Mycobiota in the Carposphere of Sour and Sweet Cherries and Antagonistic Features of Potential Biocontrol Yeasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Berries
2.2. Yeast Isolation and Culturing
2.3. Molecular Identification of Yeast Species
2.4. Determination of Sour and Sweet Cherry Fungal Microbiota by Next Generation Sequencing
2.5. Bioinformatics and Data Analysis
2.6. In Vitro Evaluation of Yeast Antagonistic Activity
3. Results
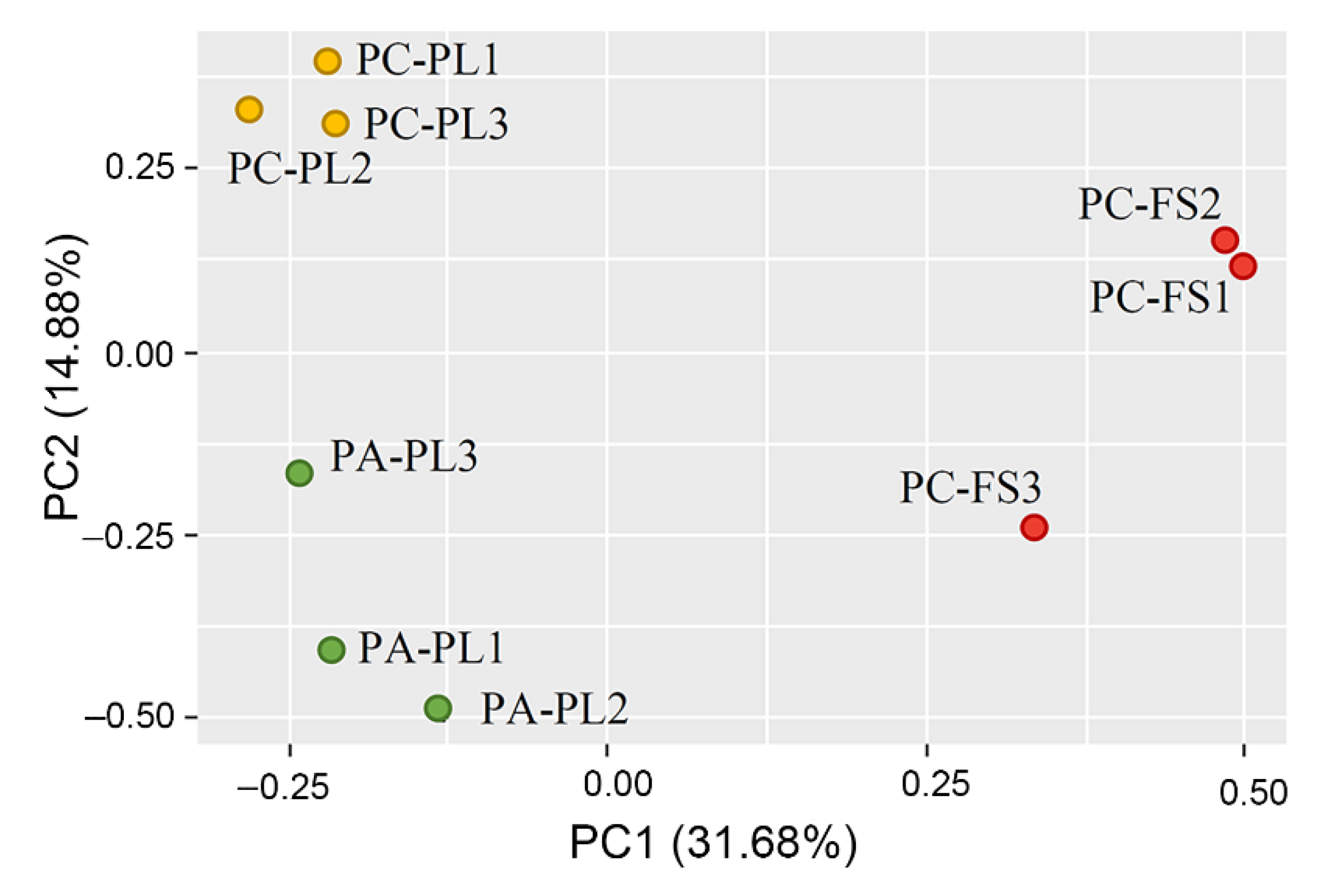
3.1. Abundance and Diversity of Fungal Microbiota on P. cerasus and P. avium
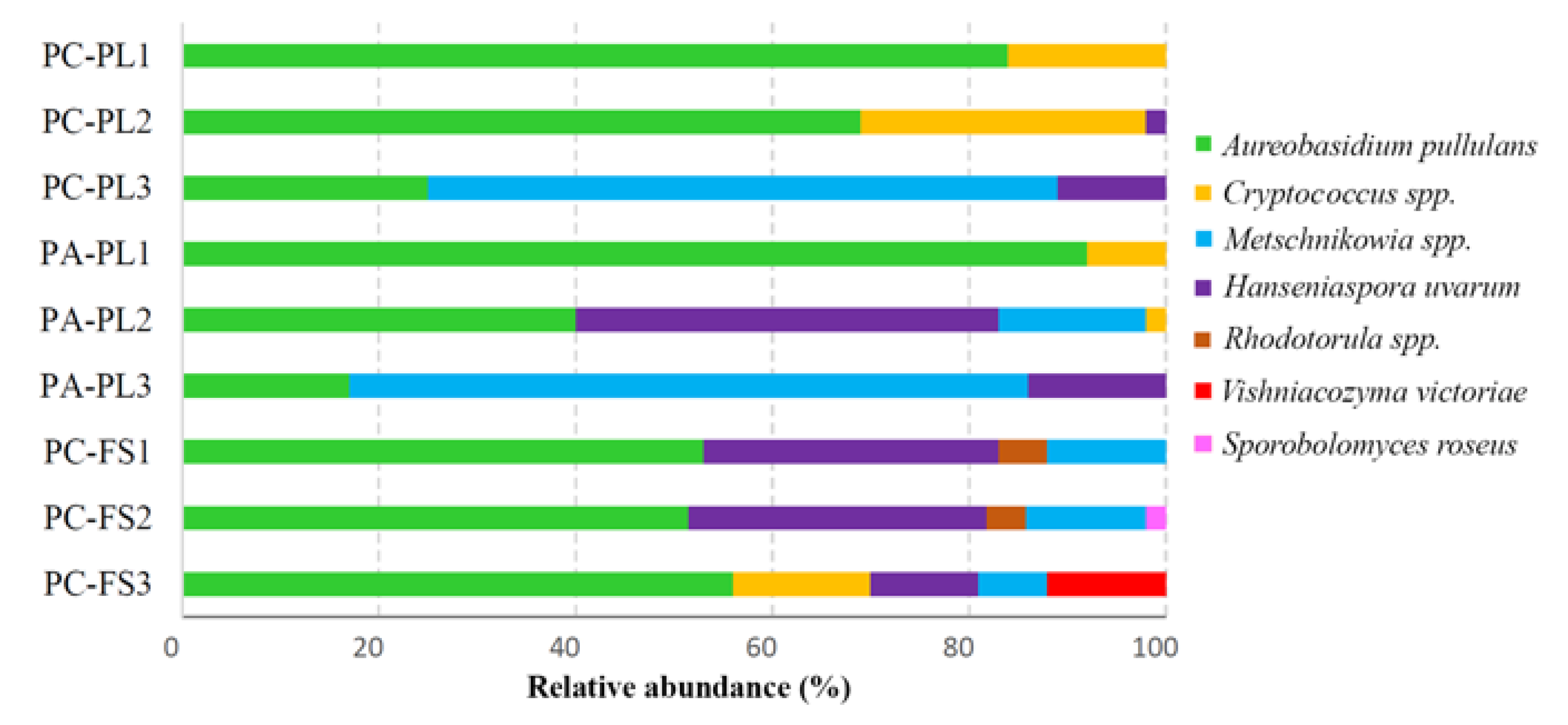
3.2. Fungal Community Profiling
3.3. Distribution of Cultivable Yeasts on P. avium and P. cerasus
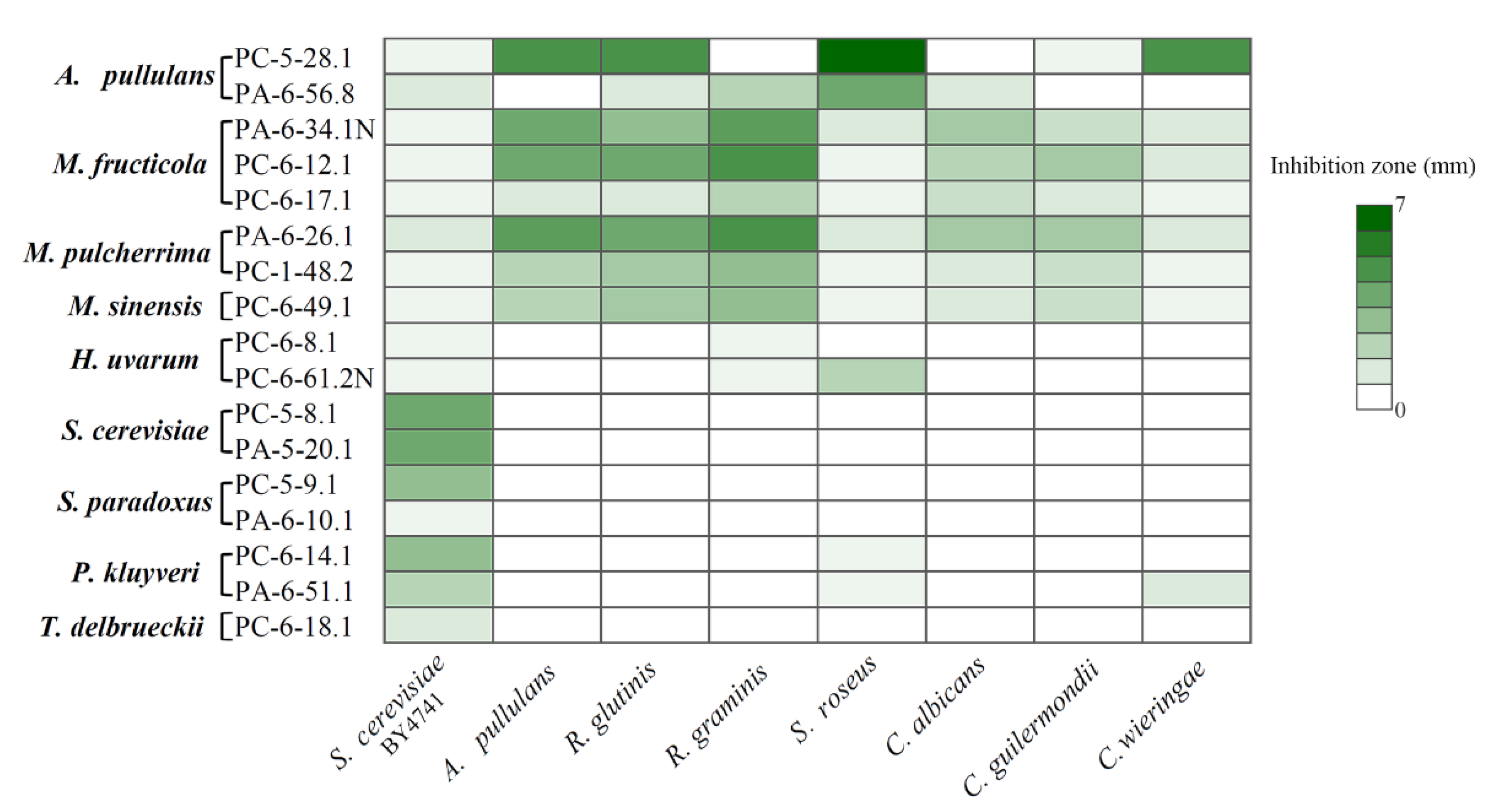
3.4. Antagonistic Activity of Sour and Sweet Cherries-Associated Yeasts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daenen, L.; Sterckx, F.; Delvaux, F.R.; Verachtert, H.; Derdelinckx, G. Evaluation of the Glycoside Hydrolase Activity of a Brettanomyces Strain on Glycosides from Sour Cherry (Prunus cerasus L.) Used in the Production of Special Fruit Beers. FEMS Yeast Res. 2008, 8, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- De Rogatis, A.; Ferrazzini, D.; Ducci, F.; Guerri, S.; Carnevale, S.; Belletti, P. Genetic Variation in Italian Wild Cherry (Prunus Avium L.) as Characterised by NSSR Markers. Forestry 2013, 2013, 1–10. [Google Scholar] [CrossRef][Green Version]
- Kelley, D.S.; Adkins, Y.; Laugero, K.D. A Review of the Health Benefits of Cherries. Nutrients 2018, 10, 368. [Google Scholar] [CrossRef] [PubMed]
- Morales-Corts, M.R.; Rodrigues, L.; Ortiz, J.; Sánchez, R. Characterization of Sour (Prunus cerasus L.) and Sweet Cherry (Prunus Avium L.) Varieties with Five Isozyme Systems. Rev. Bras. Frutic. 2008, 30. [Google Scholar] [CrossRef]
- Martinelli, I.; Micioni Di Bonaventura, M.V.; Moruzzi, M.; Amantini, C.; Maggi, F.; Gabrielli, M.G.; Fruganti, A.; Marchegiani, A.; Dini, F.; Marini, C.; et al. Effects of Prunus cerasus L. Seeds and Juice on Liver Steatosis in an Animal Modelof Diet-Induced Obesity. Nutrients 2020, 12, 1308. [Google Scholar] [CrossRef]
