Use of Oxidative Stress Responses to Determine the Efficacy of Inactivation Treatments on Cryptosporidium Oocysts
Abstract
1. Introduction
2. Materials and Methods
2.1. Parasites
2.2. Experimental Setup
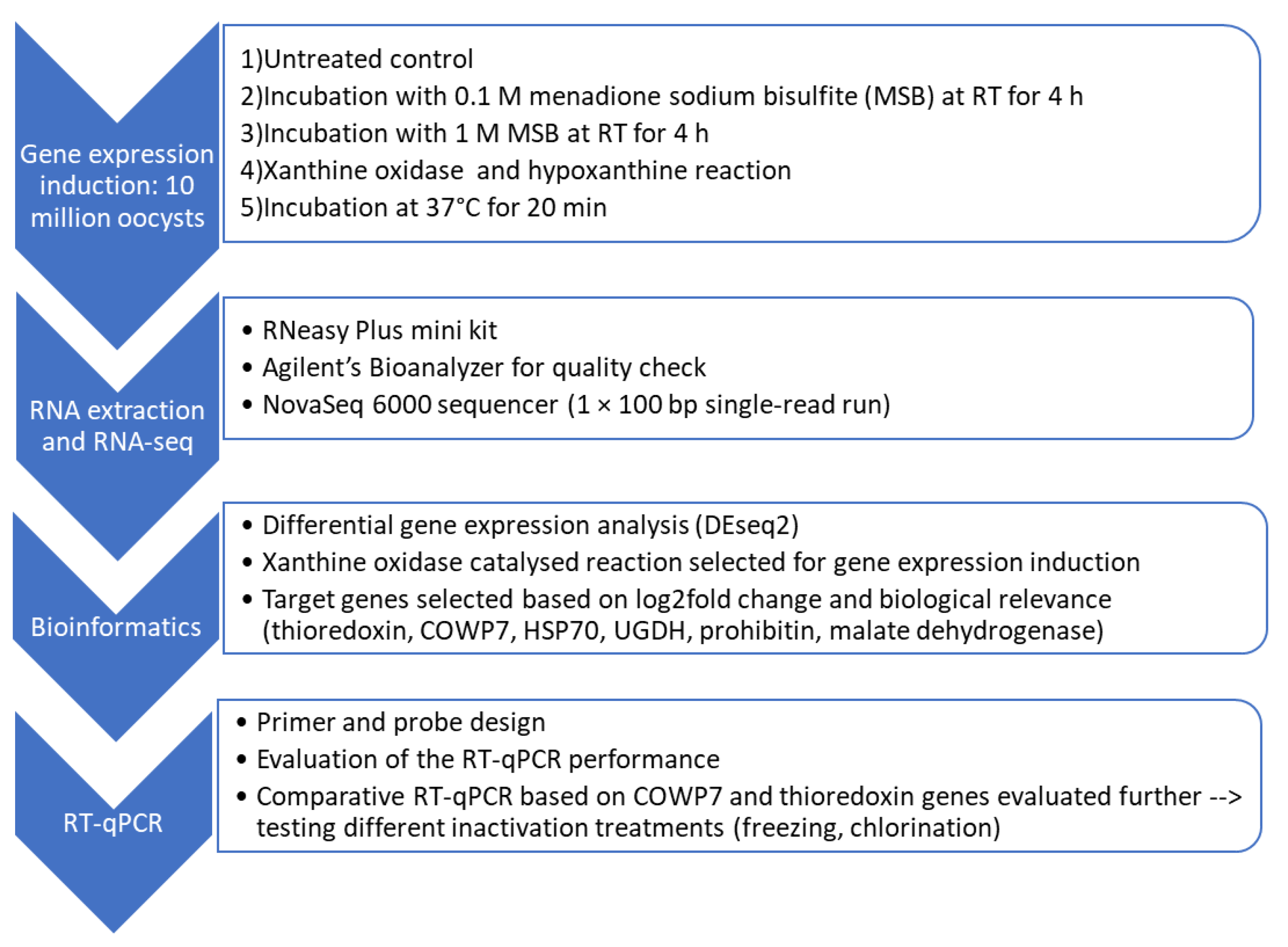
2.2.1. Identification of Inducible Target Genes Using RNA-Seq and DEG Analysis
Induction of Upregulation of Gene Expression
RNA Extraction and Quality Assessment for RNA-Seq
RNA Sequencing (RNA-Seq)
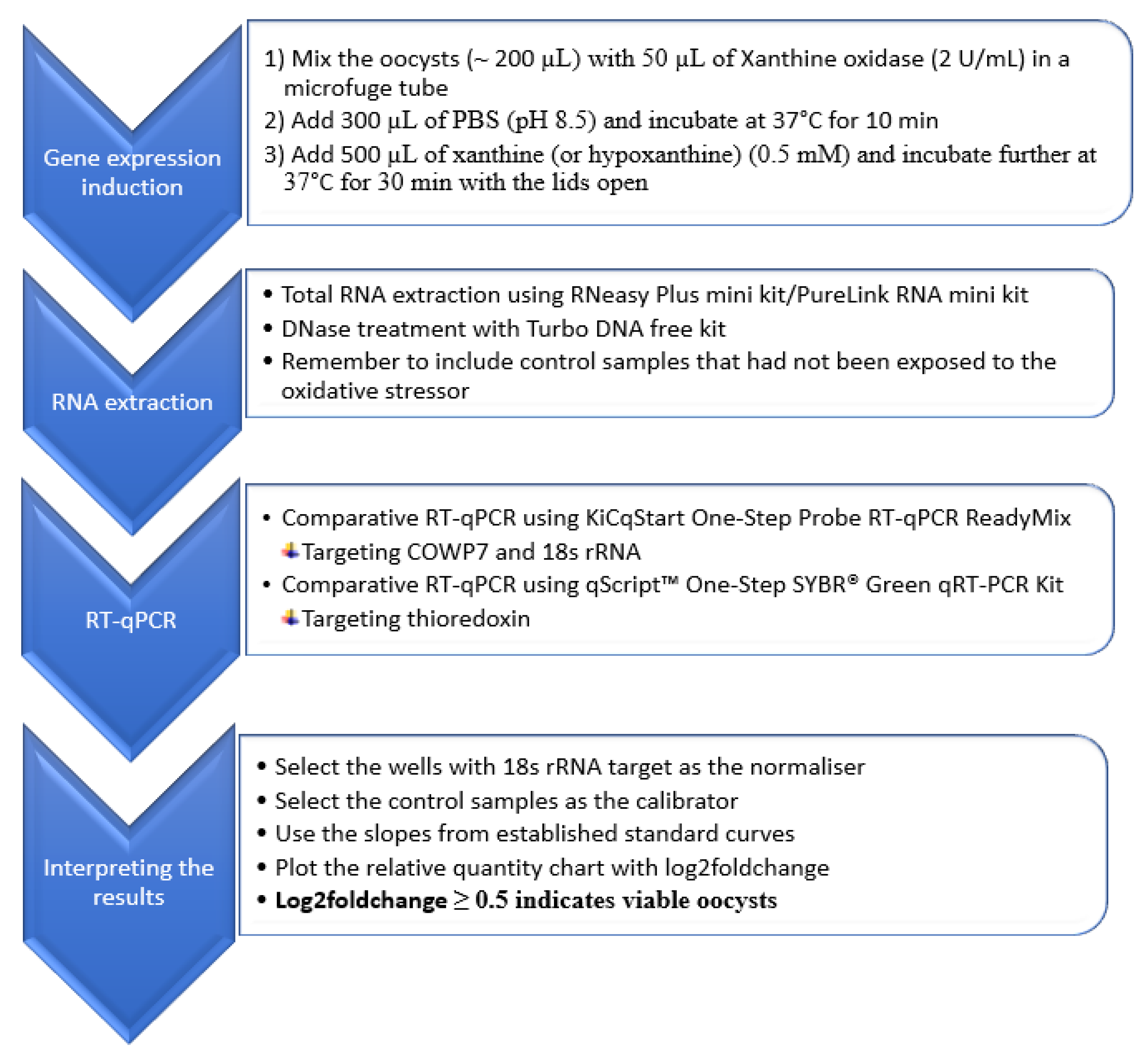
2.2.2. RT-qPCR Method Development
Target Genes and Primers
RNA Extraction for the RT-qPCR Method
Reverse Transcription Quantitative PCR (RT-qPCR)
2.2.3. Evaluation of the RT-qPCR Method
2.2.3.1. Inactivation of Cryptosporidium Oocysts
2.2.3.2. Distinguishing between Viable and Inactivated Oocysts Using the New RT-qPCR Method
2.3. Statistical Analysis
2.3.1. Data Pre-Processing and Mapping
2.3.2. Differential Gene Expression Analysis
3. Results
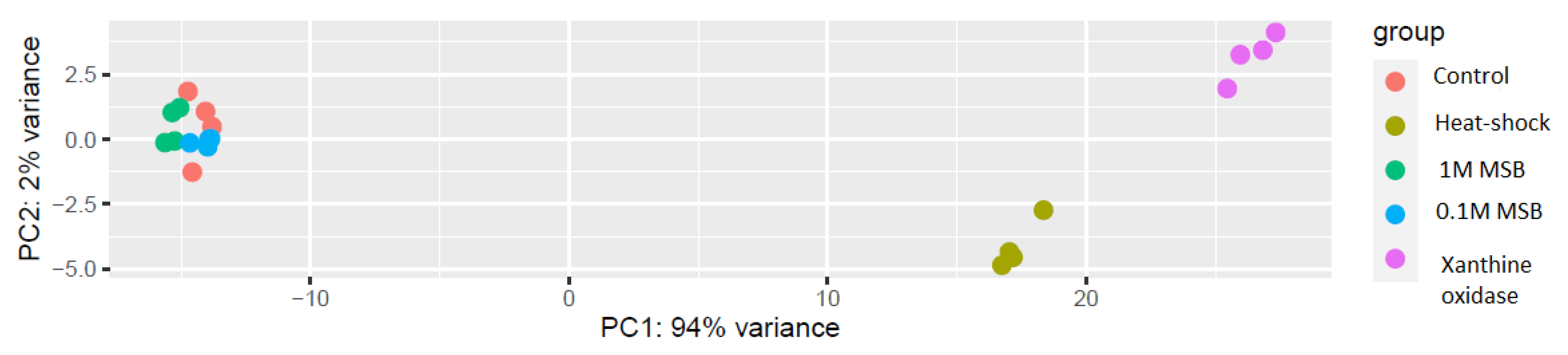
3.1. RNA-Seq Analysis
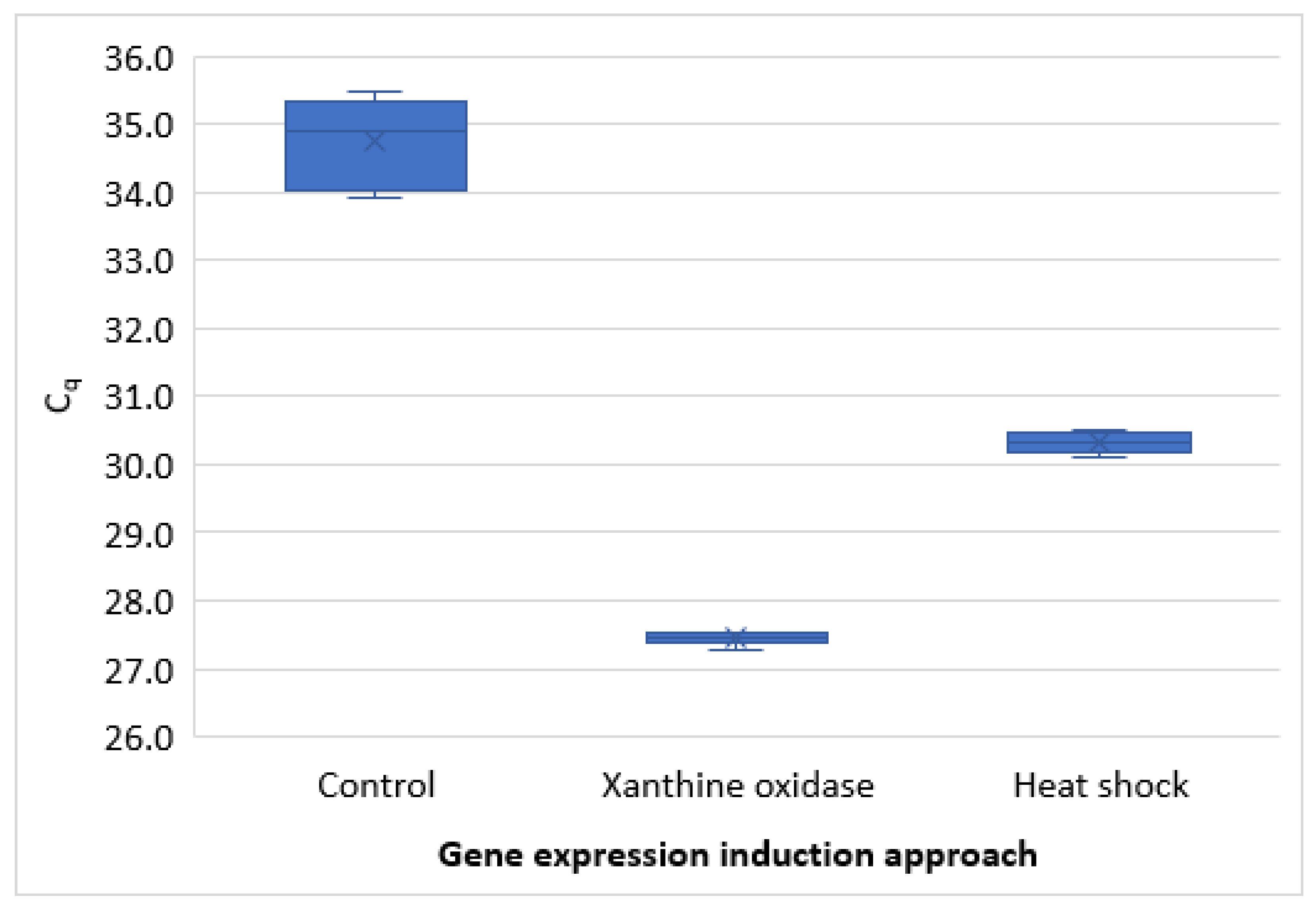
3.2. RT-qPCR Method Development
3.3. Evaluation of the RT-qPCR Method
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gerba, C.P. Environmentally transmitted pathogens. Environ. Microbiol. 2015, 509–550. [Google Scholar] [CrossRef]
