Microbiol. 2001, 67, 3264–3268. [Google Scholar] [CrossRef] [Green Version]
- Doyle, J.J. Phylogenetic perspectives on nodulation: Evolving views of plants and symbiotic bacteria. Trends Plant Sci. 1998, 3, 473–478. [Google Scholar] [CrossRef]
- Teixeira, H.; Rodríguez-Echeverría, S. Identification of symbiotic nitrogen-fixing bacteria from three african leguminous trees in Gorongosa National Park. Syst. Appl. Microbiol. 2016, 39, 350–358. [Google Scholar] [CrossRef]
- Dharni, S.; Srivastava, A.K.; Samad, A.; Patra, D.D. Impact of plant growth promoting Pseudomonas monteilii psf84 and Pseudomonas plecoglossicida psf610 on metal uptake and production of secondary metabolite (monoterpenes) by rose-scented geranium (Pelargonium graveolenscv. bourbon) grown on tannery sludge. Chemosphere 2014, 117, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Trinh, C.S.; Lee, H.; Lee, W.J.; Lee, S.J.; Chung, N.; Han, J.; Kim, J.; Hong, S.W.; Lee, H. Evaluation of the plant growth-promoting activity of Pseudomonas nitroreducens in Arabidopsis thaliana and Lactuca sativa. Plant Cell. Rep. 2018, 37, 873–885. [Google Scholar] [CrossRef]
- Natsagdorj, O.; Sakamoto, H.; Santiago, D.M.O.; Santiago, C.D.; Orikasa, Y.; Okazaki, K.; Ikeda, S.; Ohwada, T. Variovorax sp. has an optimum cell density to fully function as a plant growth promoter. Microorganisms 2019, 7, 82. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Pandey, S. ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in french bean (Phaseolus vulgaris) plants. Front. Microbiol. 2019, 10, 1506. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.M.; Pérez-Quintero, A.L.; Koebnik, R.; DuCharme, E.; Sarra, S.; Doucoure, H.; Keita, I.; Ziegle, J.; Jacobs, J.M.; Olivia, R.; et al. A Pathovar of Xanthomonas oryzae infecting wild grasses provides insight into the evolution of pathogenicity in rice agroecosystems. Front. Plant Sci. 2019, 10, 507. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Suzuki, I.; Koizumi, J.I. Balneomonas flocculans gen. nov., sp. nov., a new cellulose-producing member of the α-2 subclass of Proteobacteria. Syst. Appl. Microbiol. 2004, 27, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Ardley, J.K.; Parker, M.A.; De Meyer, S.E.; Trengove, R.D.; O’Hara, G.W.; Reeve, W.G.; Yates, R.J.; Dilworth, M.J.; Willems, A.; Howieson, J.G. Microvirga lupini sp. nov., Microvirga lotononidis sp. nov. and Microvirga zambiensis sp. nov. are alphaproteobacterial root-nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. Int. J. Syst. Evol. Microbiol. 2012, 62, 2579–2588. [Google Scholar] [CrossRef]
- Vial, L.; Chapalain, A.; Groleau, M.C.; Déziel, E. The various lifestyles of the Burkholderia cepacia complex species: A tribute to adaptation. Environ. Microbiol. 2011, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Belimov, A.A.; Dodd, I.C.; Hontzeas, N.; Theobald, J.C.; Safronova, V.I.; Davies, W.J. Rhizosphere Bacteria Containing 1-Aminocyclopropane-1-Carboxylate Deaminase Increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol. 2009, 181, 413–423. [Google Scholar] [CrossRef]
- Hamdi Hussein, Z. Rhizobia from wild legumes: Diversity, taxonomy, ecology, nitrogen fixation and biotechnology. J. Biotechnol. 2001, 91, 143–153. [Google Scholar]
- Estrada-De Los Santos, P.; Bustillos-Cristales, R.; Caballero-Mellado, J. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 2001, 67, 2790–2798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safronova, V.I.; Kuznetsova, I.G.; Sazanova, A.L.; Belimov, A.A.; Andronov, E.E.; Chirak, E.R.; Osledkin, Y.S.; Onishchuk, O.P.; Kurchak, O.N.; Shaposhnikov, A.I.; et al. Microvirga ossetica sp. nov., a species of rhizobia isolated from root nodules of the legume species Vicia alpestris Steven. Int. J. Syst. Evol. Microbiol. 2017, 67, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Hindawi Publ. Corp. Sci. 2012, 2020, 963401. [Google Scholar] [CrossRef] [Green Version]
- Braud, A.; Jézéquel, K.; Vieille, E.; Tritter, A.; Lebeau, T. Changes in extractability of Cr and Pb in a polycontaminated soil after bioaugmentation with microbial producers of biosurfactants, organic acids and siderophores. Water Air Soil Pollut. 2006, 6, 261–279. [Google Scholar] [CrossRef]
