Natural Intra- and Interclade Human Hybrid Schistosomes in Africa with Considerations on Prevention through Vaccination
Abstract
1. Schistosomiasis and Parasite Hybridization
1.1. Disease and Transmission
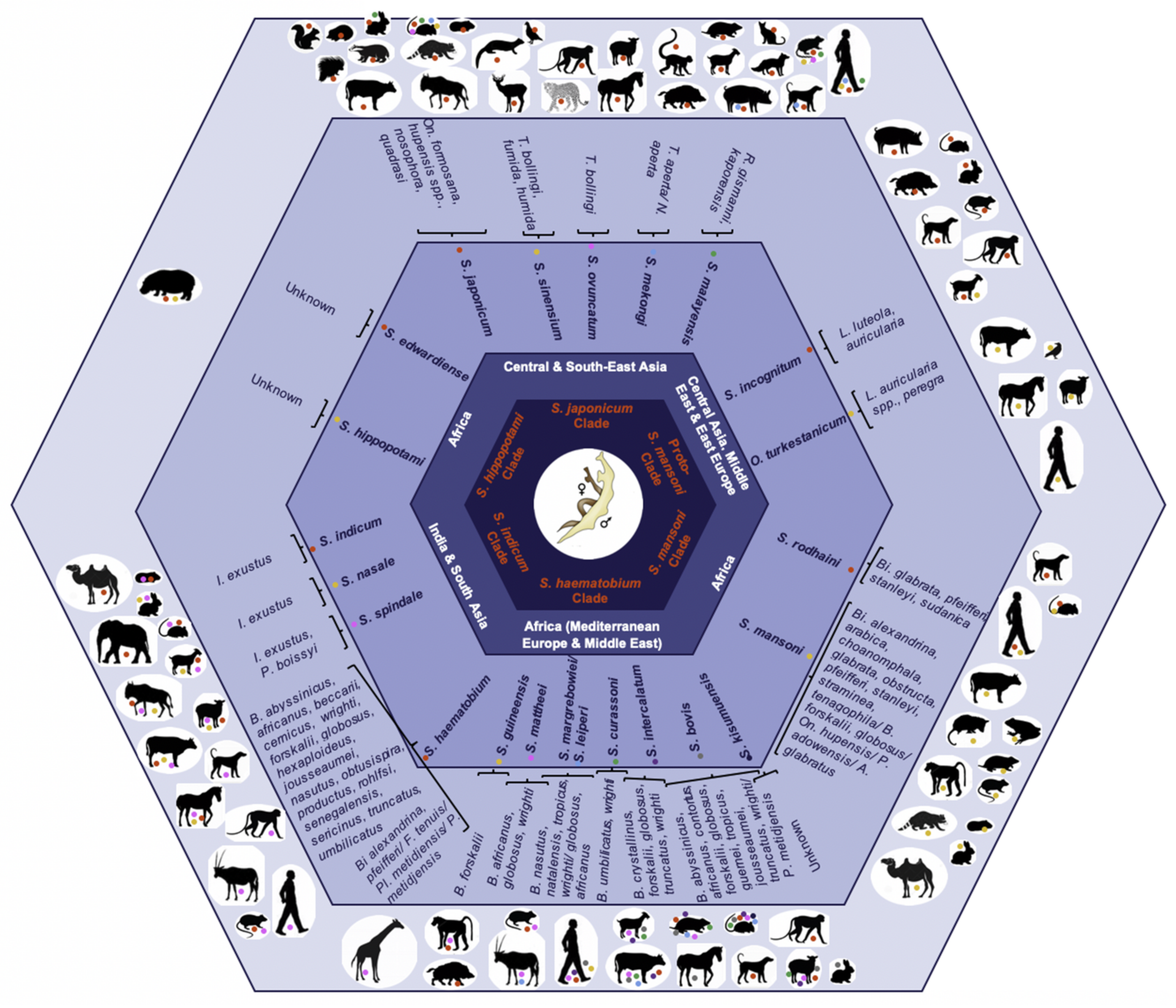
1.2. Treatment and Prevention
1.3. Instant Gene Exchange
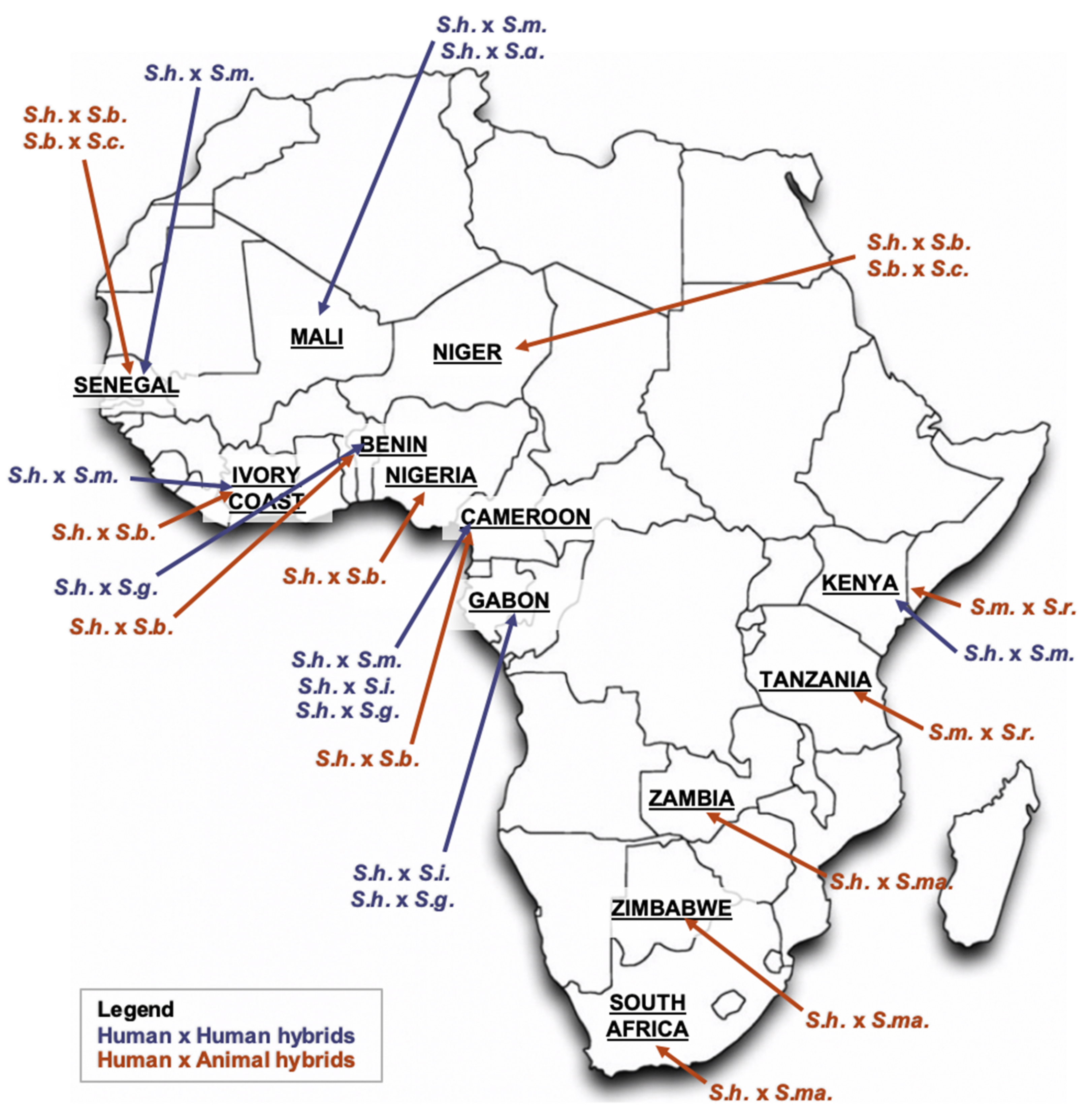
2. Schistosome Hybrids across Africa—The Tip of the Iceberg
2.1. S. haematobium/S. guineensis Hybrids
2.2. S. haematobium/S. bovis Hybrids
2.3. S. haematobium/S. mansoni Hybrids
2.4. S. mansoni/S. rodhaini Hybrids
3. Schistosomal Hybridization—Interference on Leading Vaccine Candidates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelwan, M.L. Schistosomiasis: Life Cycle, Diagnosis, and Control. Curr. Ther. Res. 2019, 91, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.P.; Neves, M.I.; Webster, B.L.; Pennance, T.; Rabone, M.; Gouvras, A.; Allan, F.; Walker, M.; Rollinson, D. Parasite Population Genetic Contributions to the Schistosomiasis Consortium for Operational Research and Evaluation within Sub-Saharan Africa. Am. J. Trop. Med. Hyg. 2020, 103, 80–91. [Google Scholar] [CrossRef]
- Colley, D.G.; Bustinduy, A.L.; Secor, W.E.; King, C.H. Human schistosomiasis. Lancet 2014, 383, 2253–2264. [Google Scholar] [CrossRef]
- Molehin, A.J. Schistosomiasis vaccine development: Update on human clinical trials. J. Biomed. Sci. 2020, 27, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Leger, E.; Webster, J.P. Hybridizations within the Genus Schistosoma: Implications for evolution, epidemiology and control. Parasitology 2017, 144, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Lawton, S.P.; Hirai, H.; Ironside, J.E.; Johnston, D.A.; Rollinson, D. Genomes and geography: Genomic insights into the evolution and phylogeography of the genus Schistosoma. Parasites Vectors 2011, 4, 131. [Google Scholar] [CrossRef]
- Webster, B.; Southgate, V.; Littlewood, D. A revision of the interrelationships of Schistosoma including the recently described Schistosoma guineensis. Int. J. Parasitol. 2006, 36, 947–955. [Google Scholar] [CrossRef]
- London NHM. Host-Parasite Database. Available online: https://www.nhm.ac.uk/research-curation/scientific-resources/taxonomy-systematics/host-parasites/ (accessed on 2 June 2021).
- Beltran, S.; Boissier, J. Schistosome monogamy: Who, how, and why? Trends Parasitol. 2008, 24, 386–391. [Google Scholar] [CrossRef]
- Russell Stothard, J.; Kayuni, S.A.; Al-Harbi, M.H.; Musaya, J.; Webster, B.L. Future schistosome hybridizations: Will all Schistosoma haematobium hybrids please stand-up! PLoS Negl. Trop. Dis. 2020, 14, e0008201. [Google Scholar] [CrossRef]
- Pearce, E.J.; Macdonald, A.S. The immunobiology of schistosomiasis. Nat. Rev. Immunol. 2002, 2, 499–511. [Google Scholar] [CrossRef]
- Loker, E.S.; Brant, S.V. Diversification, dioecy and dimorphism in schistosomes. Trends Parasitol. 2006, 22, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Steinauer, M.L.; Blouin, M.S.; Criscione, C.D. Applying evolutionary genetics to schistosome epidemiology. Infect. Genet. Evol. 2010, 10, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Vale, N.; Gouveia, M.; Rinaldi, G.; Brindley, P.J.; Gärtner, F.; da Costa, J.M.C. Praziquantel for Schistosomiasis: Single-Drug Metabolism Revisited, Mode of Action, and Resistance. Antimicrob. Agents Chemother. 2017, 61, e02582-16. [Google Scholar] [CrossRef] [PubMed]
- Chisango, T.J.; Ndlovu, B.; Vengesai, A.; Nhidza, A.F.; Sibanda, E.P.; Zhou, D.; Mutapi, F.; Mduluza, T. Benefits of annual chemotherapeutic control of schistosomiasis on the development of protective immunity. BMC Infect. Dis. 2019, 19, 219. [Google Scholar] [CrossRef]
- Driciru, E.; Koopman, J.P.R.; Cose, S.; Siddiqui, A.A.; Yazdanbakhsh, M.; Elliott, A.M.; Roestenberg, M. Immunological Considerations for Schistosoma Vaccine Development: Transitioning to Endemic Settings. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Eyoh, E.; McCallum, P.; Killick, J.; Amanfo, S.; Mutapi, F.; Astier, A.L. The anthelmintic drug praziquantel promotes human Tr1 differentiation. Immunol. Cell Biol. 2019, 97, 512–518. [Google Scholar] [CrossRef]
- Sokolow, S.H.; Wood, C.L.; Jones, I.J.; Lafferty, K.D.; Kuris, A.M.; Hsieh, M.H.; De Leo, G.A. To Reduce the Global Burden of Human Schistosomiasis, Use ‘Old Fashioned’ Snail Control. Trends Parasitol. 2018, 34, 23–40. [Google Scholar] [CrossRef]
- Merrifield, M.; Hotez, P.J.; Beaumier, C.M.; Gillespie, P.; Strych, U.; Hayward, T.; Bottazzi, M.E. Advancing a vaccine to prevent human schistosomiasis. Vaccine 2016, 34, 2988–2991. [Google Scholar] [CrossRef]
- Dumont, M.; Moné, H.; Mouahid, G.; Idris, M.A.; Shaban, M.; Boissier, J. Influence of pattern of exposure, parasite genetic diversity and sex on the degree of protection against reinfection with Schistosoma mansoni. Parasitol. Res. 2007, 101, 247–252. [Google Scholar] [CrossRef]