- Craighead, S.; Huang, R.; Chen, H.; Kniel, K.E. The Use of Pulsed Light to Inactivate Cryptosporidium parvum Oocysts on High-Risk Commodities (Cilantro, Mesclun Lettuce, Spinach, and Tomatoes). Food Control 2021, 126, 107965. [Google Scholar] [CrossRef]
- Gérard, C.; Franssen, F.; La Carbona, S.; Monteiro, S.; Cozma-Petruţ, A.; Utaaker, K.S.; Režek Jambrak, A.; Rowan, N.; Rodríguez-Lazaro, D.; Nasser, A.; et al. Inactivation of parasite transmission stages: Efficacy of treatments on foods of non-animal origin. Trends Food Sci. Technol. 2019, 91, 12–23. [Google Scholar] [CrossRef]
- Gibson, K.E.; Almeida, G.; Jones, S.L.; Wright, K.; Lee, J.A. Inactivation of bacteria on fresh produce by batch wash ozone sanitation. Food Control 2019, 106, 106747. [Google Scholar] [CrossRef]
- Pan, H.A.O.; Buenconsejo, M.; Reineke, K.F.; Shieh, Y.C. Effect of process temperature on virus inactivation during high hydrostatic pressure processing of contaminated fruit puree and juice. J. Food Prot. 2016, 79, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Morgan, U.; Upton, S.J. Epidemiology of Cryptosporidium: Transmission, detection and identification. Int. J. Parasitol. 2000, 30, 1305–1322. [Google Scholar] [CrossRef]