- Commisso, M.; Bianconi, M.; Di Carlo, F.; Poletti, S.; Bulgarini, A.; Munari, F.; Negri, S.; Stocchero, M.; Ceoldo, S.; Avesani, L.; et al. Multi-Approach Metabolomics Analysis and Artificial Simplified Phytocomplexes Reveal Cultivar-Dependent Synergy between Polyphenols and Ascorbic Acid in Fruits of the Sweet Cherry (Prunus avium L.). PLoS ONE 2017, 12, e0180889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, Q.; Qiao, G.; Qiu, Z.; Wen, Z.; Wen, X. Optimizing the Supercritical Carbon Dioxide Extraction of Sweet Cherry (Prunus avium L.) Leaves and UPLC-MS/MS Analysis. Anal. Methods 2020, 12, 3004–3013. [Google Scholar] [CrossRef]
- Fazzari, M.; Fukumoto, L.; Mazza, G.; Livrea, M.A.; Tesoriere, L.; Di Marco, L. In Vitro Bioavailability of Phenolic Compounds from Five Cultivars of Frozen SweetCherries (Prunus avium L.). J. Agric. Food Chem. 2008, 56, 3561–3568. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the Diversity of Grapevine Microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Cardoso, R.; Custódio, V.; Fernandes, J.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Wine Fermentation Microbiome: A Landscape from Different Portuguese WineAppellations. Front. Microbiol. 2015, 6, 905. [Google Scholar] [CrossRef]
- Martins, G.; Miot-Sertier, C.; Lauga, B.; Claisse, O.; Lonvaud-Funel, A.; Soulas, G.; Masneuf-Pomarède, I. Grape Berry Bacterial Microbiota: Impact of the Ripening Process and the FarmingSystem. Int. J. Food Microbiol. 2012, 158, 93–100. [Google Scholar] [CrossRef]
- Janisiewicz, W.J.; Jurick, W.M., 2nd; Peter, K.A.; Kurtzman, C.P.; Buyer, J.S. Yeasts Associated with Plums and Their Potential for Controlling Brown Rot afterHarvest. Yeast 2014, 31, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol Yeasts: Mechanisms and Applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef] [PubMed]