- Tebeje, B.M.; Harvie, M.; You, H.; Loukas, A.; McManus, D.P. Schistosomiasis vaccines: Where do we stand? Parasites Vectors 2016, 9, 1–15. [Google Scholar] [CrossRef]
- Anisuzzaman; Tsuji, N. Schistosomiasis and hookworm infection in humans: Disease burden, pathobiology and anthelmintic vaccines. Parasitol. Int. 2020, 75, 102051. [Google Scholar] [CrossRef] [PubMed]
- Moser, D.; Tendler, M.; Griffiths, G.; Klinkert, M. A 14-kDa Schistosoma mansoni polypeptide is homologous to a gene family of fatty acid binding proteins. J. Biol. Chem. 1991, 266, 8447–8454. [Google Scholar] [CrossRef]
- Becker, M.M.; Kalinna, B.H.; Waine, G.J.; McManus, D.P. Gene cloning, overproduction and purification of a functionally active cytoplasmic fatty acid-binding protein (Sj-FABPc) from the human blood fluke Schistosoma japonicum. Gene 1994, 148, 321–325. [Google Scholar] [CrossRef]
- Esteves, A.; Joseph, L.; Paulino, M.; Ehrlich, R. Remarks on the phylogeny and structure of fatty acid binding proteins from parasitic platyhelminths. Int. J. Parasitol. 1997, 27, 1013–1023. [Google Scholar] [CrossRef]
- Tendler, M.; Brito, C.A.; Vilar, M.M.; Serra-Freire, N.; Diogo, C.M.; Almeida, M.S.; Delbem, A.; Da Silva, J.F.; Savino, W.; Garratt, R.C.; et al. A Schistosoma mansoni fatty acid-binding protein, Sm14, is the potential basis of a dual-purpose anti-helminth vaccine. Proc. Natl. Acad. Sci. USA 1996, 93, 269–273. [Google Scholar] [CrossRef]
- Vilar, M.M.; Barrientos, F.; Almeida, M.; Thaumaturgo, N.; Simpson, A.; Garratt, R.; Tendler, M. An experimental bivalent peptide vaccine against schistosomiasis and fascioliasis. Vaccine 2003, 22, 137–144. [Google Scholar] [CrossRef]
- Charrin, S.; Jouannet, S.; Boucheix, C.; Rubinstein, E. Tetraspanins at a glance. J. Cell Sci. 2014, 127, 3641–3648. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.H.; Pearson, M.; Bethony, J.M.; Smyth, D.; Jones, M.; Duke, M.; A Don, T.; McManus, D.; Correa-Oliveira, R.; Loukas, A. Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nat. Med. 2006, 12, 835–840. [Google Scholar] [CrossRef]
- Zhang, W.; Li, J.; Duke, M.; Jones, M.K.; Kuang, L.; Zhang, J.; Blair, D.; Li, Y.; McManus, D. Inconsistent Protective Efficacy and Marked Polymorphism Limits the Value of Schistosoma japonicum Tetraspanin-2 as a Vaccine Target. PLoS Negl. Trop. Dis. 2011, 5, e1166. [Google Scholar] [CrossRef]
- Cupit, P.M.; Steinauer, M.L.; Tonnessen, B.W.; Agola, L.E.; Kinuthia, J.M.; Mwangi, I.N.; Mutuku, M.W.; Mkoji, G.M.; Loker, E.S.; Cunningham, C. Polymorphism associated with the Schistosoma mansoni tetraspanin-2 gene. Int. J. Parasitol. 2011, 41, 1249–1252. [Google Scholar] [CrossRef]
- Loukas, A.; Tran, M.; Pearson, M. Schistosome membrane proteins as vaccines. Int. J. Parasitol. 2007, 37, 257–263. [Google Scholar] [CrossRef]
- Jia, X.; Schulte, L.; Loukas, A.; Pickering, D.; Pearson, M.; Mobli, M.; Jones, A.; Rosengren, K.J.; Daly, N.; Gobert, G.; et al. Solution Structure, Membrane Interactions, and Protein Binding Partners of the Tetraspanin Sm-TSP-2, a Vaccine Antigen from the Human Blood Fluke Schistosoma mansoni. J. Biol. Chem. 2014, 289, 7151–7163. [Google Scholar] [CrossRef]
- Croall, D.E.; Ersfeld, K. The calpains: Modular designs and functional diversity. Genome Biol. 2007, 8, 218. [Google Scholar] [CrossRef]
- Zhang, R.; Suzuki, T.; Takahashi, S.; Yoshida, A.; Kawaguchi, H.; Maruyama, H.; Yabu, Y.; Fu, J.; Shirai, T.; Ohta, N. Cloning and molecular characterization of calpain, a calcium-activated neutral proteinase, from different strains of Schistosoma japonicum. Parasitol. Int. 2000, 48, 233–242. [Google Scholar] [CrossRef]
- Karcz, S.R.; Podesta, R.B.; Siddiqui, A.A.; Dekaban, G.A.; Strejan, G.H.; Clarke, M.W. Molecular cloning and sequence analysis of a calcium-activated neutral protease (calpain) from Schistosoma mansoni. Mol. Biochem. Parasitol. 1991, 49, 333–336. [Google Scholar] [CrossRef]
- Johnson, K.A.; Angelucci, F.; Bellelli, A.; Hervé, M.; Fontaine, J.; Tsernoglou, D.; Capron, A.; Trottein, F.; Brunori, M. Crystal Structure of the 28 kDa GlutathioneS-Transferase from Schistosoma haematobium. Biochemistry 2003, 42, 10084–10094. [Google Scholar] [CrossRef]
- Trottein, F.; Godin, C.; Pierce, R.; Sellin, B.; Taylor, M.G.; Gorillot, I.; Silva, M.S.; Lecocq, J.-P.; Capron, A. Inter-species variation of schistosome 28-kDa glutathione S-transferases. Mol. Biochem. Parasitol. 1992, 54, 63–72. [Google Scholar] [CrossRef]
- Steinauer, M.L.; Hanelt, B.; Mwangi, I.N.; Maina, G.M.; Agola, L.E.; Kinuthia, J.M.; Mutuku, M.W.; Mungai, B.N.; Wilson, W.D.; Mkoji, G.M.; et al. Introgressive hybridization of human and rodent schistosome parasites in western Kenya. Mol. Ecol. 2008, 17, 5062–5074. [Google Scholar] [CrossRef] [PubMed]