- Bouzid, M.; Hunter, P.R.; Chalmers, R.M.; Tyler, K.M. Cryptosporidium pathogenicity and virulence. Clin. Microbiol. Rev. 2013, 26, 115–134. [Google Scholar] [CrossRef]
- Gharpure, R.; Perez, A.; Miller, A.D.; Wikswo, M.E.; Silver, R.; Hlavsa, M.C. Cryptosporidiosis Outbreaks—United States, 2009–2017. Morb. Mortal. Wkly. Rep. 2019, 68, 568–572. [Google Scholar] [CrossRef]
- Ethelberg, S.; Lisby, M.; Vestergaard, L.S.; Enemark, H.L.; Olsen, K.E.P.; Stensvold, C.R.; Nielsen, H.V.; Porsbo, L.J.; Plesner, A.M.; MØLbak, K. A foodborne outbreak of Cryptosporidium hominis infection. Epidemiol. Infect. 2009, 137, 348–356. [Google Scholar] [CrossRef]
- Pönka, A.; Kotilainen, H.; Rimhanen-Finne, R.; Hokkanen, P.; Hänninen, M.L.; Kaarna, A.; Meri, T.; Kuusi, M. A foodborne outbreak due to Cryptosporidium parvum in Helsinki, November 2008. Eurosurveillance 2009, 14, 19269. [Google Scholar] [CrossRef][Green Version]
- Rousseau, A.; Carbona, S.L.; Dumètre, A.; Robertson, L.J.; Gargala, G.; Escotte-Binet, S.; Favennec, L.; Villena, I.; Gérard, C.; Aubert, D. Methods to assess viability and infectivity of Giardia duodenalis cysts, and Cryptosporidium spp., and Toxoplasma gondii oocysts: A review of the status. Parasite 2018, 25. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.L.; Arrowood, M.J. Cell Culture Infectivity to Assess Chlorine Disinfection of Cryptosporidium Oocysts in Water. In Cryptosporidium: Methods and Protocols; Mead, J.R., Arrowood, M.J., Eds.; Springer: New York, NY, USA, 2020; pp. 283–302. [Google Scholar] [CrossRef]
- Murphy, J.L.; Arrowood, M.J.; Lu, X.; Hlavsa, M.C.; Beach, M.J.; Hill, V.R. Effect of cyanuric acid on the inactivation of Cryptosporidium parvum under hyperchlorination conditions. Environ. Sci. Technol. 2015, 49, 7348–7355. [Google Scholar] [CrossRef]
- Shin, G.A.; Linden, K.G.; Arrowood, M.J.; Sobsey, M.D. Low-pressure UV inactivation and DNA repair potential of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 2001, 67, 3029–3032. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, J.L.; Slawson, R.M.; Huck, P.M. Inactivation and potential repair of Cryptosporidium parvum following low- and medium-pressure ultraviolet irradiation. Water Res. 2003, 37, 3517–3523. [Google Scholar] [CrossRef]
- Morita, S.; Namikoshi, A.; Hirata, T.; Oguma, K.; Katayama, H.; Ohgaki, S.; Motoyama, N.; Fujiwara, M. Efficacy of UV irradiation in inactivating Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 2002, 68, 5387–5393. [Google Scholar] [CrossRef]
- Yanta, C.A.; Bessonov, K.; Robinson, G.; Troell, K.; Guy, R.A. CryptoGenotyper: A new bioinformatics tool for rapid Cryptosporidium identification. Food Waterborne Parasitol. 2021, 23, e00115. [Google Scholar] [CrossRef]
- UKWIR. Cryptosporidium: Enhancing the Water Industry’s Capability to Respond; Report Ref. No. 20/DW/06/22; UKWIR Limited: Westminster, UK; London, UK, 2020. [Google Scholar]
- Fayer, R.; Nerad, T. Effects of low temperatures on viability of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 1996, 62, 1431–1433. [Google Scholar] [CrossRef]
- Travaillé, E.; La Carbona, S.; Gargala, G.; Aubert, D.; Guyot, K.; Dumètre, A.; Villena, I.; Houssin, M. Development of a qRT-PCR method to assess the viability of Giardia intestinalis cysts, Cryptosporidium spp. and Toxoplasma gondii oocysts. Food Control 2016, 59, 359–365. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Aziz, N.; Jamil, R. Biochemistry, Xanthine Oxidase. Available online: https://www.ncbi.nlm.nih.gov/books/NBK545245/ (accessed on 12 March 2021).