- Lukša, J.; Vepštaitė-Monstavičė, I.; Apšegaitė, V.; Blažytė-Čereškienė, L.; Stanevičienė, R.; Strazdaitė-Žielienė, Ž.; Ravoitytė, B.; Aleknavičius, D.; Būda, V.; Mozūraitis, R.; et al. Fungal Microbiota of Sea Buckthorn Berries at Two Ripening Stages and VolatileProfiling of Potential Biocontrol Yeasts. Microorganisms 2020, 8, 456. [Google Scholar] [CrossRef]
- Coutinho, T.A.; Venter, S.N. Pantoea Ananatis: An Unconventional Plant Pathogen. Mol. Plant Pathol. 2009, 10, 325–335. [Google Scholar] [CrossRef]
- Lukša, J.; Vepštaitė-Monstavičė, I.; Yurchenko, V.; Serva, S.; Servienė, E. High Content Analysis of Sea Buckthorn, Black Chokeberry, Red and White CurrantsMicrobiota—A Pilot Study. Food Res. Int. 2018, 111, 597–606. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Wisniewski, M.; Li Destri Nicosia, M.G.; Cacciola, S.O.; Schena, L. Metagenomic Analysis of Fungal Diversity on Strawberry Plants and the Effect of Management Practices on the Fungal Community Structure of Aerial Organs. PLoS ONE 2016, 11, e0160470. [Google Scholar] [CrossRef]
- Pintér, S.; Bata-Vidács, I.; Beczner, J. Epiphytic Microbiota of Sour Cherry (Prunus cerasus L.) in Integrated and Organic Growing. Acta Aliment. 2013, 42, 618–630. [Google Scholar] [CrossRef]
- Liang, S.; Liu, H.; Wu, S.; Xu, S.; Jin, D.; Faiola, F.; Zhuang, X.; Zhuang, G.; Qu, D.; Fan, H.; et al. Genetic Diversity of Diazotrophs and Total Bacteria in the Phyllosphere of Pyrus serotina, Prunus armeniaca, Prunus avium, and Vitis vinifera. Can. J. Microbiol. 2019, 65, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Toledo Del Árbol, J.; Pérez Pulido, R.; La Storia, A.; Grande Burgos, M.J.; Lucas, R.; Ercolini, D.; Gálvez, A. Microbial Diversity in Pitted Sweet Cherries (Prunus Avium L.) as Affected byHigh-Hydrostatic Pressure Treatment. Food Res. Int. 2016, 89, 790–796. [Google Scholar] [CrossRef]
- Abdollahi Aghdam, S.; Fotouhifar, K.-B. New Reports of Endophytic Fungi Associated with Cherry (Prunus avium) and Sour Cherry (Prunus cerasus) Trees in Iran. Mycol. Iran. 2016, 3, 75–85. [Google Scholar] [CrossRef]