- King, K.; Stelkens, R.B.; Webster, J.P.; Smith, D.F.; Brockhurst, M.A. Hybridization in Parasites: Consequences for Adaptive Evolution, Pathogenesis, and Public Health in a Changing World. PLoS Pathog. 2015, 11, e1005098. [Google Scholar] [CrossRef]
- Arnold, M.L. Natural hybridization and the evolution of domesticated, pest and disease organisms. Mol. Ecol. 2004, 13, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Detwiler, J.T.; Criscione, C.D. An Infectious Topic in Reticulate Evolution: Introgression and Hybridization in Animal Parasites. Genes 2010, 1, 102–123. [Google Scholar] [CrossRef]
- Léger, E.; Garba, A.; Hamidou, A.A.; Webster, B.; Pennance, T.; Rollinson, D.; Webster, J.P. Introgressed Animal Schistosomes Schistosoma curassoni and S. bovis Naturally Infecting Humans. Emerg. Infect. Dis. 2016, 22, 2212–2214. [Google Scholar] [CrossRef] [PubMed]
- Huyse, T.; Webster, B.L.; Geldof, S.; Stothard, J.R.; Diaw, O.T.; Polman, K.; Rollinson, D. Bidirectional introgressive hybridization between a cattle and human schistosome species. PLoS Pathog. 2009, 5, e1000571. [Google Scholar] [CrossRef]
- Webster, B.; Diaw, O.T.; Seye, M.M.; Webster, J.P.; Rollinson, D. Introgressive Hybridization of Schistosoma haematobium Group Species in Senegal: Species Barrier Break Down between Ruminant and Human Schistosomes. PLoS Negl. Trop. Dis. 2013, 7, e2110. [Google Scholar] [CrossRef]
- Crego-Vicente, B.; Fernández-Soto, P.; Febrer-Sendra, B.; Diego, J.G.-B.; Boissier, J.; Angora, E.; Oleaga, A.; Muro, A. Application of a Genus-Specific LAMP Assay for Schistosome Species to Detect Schistosoma haematobium × Schistosoma bovis Hybrids. J. Clin. Med. 2021, 10, 1308. [Google Scholar] [CrossRef] [PubMed]
- Depaquit, J.; Akhoundi, M.; Haouchine, D.; Mantelet, S.; Izri, A. No limit in interspecific hybridization in schistosomes: Observation from a case report. Parasite 2019, 26, 10. [Google Scholar] [CrossRef] [PubMed]
- Boon, N.A.; Van Den Broeck, F.; Faye, D.; Volckaert, F.A.; Mboup, S.; Polman, K.; Huyse, T. Barcoding hybrids: Heterogeneous distribution of Schistosoma haematobium × Schistosoma bovis hybrids across the Senegal River Basin. Parasitology 2018, 145, 634–645. [Google Scholar] [CrossRef]
- Rey, O.; Toulza, E.; Chaparro, C.; Allienne, J.-F.; Kincaid-Smith, J.; Mathieu-Begné, E.; Allan, F.; Rollinson, D.; Webster, B.L.; Boissier, J. Diverging patterns of introgression from Schistosoma bovis across S. haematobium African lineages. PLoS Pathog. 2021, 17, e1009313. [Google Scholar] [CrossRef]
- Moné, H.; Minguez, S.; Ibikounlé, M.; Allienne, J.F.; Massougbodji, A.; Mouahid, G. Natural interactions between S. haematobium and S. guineensis in the Republic of Benin. Sci. World J. 2012, 2012, 793420. [Google Scholar] [CrossRef]
- Webster, B.L.; Tchuenté, L.A.T.; Southgate, V.R. A single-strand conformation polymorphism (SSCP) approach for investigating genetic interactions of Schistosoma haematobium and Schistosoma guineensis in Loum, Cameroon. Parasitol. Res. 2007, 100, 739–745. [Google Scholar] [CrossRef]
- Rollinson, D. Biochemical genetics in the study of schistosomes and their intermediate hosts. Parassitologia 1985, 27, 123–139. [Google Scholar]
- Wright, C.; Ross, G. Hybrids between Schistosoma haematobium and S. mattheei and their identification by isoelectric focusing of enzymes. Trans. R. Soc. Trop. Med. Hyg. 1980, 74, 326–332. [Google Scholar] [CrossRef]
- Catalano, S.; Sène, M.; Diouf, N.D.; Fall, C.B.; Borlase, A.; Léger, E.; Bâ, K.; Webster, J.P. Rodents as natural hosts of zoonotic schistosoma species and hybrids: An epidemiological and evolutionary perspective from West Africa. J. Infect. Dis. 2018, 218, 429–433. [Google Scholar] [CrossRef]
- Wang, S.; Zhu, X.-Q.; Cai, X. Gene Duplication Analysis Reveals No Ancient Whole Genome Duplication but Extensive Small-Scale Duplications during Genome Evolution and Adaptation of Schistosoma mansoni. Front. Cell. Infect. Microbiol. 2017, 7, 412. [Google Scholar] [CrossRef]
- Boon, N.A.M.; Fannes, W.; Rombouts, S.; Polman, K.; Volckaert, F.A.M.; Huyse, T. Detecting hybridization in African schistosome species: Does egg morphology complement molecular species identification? Parasitology 2017, 144, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.A.T.; DeJong, R.J.; Lwambo, N.J.S.; Mungai, B.N.; Mkoji, G.M.; Loker, E.S. First Report of a Natural Hybrid Between Schistosoma mansoni and S. rodhaini. J. Parasitol. 2003, 89, 416–418. [Google Scholar] [CrossRef]
- Walker, T.K.; Rollinson, D.; Simpson, A.J. Differentiation of Schistosoma haematobium from related species using cloned ribosomal RNA gene probes. Mol. Biochem. Parasitol. 1986, 20, 123–131. [Google Scholar] [CrossRef]