- Lippuner, C.; Ramakrishnan, C.; Basso, W.U.; Schmid, M.W.; Okoniewski, M.; Smith, N.C.; Hässig, M.; Deplazes, P.; Hehl, A.B. RNA-Seq analysis during the life cycle of Cryptosporidium parvum reveals significant differential gene expression between proliferating stages in the intestine and infectious sporozoites. Int. J. Parasitol. 2018, 48, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Matos, L.V.S.; McEvoy, J.; Tzipori, S.; Bresciani, K.D.S.; Widmer, G. The transcriptome of Cryptosporidium oocysts and intracellular stages. Sci. Rep. 2019, 9, 7856. [Google Scholar] [CrossRef] [PubMed]
- Tandel, J.; English, E.D.; Sateriale, A.; Gullicksrud, J.A.; Beiting, D.P.; Sullivan, M.C.; Pinkston, B.; Striepen, B. Life cycle progression and sexual development of the apicomplexan parasite Cryptosporidium parvum. Nat. Microbiol. 2019, 4, 2226–2236. [Google Scholar] [CrossRef]
- Templeton, T.J.; Lancto, C.A.; Vigdorovich, V.; Liu, C.; London, N.R.; Hadsall, K.Z.; Abrahamsen, M.S. The Cryptosporidium oocyst wall protein is a member of a multigene family and has a homolog in Toxoplasma. Infect. Immun. 2004, 72, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Nishinaka, Y.; Masutani, H.; Nakamura, H.; Yodoi, J. Regulatory roles of thioredoxin in oxidative stress-induced cellular responses. Redox Rep. 2001, 6, 289–295. [Google Scholar] [CrossRef]
- Bajszár, G.; Dekonenko, A. Stress-induced Hsp70 Gene expression and inactivation of Cryptosporidium parvum oocysts by chlorine-based oxidants. Appl. Environ. Microbiol. 2010, 76, 1732. [Google Scholar] [CrossRef]
- Garcés-Sanchez, G.; Wilderer, P.A.; Horn, H.; Munch, J.-C.; Lebuhn, M. Assessment of the viability of Cryptosporidium parvum oocysts with the induction ratio of hsp70 mRNA production in manure. J. Microbiol. Methods 2013, 94, 280–289. [Google Scholar] [CrossRef]
- Lee, S.-U.; Joung, M.; Ahn, M.-H.; Huh, S.; Song, H.; Park, W.-Y.; Yu, J.-R. CP2 gene as a useful viability marker for Cryptosporidium parvum. Parasitol. Res. 2008, 102, 381–387. [Google Scholar] [CrossRef]
- Chambers, C.D. Development and application of a molecular viability assay for Cryptosporidium parvum based on heat shock protein 70 gene expression: A thesis presented in partial fulfilment of the requirements for the degree of Master of Science in Microbiology at Massey University. Ph.D. Thesis, Massey University, Auckland, New Zealand, 2005. [Google Scholar]
- Widmer, G.; Orbacz, E.A.; Tzipori, S. Beta-tubulin mRNA as a marker of Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 1999, 65, 1584–1588. [Google Scholar] [CrossRef] [PubMed]
- Chauret, C.P.; Radziminski, C.Z.; Lepuil, M.; Creason, R.; Andrews, R.C. Chlorine dioxide inactivation of Cryptosporidium parvum oocysts and bacterial spore indicators. Appl. Environ. Microbiol. 2001, 67, 2993–3001. [Google Scholar] [CrossRef] [PubMed]
- Korich, D.G.; Mead, J.R.; Madore, M.S.; Sinclair, N.A.; Sterling, C.R. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 1990, 56, 1423–1428. [Google Scholar] [CrossRef] [PubMed]