- Bien, S.; Damm, U. Prunus Trees in Germany—A Hideout of Unknown Fungi? Mycol. Prog. 2020, 19, 667–690. [Google Scholar] [CrossRef]
- Vepštaitė-Monstavičė, I.; Lukša, J.; Stanevičienė, R.; Strazdaitė-Žielienė, Ž.; Yurchenko, V.; Serva, S.; Servienė, E. Distribution of Apple and Blackcurrant Microbiota in Lithuania and the CzechRepublic. Microbiol. Res. 2018, 206, 1–8. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science UsingQIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’sQ2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Horikoshi, M.; Tang, Y. Ggfortify: Data Visualization Tools for Statistical Analysis Results. 2018. Available online: https://CRAN.R-project.org/package=ggfortify (accessed on 22 February 2021).
- Warnes, G.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Mächler, M.; Magnusson, A.; Möller, S. Gplots: Various R Programming Tools for Plotting Data. 2005. Available online: https://CRAN.R-project.org/package=gplots (accessed on 24 February 2021).
- Martorell, P.; Fernández-Espinar, M.T.; Querol, A. Molecular Monitoring of Spoilage Yeasts during the Production of Candied FruitNougats to Determine Food Contamination Sources. Int. J. Food Microbiol. 2005, 101, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Martinez, L.; Gomez-Lopez, A.; Castelli, M.V.; Mesa-Arango, A.C.; Zaragoza, O.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Susceptibility Profile of Clinical Isolates of Non-Cryptococcus neoformans/Non-Cryptococcus gattii Cryptococcus Species and Literature Review. Med. Mycol. 2010, 48, 90–96. [Google Scholar] [CrossRef] [PubMed]
- McNicholas, S.; McDermott, H.; Power, L.; Johnson, E.M.; Moroney, J.; Humphreys, H.; Smyth, E.G. Sporobolomyces roseus in the Cerebrospinal Fluid of an Immunocompetent Patient--toTreat or Not to Treat? J. Med. Microbiol. 2012, 61, 295–296. [Google Scholar] [CrossRef]