- Rodrigues, M.S.; Morelli, K.A.; Jansen, A.M. Cytochrome c oxidase subunit 1 gene as a DNA barcode for discriminating Trypanosoma cruzi DTUs and closely related species. Parasites Vectors 2017, 10, 1–18. [Google Scholar] [CrossRef]
- Zhao, Y.; Gentekaki, E.; Yi, Z.; Lin, X. Genetic Differentiation of the Mitochondrial Cytochrome Oxidase c Subunit I Gene in Genus Paramecium (Protista, Ciliophora). PLoS ONE 2013, 8, e77044. [Google Scholar] [CrossRef] [PubMed]
- Schols, R.; Carolus, H.; Hammoud, C.; Mulero, S.; Mudavanhu, A.; Huyse, T. A rapid diagnostic multiplex PCR approach for xenomonitoring of human and animal schistosomiasis in a ‘One Health’ context. Trans. R. Soc. Trop. Med. Hyg. 2019, 113, 722–729. [Google Scholar] [CrossRef]
- Webster, B.; Tchuenté, L.T.; Jourdane, J.; Southgate, V. The interaction of Schistosoma haematobium and S. guineensis in Cameroon. J. Helminthol. 2005, 79, 193–197. [Google Scholar] [CrossRef]
- Wright, C.A.; Southgate, V.R.; Van Wijk, H.B.; Moore, P.J. Letter: Hybrids between Schistosoma haematobium and S. intercalatum in Cameroon. Trans. R. Soc. Trop. Med. Hyg. 1974, 68, 413–414. [Google Scholar] [CrossRef]
- Webster, B.; Southgate, V.; Tchuenté, L.-A.T. Isoenzyme analysis of Schistosoma haematobium, S. intercalatum and their hybrids and occurrences of natural hybridization in Cameroon. J. Helminthol. 2003, 77, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Southgate, V.R.; van Wijk, H.B.; Wright, C.A. Schistosomiasis at Loum, Cameroun; Schistosoma haematobium, S. intercalatum and their natural hybrid. Parasitol. Res. 1976, 49, 145–159. [Google Scholar] [CrossRef]
- Tchuem Tchuenté, L.A.; Southgate, V.R.; Njiokou, F.; Njiné, T.; Kouemeni, L.E.; Jourdane, J. The evolution of schistosomiasis at Loum, Cameroon: Replacement of Schistosoma intercalatum by S. haematobium through introgressive hybridization. Trans. R. Soc. Trop. Med. Hyg. 1997, 91, 664–665. [Google Scholar] [CrossRef]
- Southgate, V.R. On factors possibly restricting the distribution of Schistosoma intercalatum Fisher, 1934. Parasitol. Res. 1978, 56, 183–193. [Google Scholar] [CrossRef]
- Cosgrove, C.L.; Southgate, V.R. Competitive mating interactions between Schistosoma haematobium and S. intercalatum (Lower Guinea strain). Parasitol. Res. 2003, 89, 238–241. [Google Scholar] [CrossRef]
- Webster, B.L.; Southgate, V.R. Mating interactions of Schistosoma haematobium and S. intercalatum with their hybrid offspring. Parasitology 2003, 126, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Tchuem Tchuenté, L.A.; Southgate, V.R.; Vercruysse, J.; Kaukas, A.; Kane, R.; Mulumba, M.P.; Pagès, J.R.; Jourdane, J. Epidemiological and genetic observations on human schistosomiasis in Kinshasa, Zaire. Trans. R. Soc. Trop. Med. Hyg. 1997, 91, 263–269. [Google Scholar] [CrossRef]
- Vercruysse, J.; Southgate, V.R.; Rollinson, D.; De Clercq, D.; Sacko, M.; De Bont, J.; Mungomba, L.M. Studies on transmission and schistosome interactions in Senegal, Mali and Zambia. Trop. Geogr. Med. 1994, 46, 220–226. [Google Scholar]
- Mintsa Nguema, R.; Mengue Ngou Milama, K.; Kombila, M.; Richard-Lenoble, D.; Tisseyre, P.; Ibikounlé, M.; Moné, H.; Mouahid, G. Morphometric and molecular characterizations of schistosome populations in Estuaire province Gabon. J. Helminthol. 2010, 84, 81–85. [Google Scholar] [CrossRef]
- Burchard, G.D.; Kern, P. Probable hybridization between S. intercalatum and S. haematobium in western Gabun. Trop. Geogr. Med. 1985, 37, 119–123. [Google Scholar] [PubMed]
- Zwingenberger, K.; Feldmeier, H.; Bienzle, U.; Steiner, A. Mixed Schistosoma haematobium/Schistosoma intercalatum infection. Ann. Trop. Med. Parasitol. 1990, 84, 85–87. [Google Scholar] [CrossRef]
- Moné, H.; Holtfreter, M.C.; Allienne, J.-F.; Mintsa-Nguéma, R.; Ibikounle, M.; Boissier, J.; Berry, A.; Mitta, G.; Richter, J.; Mouahid, G. Introgressive hybridizations of Schistosoma haematobium by Schistosoma bovis at the origin of the first case report of schistosomiasis in Corsica (France, Europe). Parasitol. Res. 2015, 114, 4127–4133. [Google Scholar] [CrossRef] [PubMed]
- Huyse, T.; Broeck, F.V.D.; Hellemans, B.; Volckaert, F.; Polman, K. Hybridisation between the two major African schistosome species of humans. Int. J. Parasitol. 2013, 43, 687–689. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, D.; Rollinson, D.; Diarra, A.; Sacko, M.; Coulibaly, G.; Landouré, A.; Traoré, M.; Southgate, V.R.; Kaukas, A.; Vercruysse, J. Schistosomiasis in Dogon country, Mali: Identification and prevalence of the species responsible for infection in the local community. Trans. R. Soc. Trop. Med. Hyg. 1994, 88, 653–656. [Google Scholar] [CrossRef]