| Treatment | Brief Description |
|---|---|
| 1 M MSB | 200 µL of oocysts (ca. 10 million) were mixed with 500 µL of 1 M MSB, vortexed, then held at room temperature for 4 h. The suspension was then washed 3 times with water before total RNA extraction |
| 0.1 M MSB | 200 µL of oocysts (ca. 10 million) were mixed with 500 µL of 0.1 M MSB, vortexed, then held at room temperature for 4 h. The suspension was then washed 3 times with water before total RNA extraction |
| Xanthine oxidase and hypoxanthine | 200 µL of oocysts (ca. 10 million) were vortexed and 50 µL of xanthine oxidase (20 U/mL) added to the suspension. The suspension was brought up to 500 µL with PBS (pH 8.5) and incubated at 37 °C for 10 min. Thereafter, 500 µL of 0.5 mM hypoxanthine was added to the mixture, briefly vortexed and further incubated at 37 °C for 30 min with the lids open. The sample was then washed 3 times with water before total RNA extraction. |
| Heat shock | 200 µL of oocysts (ca. 10 million) were incubated at 37 °C for 20 min. The sample was then washed 3 times with water before total RNA extraction. |
| Heat shock | 200 µL of oocysts (ca. 10 million) were incubated at 45 °C for 20 min. The sample was then washed 3 times with water before total RNA extraction. |
| Control | 200 µL of oocysts (ca. 10 million) stored at refrigeration temperature were washed five times with water before total RNA extraction. |
| Target Gene | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) | Probe (5′ → 3′) | Product Size (bp) | Locus Tag (Ref.) |
|---|---|---|---|---|---|
| COWP7 | CTATGGGATTCAATTTCGAAGTTCC | CCCAATACAAAATCTGCTACTTCCA | ATGGAATATCATCATCCCCTCAGCAA | 97 | cgd4_500 |
| Thioredoxin | GAAAAGCTGAACCTCGCATTCG | CGTCCCGTGGTCAATGCAATAA | NA | 134 | cgd7_4080 |
| Prohibitin | CCTTTTAGGTGCAATCGGAACA | CATGGGAGGAAGAAGTGGGTAC | NA | 141 | cgd7_4240 |
| MDH | TCCTCTAGATGCGATGGTTTACTAC | CCACCTACAACAATGGCTGATACA | NA | 162 | cgd7_470 |
| UGDH | CCTCCAACATTATCAGCTTTTTGAG | TGCATTTTAGAGTGAACCGCTT | NA | 141 | cgd8_920 |
| HSP70 | AGCCCGTATGAGTACAGAAGACT | GCCTGTGCCAAGAACCCTAAGA | NA | 168 | cgd4_3270 |
| 18S rRNA | JF1: AAGCTCGTAGTTggatTTCTGJF2: AAGCTCGTAGTTaatcTTCTG | JR: TAAGGTGCTGAAGGAGTAAGG | JT2: TCAGATACCGTCGTAGTCT | 434 | [18] |
| Target Gene | Test Samples (Cq) a | Control Samples (Cq) | Log2FC b | |||||
|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | Mean | C1 | C2 | Mean | ||
| COWP7 | 28.7 | 28.9 | 28.3 | 28.6 | 34.6 | 35.1 | 34.8 | 6.34 |
| Thioredoxin | 23.4 | 23.5 | 23.2 | 23.3 | 27.3 | 27.3 | 27.3 | 4.08 |
| UGDH | 25.1 | 25.2 | 25.1 | 25.1 | 29.6 | 29.4 | 29.5 | 4.49 |
| MDH | 22.7 | 22.6 | 22.6 | 22.6 | 26.5 | 26.7 | 26.6 | 4.11 |
| HSP70 | 24.6 | 24.0 | 24.5 | 24.3 | 28.9 | 28.6 | 28.7 | 4.54 |
| Prohibitin | 26.0 | 25.8 | 25.9 | 25.9 | 30.0 | 30.1 | 30.0 | 4.28 |
| Sample Preparation | Induction Treatment Group | ||||