- Liu, D.; Ma, L.; Shi, Y.; Wang, A.; Liu, C. Molecular Diagnosis and Source Tracing of an Infection of Aureobasidium pullulans. J. Infect. Dev. Ctries. 2019, 13, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Talapko, J.; Juzbašić, M.; Matijević, T.; Pustijanac, E.; Bekić, S.; Kotris, I.; Škrlec, I. Candida Albicans-The Virulence Factors and Clinical Manifestations of Infection. J. Fungi 2021, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Wirth, F.; Goldani, L.Z. Epidemiology of Rhodotorula: An Emerging Pathogen. Interdiscip. Perspect. Infect. Dis. 2012, 465717. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Freilich, S.; Bartuv, R.; Zhimo, V.Y.; Kumar, A.; Biasi, A.; Salim, S.; Feygenberg, O.; Burchard, E.; Dardick, C.; et al. Global Analysis of the Apple Fruit Microbiome: Are All Apples the Same? Environ. Microbiol. 2021. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. Changes in Sour Rotten Grape Berry Microbiota during Ripening and Wine Fermentation. Int. J. Food Microbiol. 2012, 154, 152–161. [Google Scholar] [CrossRef]
- Pawlikowska, E.; James, S.A.; Breierova, E.; Antolak, H.; Kregiel, D. Biocontrol Capability of Local Metschnikowia sp. Isolates. Antonie Van Leeuwenhoek 2019, 112, 1425–1445. [Google Scholar] [CrossRef]
- Moura, V.S.; Pollettini, F.L.; Ferraz, L.P.; Mazzi, M.V.; Kupper, K.C. Purification of a Killer Toxin from Aureobasidium pullulans for the Biocontrol of Phytopathogens. J. Basic Microbiol. 2021, 61, 77–87. [Google Scholar] [CrossRef]
- Tilocca, B.; Cao, A.; Migheli, Q. Scent of a Killer: Microbial Volatilome and Its Role in the Biological Control of Plant Pathogens. Front. Microbiol. 2020, 11, 41. [Google Scholar] [CrossRef]
- Parafati, L.; Vitale, A.; Restuccia, C.; Cirvilleri, G. Biocontrol Ability and Action Mechanism of Food-Isolated Yeast Strains against Botrytis cinerea Causing Post-Harvest Bunch Rot of Table Grape. Food Microbiol. 2015, 47, 85–92. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Droby, S. Metschnikowia fructicola, a New Ascosporic Yeast with Potential for Biocontrol of Postharvest Fruit Rots. Syst. Appl. Microbiol. 