- Picquet, M.; Ernould, J.C.; Vercruysse, J.; Southgate, V.R.; Mbaye, A.; Sambou, B.; Niang, M.; Rollinson, D. Royal Society of Tropical Medicine and Hygiene Meeting at Manson House, London, 18 May 1995. The epidemiology of human schistosomiasis in the Senegal river basin. Trans. R. Soc. Trop. Med. Hyg. 1996, 90, 340–346. [Google Scholar] [CrossRef]
- Southgate, V. Schistosomiasis in the Senegal River Basin: Before and after the construction of the dams at Diama, Senegal and Manantali, Mali and future prospects. J. Helminthol. 1997, 71, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Van den Broeck, F.; Maes, G.E.; Larmuseau, M.H.D.; Rollinson, D.; Sy, I.; Faye, D.; Volckaert, F.A.M.; Polman, K.; Huyse, T. Reconstructing colonization dynamics of the human parasite Schistosoma mansoni following anthropogenic environmental changes in Northwest Senegal. PLoS Negl. Trop. Dis. 2015, 9, e0003998. [Google Scholar] [CrossRef]
- Boon, N.A.; Mbow, M.; Paredis, L.; Moris, P.; Sy, I.; Maes, T.; Webster, B.; Sacko, M.; Volckaert, F.A.; Polman, K.; et al. No barrier breakdown between human and cattle schistosome species in the Senegal River Basin in the face of hybridisation. Int. J. Parasitol. 2019, 49, 1039–1048. [Google Scholar] [CrossRef]
- Platt, R.; McDew-White, M.; Le Clec’H, W.; Chevalier, F.; Allan, F.; Emery, A.M.; Garba, A.; Hamidou, A.A.; Ame, S.M.; Webster, J.P.; et al. Ancient Hybridization and Adaptive Introgression of an Invadolysin Gene in Schistosome Parasites. Mol. Biol. Evol. 2019, 36, 2127–2142. [Google Scholar] [CrossRef]
- Kruger, F.J.; Evans, A.C. Do all human urinary infections with Schistosoma mattheei Represent hybridization between S. haematobium and S. mattheei? J. Helminthol. 1990, 64, 330–332. [Google Scholar] [CrossRef]
- Léger, E.; Borlase, A.; Fall, C.B.; Diouf, N.D.; Diop, S.D.; Yasenev, L.; Catalano, S.; Thiam, C.T.; Ndiaye, A.; Emery, A.; et al. Prevalence and distribution of schistosomiasis in human, livestock, and snail populations in northern Senegal: A One Health epidemiological study of a multi-host system. Lancet Planet. Heal. 2020, 4, e330–e342. [Google Scholar] [CrossRef]
- Sene, M.; Marchand, B.; Rollinson, D.; Webster, B.L. Urogenital schistosomiasis and hybridization between Schistosoma haematobium and Schistosoma bovis in adults living in Richard-Toll, Senegal. Parasitology 2018, 145, 1723–1726. [Google Scholar] [CrossRef]
- Boissier, J.; Grech-Angelini, S.; Webster, B.; Allienne, J.-F.; Huyse, T.; Mas-Coma, S.; Toulza, E.; Barré-Cardi, H.; Rollinson, D.; Kincaid-Smith, J.; et al. Outbreak of urogenital schistosomiasis in Corsica (France): An epidemiological case study. Lancet Infect. Dis. 2016, 16, 971–979. [Google Scholar] [CrossRef]
- Holtfreter, M.C.; Moné, H.; Müller-Stöver, I.; Mouahid, G.; Richter, J. Schistosoma haematobium infections acquired in Corsica, France, August 2013. Eurosurveillance 2014, 19, 20821. [Google Scholar] [CrossRef] [PubMed]
- Moné, H.; Holtfreter, M.C.; Mouahid, G.; Richter, J. Difficulties in Schistosomiasis Assessment, Corsica, France. Emerg. Infect. Dis. 2016, 22, 762–765. [Google Scholar] [CrossRef]
- Berry, A.; Paris, L.; Boissier, J.; Caumes, E. Schistosomiasis Screening of Travelers to Corsica, France. Emerg. Infect. Dis. 2016, 22, 159. [Google Scholar] [CrossRef] [PubMed]
- Berry, A.; Moné, H.; Iriart, X.; Mouahid, G.; Abbo, O.; Boissier, J.; Fillaux, J.; Cassaing, S.; Debuisson, C.; Valentin, A.; et al. Schistosomiasis Haematobium, Corsica, France. Emerg. Infect. Dis. 2014, 20, 1595–1597. [Google Scholar] [CrossRef]
- Gautret, P.; Mockenhaupt, F.; Von Sonnenburg, F.; Rothe, C.; Libman, M.; Van De Winkel, K.; Bottieau, E.; Grobusch, M.P.; Hamer, D.; Esposito, D.H.; et al. Local and International Implications of Schistosomiasis Acquired in Corsica, France. Emerg. Infect. Dis. 2015, 21, 1865–1868. [Google Scholar] [CrossRef]
- Berry, A.; Fillaux, J.; Martin-Blondel, G.; Boissier, J.; Iriart, X.; Marchou, B.; Magnaval, J.F.; Delobel, P. Evidence for a permanent presence of schistosomiasis in Corsica, France, 2015. Eurosurveillance 2016, 21. [Google Scholar] [CrossRef]
- Oleaga, A.; Rey, O.; Polack, B.; Grech-Angelini, S.; Quilichini, Y.; Pérez-Sánchez, R.; Boireau, P.; Mulero, S.; Brunet, A.; Rogon, A.; et al. Epidemiological surveillance of schistosomiasis outbreak in Corsica (France): Are animal reservoir hosts implicated in local transmission? PLoS Negl. Trop. Dis. 2019, 13, e0007543. [Google Scholar] [CrossRef] [PubMed]
- Boissier, J.; Moné, H.; Mitta, G.; Bargues, M.D.; Molyneux, D.; Mas-Coma, S. Schistosomiasis reaches Europe. Lancet Infect. Dis. 2015, 15, 757–758. [Google Scholar] [CrossRef]
- Bisoffi, Z.; Buonfrate, D.; Beltrame, A. Schistosomiasis transmission in Europe. Lancet Infect. Dis. 2016, 16, 878–880. [Google Scholar] [CrossRef]