- Belimov, A.A.; Hontzeas, N.; Safronova, V.I.; Demchinskaya, S.V.; Piluzza, G.; Bullitta, S.; Glick, B.R. Cadmium-tolerant plant growth-promoting bacteria associated with the roots of indian mustard (Brassica Juncea L. Czern.). Soil. Biol. Biochem. 2005, 37, 241–250. [Google Scholar] [CrossRef]
- Burd, G.I.; Dixon, D.G.; Glick, B.R. A plant growth-promoting bacterium that decreases nickel toxicity in seedlings. Appl. Environ. Microbiol. 1998, 64, 3663–3668. [Google Scholar] [CrossRef] [Green Version]
- Burd, G.I.; Dixon, D.G.; Glick, B.R. Plant growth-promoting bacteria that decrease heavy metal toxicity in plants. Can. J. Microbiol. 2000, 46, 237–245. [Google Scholar] [CrossRef]
- Jiang, C.; Sheng, X.F.; Qian, M.; Wang, Q. Isolation and characterization of a heavy metal-resistant Burkholderia sp. from heavy metal-contaminated paddy field soil and its potential in promoting plant growth and heavy metal accumulation in metal-polluted soil. Chemosphere 2008, 72, 157–164. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 1980, 286, 885–886. [Google Scholar] [CrossRef]
- Ghio, S.; Di Lorenzo, G.S.; Lia, V.; Talia, P.; Cataldi, A.; Grasso, D.; Campos, E. Isolation of Paenibacillus Sp. and Variovorax Sp. strains from decaying woods and characterization of their potential for cellulose deconstruction. Int. J. Biochem. Mol. Biol. 2012, 3, 352–364. [Google Scholar] [PubMed]
- Jadhav, H.P.; Sayyed, R.Z. Hydrolytic enzymes of rhizospheric microbes in crop protection. MOJ Cell Sci. Rep. 2016, 3, 135–136. [Google Scholar] [CrossRef]


| Soil Type | Fire Frequency | Code |
|---|---|---|
| Brownish-gray sandy soils | Low | SL |
| High | SH | |
| Red soils of medium texture | Low | RL |
| High | RH | |
| Red oxic soils with medium texture | Low | OL |
| High | OH |
| Soil Type | Fire Frequency | Isolate | Most Related 16S rRNA Gene Sequence | GenBank Accession Number | % Identity | Growth in N-Free Media | Phosphate Solubilization | Indole Acetic Acid Production (1) | Siderophore Production | Hydrolysis of Cellulose |
|---|---|---|---|---|---|---|---|---|---|---|
| Sandy soils | Low | 10SLA | Microvirga sp. | MZ571264 | 96.54% | + | - | + | - | - |
| Red soils | High | 4RHB | Caballeronia zhejiangensis strain ND-B | MZ571257 | 96.90% | + | - | - | + | - |
| 5RHB | Burkholderia sp. | MZ571258 | 96.27% | + | - | - | + | - | ||
| 6RHB | Burkholderia sp. clone P4s-284 | MZ571259 | 72.58% | + | - | + | - | - | ||
| Oxi-soils | Low | 4OLA | Rhizobium sp. NA11036 | MZ571262 | 90.95% | + | - | + | - | - |
| 5OLA | Rhizobium altiplani strain BR 10423 | MZ571263 | 89.23% | + | - | + | - | - | ||
| High | 5OHA | Rhizobium sp. isolate Moz93 | MZ571260 | 89.70% | + | - | + | - | - | |
| 10OHA | Variovorax defluvii strain 2C1-21 | MZ571261 | 97.88% | + | - | - | - | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maquia, I.S.A.; Fareleira, P.; Videira e. Castro, I.; Soares, R.; Brito, D.R.A.; Mbanze, A.A.; Chaúque, A.; Máguas, C.; Ezeokoli, O.T.; Ribeiro, N.S.; et al. The Nexus between Fire and Soil Bacterial Diversity in the African Miombo Woodlands of Niassa Special Reserve, Mozambique. Microorganisms 2021, 9, 1562. https://doi.org/10.3390/microorganisms9081562
Maquia ISA, Fareleira P, Videira e. Castro I, Soares R, Brito DRA, Mbanze AA, Chaúque A, Máguas C, Ezeokoli OT, Ribeiro NS, et al. The Nexus between Fire and Soil Bacterial Diversity in the African Miombo Woodlands of Niassa Special Reserve, Mozambique. Microorganisms. 2021; 9(8):1562. https://doi.org/10.3390/microorganisms9081562
Chicago/Turabian StyleMaquia, Ivete Sandra Alberto, Paula Fareleira, Isabel Videira e. Castro, Ricardo Soares, Denise R. A. Brito, Aires Afonso Mbanze, Aniceto Chaúque, Cristina Máguas, Obinna T. Ezeokoli, Natasha Sofia Ribeiro, and et al. 2021. "The Nexus between Fire and Soil Bacterial Diversity in the African Miombo Woodlands of Niassa Special Reserve, Mozambique" Microorganisms 9, no. 8: 1562. https://doi.org/10.3390/microorganisms9081562
APA StyleMaquia, I. S. A., Fareleira, P., Videira e. Castro, I., Soares, R., Brito, D. R. A., Mbanze, A. A., Chaúque, A., Máguas, C., Ezeokoli, O. T., Ribeiro, N. S., Marques, I., & Ribeiro-Barros, A. I. (2021). The Nexus between Fire and Soil Bacterial Diversity in the African Miombo Woodlands of Niassa Special Reserve, Mozambique. Microorganisms, 9(8), 1562. https://doi.org/10.3390/microorganisms9081562