|---|---|---|---|---|---|
| Viable (%) | Inactivated (%) | Control Cq Value | Oxidative Stress Cq Value | ΔCq | |
| Sample 1 | 100 | 0 | 29.9 | 25.6 | 4.3 |
| 100 | 0 | 30.0 | 25.8 | 4.2 | |
| Sample 2 | 10 | 90 | 34.9 | 29.6 | 5.3 |
| 10 | 90 | 33.6 | 28.6 | 5 | |
| Sample 3 | 1 | 99 | No Cq | 32.0 | NA |
| 1 | 99 | 36.4 | 31.5 | 4.9 | |
| Sample 4 | 0 | 100 | No Cq | No Ct | NA |
| 0 | 100 | 35.9 | 36.0 | −0.1 | |
| Treatment Condition | Cq Value COWP7 | Cq Value Thioredoxin | |||
|---|---|---|---|---|---|
| No Oxidative Stress | Oxidative Stress | No Oxidative Stress | Oxidative Stress | ||
| −20 °C for 2 h | Replicate 1 | 34.1 | 28.4 | 26.9 | 24.6 |
| Replicate 2 | 34.3 | 28.5 | 26.7 | 23.6 | |
| Replicate 3 | 33.7 | 28.4 | 26.9 | 23.9 | |
| Mean Cq ± SD | 34 ± 0.3 | 28.5 ± 0.1 | 26.8 ± 0.1 | 24.0 ± 0.5 | |
| −20 °C for 24 h | Replicate 1 | 33.7 | 35.3 | 30.2 | 30.2 |
| Replicate 2 | 33.9 | 34.7 | 30.4 | 30.0 | |
| Replicate 3 | 33.6 | 33.7 | 30.2 | 29.1 | |
| Mean Cq ± SD | 33.7 ± 0.2 | 34.6 ± 0.8 | 30.2 ± 0.1 | 29.7 ± 0.6 | |
| −20 °C for 48 h | Replicate 1 | 36.6 | 37.8 | 30.7 | 30.8 |
| Replicate 2 | 37.9 | 37.6 | 30.7 | 32.8 | |
| Replicate 3 | 35.1 | 36.4 | 29.9 | 31.2 | |
| Mean Cq ± SD | 36.5 ± 1.4 | 37.3 ± 0.7 | 30.4 ± 0.5 | 31.6 ± 1.1 | |
| 60 °C for 2 min | Replicate 1 | 37.8 | 36.7 | 31.8 | 30.8 |
| Replicate 2 | 37.7 | 35.1 | 32.9 | 31.0 | |
| Replicate 3 | No Cq | 35.7 | 31.7 | 30.8 | |
| Mean Cq ± SD | 37.8 ± 0.1 | 35.8 ± 0.8 | 32.1 ± 0.7 | 30.9 ± 0.1 | |
| 80 °C for 3 min | Replicate 1 | No Cq | No Cq | No Cq | No Cq |
| Replicate 2 | No Cq | No Cq | 35.9 | 36.0 | |
| Mean Cq ± SD | NA | NA | NA | NA | |
| Control a | Replicate 1 | 34.6 | 28.7 | 27.3 | 23.4 |
| Replicate 2 | 35.1 | 28.9 | 27.3 | 23.5 | |
| Replicate 3 | ND | 28.3 | ND | 23.2 | |
| Mean Cq ± SD | 34.9 ± 0.4 | 28.6 ± 0.3 | 27.3 | 23.4 ± 0.2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Temesgen, T.T.; Tysnes, K.R.; Robertson, L.J. Use of Oxidative Stress Responses to Determine the Efficacy of Inactivation Treatments on Cryptosporidium Oocysts. Microorganisms 2021, 9, 1463. https://doi.org/10.3390/microorganisms9071463
Temesgen TT, Tysnes KR, Robertson LJ. Use of Oxidative Stress Responses to Determine the Efficacy of Inactivation Treatments on Cryptosporidium Oocysts. Microorganisms. 2021; 9(7):1463. https://doi.org/10.3390/microorganisms9071463
Chicago/Turabian StyleTemesgen, Tamirat Tefera, Kristoffer Relling Tysnes, and Lucy Jane Robertson. 2021. "Use of Oxidative Stress Responses to Determine the Efficacy of Inactivation Treatments on Cryptosporidium Oocysts" Microorganisms 9, no. 7: 1463. https://doi.org/10.3390/microorganisms9071463
APA StyleTemesgen, T. T., Tysnes, K. R., & Robertson, L. J. (2021). Use of Oxidative Stress Responses to Determine the Efficacy of Inactivation Treatments on Cryptosporidium Oocysts. Microorganisms, 9(7), 1463. https://doi.org/10.3390/microorganisms9071463