- Duplantier, J.; Sène, M. Rodents as reservoir hosts in the transmission of Schistosoma mansoni in Richard-Toll, Senegal, West Africa. J. Helminthol. 2000, 74, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Savassi, B.A.E.S.; Dobigny, G.; Etougbétché, J.R.; Avocegan, T.T.; Quinsou, F.T.; Gauthier, P.; Ibikounlé, M.; Moné, H.; Mouahid, G. Mastomys natalensis (Smith, 1834) as a natural host for Schistosoma haematobium (Bilharz, 1852) Weinland, 1858 × Schistosoma bovis Sonsino, 1876 introgressive hybrids. Parasitol. Res. 2021, 120, 1755–1770. [Google Scholar] [CrossRef] [PubMed]
- Savassi, B.A.; Mouahid, G.; Lasica, C.; Mahaman, S.-D.K.; Garcia, A.; Courtin, D.; Allienne, J.-F.; Ibikounlé, M.; Moné, H. Cattle as natural host for Schistosoma haematobium (Bilharz, 1852) Weinland, 1858 × Schistosoma bovis Sonsino, 1876 interactions, with new cercarial emergence and genetic patterns. Parasitol. Res. 2020, 119, 2189–2205. [Google Scholar] [CrossRef]
- Djuikwo-Teukeng, F.F.; Simo, A.K.; Allienne, J.-F.; Rey, O.; Ngapagna, A.N.; Tchuem-Tchuente, L.A.; Boissier, J. Population genetic structure of Schistosoma bovis in Cameroon. Parasites Vectors 2019, 12, 56. [Google Scholar] [CrossRef]
- Rothe, C.; Zimmer, T.; Schunk, M.; Wallrauch, C.; Helfrich, K.; Gültekin, F.; Bretzel, G.; Allienne, J.F.; Boissier, J. Developing endemicity of schistosomiasis, Corsica, France. Emerg. Infect. Dis. 2021, 27. [Google Scholar] [CrossRef]
- Mulero, S.; Rey, O.; Arancibia, N.; Mas-Coma, S.; Boissier, J. Persistent establishment of a tropical disease in Europe: The preadaptation of schistosomes to overwinter. Parasites Vectors 2019, 12, 379–410. [Google Scholar] [CrossRef]
- Oey, H.; Zakrzewski, M.; Gravermann, K.; Young, N.D.; Korhonen, P.K.; Gobert, G.N.; Nawaratna, S.; Hasan, S.; Martínez, D.M.; You, H.; et al. Whole-genome sequence of the bovine blood fluke Schistosoma bovis supports interspecific hybridization with S. haematobium. PLoS Pathog. 2019, 15, e1007513. [Google Scholar] [CrossRef]
- Angora, E.K.; Allienne, J.F.; Rey, O.; Menan, H.; Touré, A.O.; Coulibaly, J.T.; Raso, G.; Yavo, W.; N’Goran, E.K.; Utzinger, J.; et al. High prevalence of Schistosoma haematobium × Schistosoma bovis hybrids in schoolchildren in Cote d’Ivoire. Parasitology 2019, 147, 287–294. [Google Scholar] [CrossRef]
- Webster, B.L.; Alharbi, M.H.; Kayuni, S.; Makaula, P.; Halstead, F.; Christiansen, R.; Juziwelo, L.; Stanton, M.C.; LaCourse, J.; Rollinson, D.; et al. Schistosome Interactions within the Schistosoma haematobium Group, Malawi. Emerg. Infect. Dis. 2019, 25, 1245–1247. [Google Scholar] [CrossRef]
- Pitchford, R. Observations on a possible hybrid between the two schistosomes S. haematobium and S. mattheei. Trans. R. Soc. Trop. Med. Hyg. 1961, 55, 44–51. [Google Scholar] [CrossRef]
- Van Wyk, J.A. The importance of animals in human schistosomiasis in South Africa. S. Afr. Med. J. 1983, 63, 201–203. [Google Scholar] [PubMed]
- Kruger, F.J.; Schutte, C.H.; Visser, P.S.; Evans, A.C. Phenotypic differences in Schistosoma mattheei ova from populations sympatric and allopatric to S. haematobium. Onderstepoort J. Vet. Res. 1986, 53, 103–107. [Google Scholar] [PubMed]
- Kruger, F.J.; Hamilton-Attwell, V.L. Scanning electron microscope studies of miracidia suggest introgressive hybridization between Schistosoma haematobium and S. haematobium × S. mattheei in the Eastern Transvaal. J. Helminthol. 1988, 62, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Cunin, P.; Tchuenté, L.-A.T.; Poste, B.; Djibrilla, K.; Martin, P.M.V. Interactions between Schistosoma haematobium and Schistosoma mansoni in humans in north Cameroon. Trop. Med. Int. Health 2003, 8, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Meurs, L.; Mbow, M.; Vereecken, K.; Menten, J.; Mboup, S.; Polman, K. Epidemiology of mixed Schistosoma mansoni and Schistosoma haematobium infections in northern Senegal. Int. J. Parasitol. 2012, 42, 305–311. [Google Scholar] [CrossRef]
- Le Govic, Y.; Kincaid-Smith, J.; Allienne, J.F.; Rey, O.; de Gentile, L.; Boissier, J. Schistosoma haematobium-Schistosoma mansoni hybrid parasite in migrant boy, france, 2017. Emerg. Infect. Dis. 2019, 25, 365–367. [Google Scholar] [CrossRef]
- Norton, A.J.; Webster, J.P.; Kane, R.A.; Rollinson, D. Inter-specific parasite competition: Mixed infections of Schistosoma mansoni and S. rodhaini in the definitive host. Parasitology 2008, 135, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Thèron, A. Hybrids between Schistosoma mansoni and S. rodhaini: Characterization by cercarial emergence rhythms. Parasitology 1989, 99, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Simões, M.; Bahia, D.; Zerlotini, A.; Torres, K.; Artiguenave, F.; Neshich, G.; Kuser, P.; Oliveira, G. Single nucleotide polymorphisms identification in expressed genes of Schistosoma mansoni. Mol. Biochem. Parasitol. 2007, 154, 134–140. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ramos, C.R.R.; Figueredo, R.C.R.; Pertinhez, T.A.; Vilar, M.M.; Nascimento, A.L.T.O.D.; Tendler, M.; Raw, I.; Spisni, A.; Ho, P.L. Gene Structure and M20T Polymorphism of the Schistosoma mansoni Sm14 Fatty Acid-binding Protein. J. Biol. Chem. 2003, 278, 12745–12751. [Google Scholar] [CrossRef]
- Fukushige, M.; Mutapi, F.; Woolhouse, M.E. Population level changes in schistosome-specific antibody levels following chemotherapy. Parasite Immunol. 2019, 41, e12604. [Google Scholar] [CrossRef]
- McManus, D.P.; Bergquist, R.; Cai, P.; Ranasinghe, S.; Tebeje, B.M.; You, H. Schistosomiasis—From immunopathology to vaccines. Semin. Immunopathol. 2020, 42, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Panzner, U.; Excler, J.L.; Kim, J.H.; Marks, F.; Carter, D.; Siddiqui, A.A. Recent advances and methodological considerations on vaccine candidates for human schistosomiasis. Front. Trop. Dis. 2021. submitted. [Google Scholar]
- Tendler, M.; Almeida, M.S.; Vilar, M.M.; Pinto, P.M.; Limaverde-Sousa, G. Current Status of the Sm14/GLA-SE Schistosomiasis Vaccine: Overcoming Barriers and Paradigms towards the First Anti-Parasitic Human (itarian) Vaccine. Trop. Med. Infect. Dis. 2018, 3, 121. [Google Scholar] [CrossRef]
- Ramos, C.R.R.; Spisni, A.; Oyama, S.; Sforça, M.; Ramos, H.R.; Vilar, M.M.; Alves, A.C.; Figueredo, R.C.R.; Tendler, M.; Zanchin, N.; et al. Stability improvement of the fatty acid binding protein Sm14 from S. mansoni by Cys replacement: Structural and functional characterization of a vaccine candidate. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 655–662. [Google Scholar] [CrossRef]
- Riveau, G.; Schacht, A.-M.; Dompnier, J.-P.; Deplanque, D.; Seck, M.; Waucquier, N.; Senghor, S.; Delcroix-Genete, D.; Hermann, E.; Idris-Khodja, N.; et al. Safety and efficacy of the rSh28GST urinary schistosomiasis vaccine: A phase 3 randomized, controlled trial in Senegalese children. PLOS Neglected Trop. Dis. 2018, 12, e0006968. [Google Scholar] [CrossRef]
- Medicine/ClinicalTrials.gov USNLo. Efficacy and Safety Evaluation of the Therapeutic Vaccine Candidate Sh28GST in Association with Praziquantel (PZQ) for Prevention of Clinical and Parasitological Recurrences of S. haematobium Infection in Children [NCT00870649]. 2012. Available online: https://clinicaltrials.gov/ct2/show/NCT00870649?term=sh28GST&cond=Schistosomiasis&draw=2&rank=1 (accessed on 19 June 2021).
- Medicine/ClinicalTrials.gov USNLo. Safety and Immunogenicity Evaluation of the Vaccine Candidate Sm14 in Combination with the Adjuvant Glucopyranosyl Lipid A (GLA-SE) in Adults Living in Endemic Regions for S. Mansoni and S. haematobium in Senegal. A Comparative, Randomized, Open-Label Trial [NCT03041766]. 2016. Available online: https://clinicaltrials.gov/ct2/show/NCT03041766?term=Sm14&cond=Schistosomiasis&rank=3 (accessed on 19 June 2021).
- Medicine/ClinicalTrials.gov USNLo. Safety and Immunogenicity Evaluation of the Vaccine Candidate Sm14 against Schistosomiasis in Senegalese School Children Healthy or Infected with S. Mansoni and/or S. haematobium. A Comparative, Randomized, Controlled, Open-label Trial [NCT03799510]. 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT03799510?term=Sm14&cond=Schistosomiasis&rank=2 (accessed on 19 June 2021).
- Hughes, A.L. Rates of amino acid evolution in the 26- and 28-kDa glutathione S-transferases of Schistosoma. Mol. Biochem. Parasitol. 1993, 58, 43–52. [Google Scholar] [CrossRef]
- Philippsen, G.S.; Wilson, R.A.; DeMarco, R. Accelerated evolution of schistosome genes coding for proteins located at the host-parasite interface. Genome Biol. Evol. 2015, 7, 431–443. [Google Scholar] [CrossRef]
- Hughes, A. Conserved proteins as immunogens: Glutathione S-transferase of Schistosoma. Parasitol. Today 1994, 10, 149–151. [Google Scholar] [CrossRef]
- Molehin, A.J.; Sennoune, S.R.; Zhang, W.; Rojo, J.U.; Siddiqui, A.J.; Herrera, K.A.; Johnson, L.; Sudduth, J.; May, J.; Siddiqui, A.A. Cross-species prophylactic efficacy of Sm-p80-based vaccine and intracellular localization of Sm-p80/Sm-p80 ortholog proteins during development in Schistosoma mansoni, Schistosoma japonicum, and Schistosoma haematobium. Parasitol. Res. 2017, 116, 3175–3188. [Google Scholar] [CrossRef]
- Karmakar, S.; Zhang, W.; Ahmad, G.; Torben, W.; Alam, M.U.; Le, L.; Damian, R.T.; Wolf, R.F.; White, G.L.; Carey, D.W.; et al. Cross-species protection: Schistosoma mansoni Sm-p80 vaccine confers protection against Schistosoma haematobium in hamsters and baboons. Vaccine 2014, 32, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Medicine/ClinicalTrials.gov USNLo. Sm-TSP-2 Schistosomiasis Vaccine in Healthy Ugandan Adults [NCT03910972]. 2019. Available online: https://clinicaltrials.gov/ct2/show/NCT03910972?term=TSP&cond=Schistosomiasis&rank=2 (accessed on 19 June 2021).
- Pitchford, R.J. A check list of definitive hosts exhibiting evidence of the genus Schistosoma Weinland, 1858 acquired naturally in Africa and the Middle East. J. Helminthol. 1977, 51, 229–251. [Google Scholar] [CrossRef] [PubMed]
- Rollinson, D.; Southgate, V.R. The genus Schistosoma: A taxonomic appraisal. In The Biology of Schistosomesl; Academic Press: Cambridge, MA, USA, 1987; pp. 1–26. [Google Scholar]
- Pitchford, R.J.; Visser, P.S. The role of naturally infected wild rodents in the epidemiology of schistosomiasis in the Eastern Transvaal. Trans. R. Soc. Trop. Med. Hyg. 1962, 56, 126–135. [Google Scholar] [CrossRef]
- Rey, L. Non-Human vertebrate hosts of Schistosoma mansoni and schistosomiaisis transmission in Brazil. Res. Rev. Parasitol. 1992, 53, 13–25. [Google Scholar]
- McMahon, J.E.; Baalawy, S.S. A search for animal reservoirs of Schistosoma mansoni in the Mwanza area of Tanzania. East. Afr. Med. J. 1967, 44, 325–326. [Google Scholar]
- Lapierre, J.; Tourte-Schaefer, C.; Dupouy-Camet, J.; Cot, M.; Heyer, F.; Faurant, C. Complement to the epidemiologic study of the focus of Schistosoma mansoni bilharziasis in Kara (northern Togo). Bull. Société de Pathol. Exot. 1992, 85, 232–237. [Google Scholar]
- Dias, L.; Ávila-Pires, F.; Pinto, A. Parasitological and ecological aspects of schistosomiasis mansoni in the valley of the Paraíba do Sul river (São Paulo State, Brazil) I. Natural infection of small mammals with Schistosoma mansoni. Trans. R. Soc. Trop. Med. Hyg. 1978, 72, 496–500. [Google Scholar] [CrossRef]
- Silva, T.M.; Andrade, Z.A. Natural infection of wild rodents by Schistosoma mansoni. Mem. Inst. Oswaldo Cruz 1989, 84, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Mac Pherson, C.N.L.; Craig, P.S. Parasitic Helminths and Zoonoses in Africa; Unwin Hyman: London, UK, 1991; p. 270. [Google Scholar]
- Nelson, G.S.; Teesdale, C.; Highton, R.B. The role of animals as reservoirs in Africa. In CIBA Foundation Symposium on Bilharziasis; Wolstenhome, G.E.W., O’Connor, M., Eds.; Churchill: London, UK, 1962; pp. 149–227. [Google Scholar]
- Zahed, N.Z.; Ghandour, A.M.; Banaja, A.A.; Banerjee, R.K.; Dehlawi, M.S. Hamadryas baboons Papio hamadryas as maintenance hosts of Schistosoma mansoni in Saudi Arabia. Trop. Med. Int. Health 1996, 1, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Hsü, S.Y.; Hsü, H.F.; Majumdar, A.N.; Wadsworth, G. A chimpanzee naturally infected with Schistosoma mansoni; Its resistance against a challenge infection of, s. japonicum. Trans. R. Soc. Trop. Med. Hyg. 1968, 62, 901–902. [Google Scholar] [CrossRef]
- Rollinson, D.; Imbert-Establet, D.; Ross, G.C. Schistosoma mansoni from naturally infected Rattus rattus in Guadeloupe: Identification, prevalence and enzyme polymorphism. Parasitology 1986, 93, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, M.C. Some observations on schistosomiasis in Jabalpur area. Livestock Adviser Bangalore 1981, 6, 53–55. [Google Scholar]
- Agrawal, M.C.; Shah, H.L. A review on Schistosoma incognitum Chandler 1926. Helminthol. Abstr. 1989, 58, 239–251. [Google Scholar]
- Banerjee, P.S.; Agrawal, M.C. Prevalence of Schistosoma nasale Rao 1933 in Jabalpur. Indian J. Anim. Sci. 1991, 61, 789–791. [Google Scholar]
- Agrawal, M.C.; Alwar, G.S. Nasal schistosomiasis: A review. Helminthol. Abstracts 1992, 61, 373–384. [Google Scholar]
- Hiregoudar, L.S. Spirometra and Schistosoma infection among lions of Gir forest. India Curr. Res. 1975, 4, 134–135. [Google Scholar]
- Singh, K.I.; Krishnasamy, M.; Ambu, S.; Rasul, R.; Chong, N.L. Studies on animal schistosomes in Peninsular Malaysia: Record of naturally infected animals and additional hosts of Schistosoma spindale. Southeast Asian J. Trop. Med. Public Heal. 1997, 28, 303–307. [Google Scholar]
- Jordan, P.; Webbe, G.; Sturrock, R.F. Human Schistosomiasis; CAB International: Oxford, UK, 1993; p. 442. [Google Scholar]
- He, Y.-X.; Salafsky, B.; Ramaswamy, K. Host–parasite relationships of Schistosoma japonicum in mammalian hosts. Trends Parasitol. 2001, 17, 320–324. [Google Scholar] [CrossRef]
- Jordan, P.; Webbe, G. Schistosomiasis. Epidemiology, Treatment and Control; William Heinemann Medical Books: London, UK, 1982; p. 345. [Google Scholar]
- Xia, M. Contribution à l’étude du développement et de la variabilité génétique de Schistosoma japonicum. Ph.D. Thesis, Université de Perpignan, Perpignan, France, 1990; p. 243. [Google Scholar]
- Kumar, V.; de Burbure, G. Schistosomes of animals and man in Asia. Helminthol. Abstr. 1986, 55, 474–480. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panzner, U.; Boissier, J. Natural Intra- and Interclade Human Hybrid Schistosomes in Africa with Considerations on Prevention through Vaccination. Microorganisms 2021, 9, 1465. https://doi.org/10.3390/microorganisms9071465
Panzner U, Boissier J. Natural Intra- and Interclade Human Hybrid Schistosomes in Africa with Considerations on Prevention through Vaccination. Microorganisms. 2021; 9(7):1465. https://doi.org/10.3390/microorganisms9071465
Chicago/Turabian StylePanzner, Ursula, and Jerome Boissier. 2021. "Natural Intra- and Interclade Human Hybrid Schistosomes in Africa with Considerations on Prevention through Vaccination" Microorganisms 9, no. 7: 1465. https://doi.org/10.3390/microorganisms9071465
APA StylePanzner, U., & Boissier, J. (2021). Natural Intra- and Interclade Human Hybrid Schistosomes in Africa with Considerations on Prevention through Vaccination. Microorganisms, 9(7), 1465. https://doi.org/10.3390/microorganisms9071465