2001, 24, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Vadkertiová, R.; Molnárová, J.; Vránová, D.; Sláviková, E. Yeasts and Yeast-like Organisms Associated with Fruits and Blossoms of Different Fruit Trees. Can. J. Microbiol. 2012, 58, 1344–1352. [Google Scholar] [CrossRef]
- Lachance, M.A.; Starmer, W.T.; Rosa, C.A.; Bowles, J.M.; Barker, J.S.; Janzen, D.H. Biogeography of the Yeasts of Ephemeral Flowers and Their Insects. FEMS Yeast Res. 2001, 1, 1–8. [Google Scholar] [CrossRef]
- Büyüksırıt Bedir, T.; Kuleaşan, H. A Natural Approach, the Use of Killer Toxin Produced by Metschnikowia pulcherrima in Fresh Ground Beef Patties for Shelf Life Extention. Int. J. Food Microbiol. 2021, 345, 109154. [Google Scholar] [CrossRef]
- Janisiewicz, W.J.; Tworkoski, T.J.; Kurtzman, C.P. Biocontrol Potential of Metchnikowia Pulcherrima Strains Against Blue Mold of Apple. Phytopathology 2001, 91, 1098–1108. [Google Scholar] [CrossRef] [PubMed]
- Piombo, E.; Sela, N.; Wisniewski, M.; Hoffmann, M.; Gullino, M.L.; Allard, M.W.; Levin, E.; Spadaro, D.; Droby, S. Genome Sequence, Assembly and Characterization of Two Metschnikowia fructicola Strains Used as Biocontrol Agents of Postharvest Diseases. Front. Microbiol. 2018, 9, 593. [Google Scholar] [CrossRef]
- Sipiczki, M. Metschnikowia Strains Isolated from Botrytized Grapes Antagonize Fungal and Bacterial Growth by Iron Depletion. Appl. Environ. Microbiol. 2006, 72, 6716–6724. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Wang, L.; Xue, Y.; Lin, S.; Yu, G.; Yang, S. Purification and Molecular Characterization of a Metschnikowia saccharicola Killer Toxin Lethal to a Crab Pathogenic Yeast. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Vicente, J.; Ruiz, J.; Belda, I.; Benito-Vázquez, I.; Marquina, D.; Calderón, F.; Santos, A.; Benito, S. The Genus Metschnikowia in Enology. Microorganisms 2020, 8, 1038. [Google Scholar] [CrossRef]
- Graça, A.; Santo, D.; Esteves, E.; Nunes, C.; Abadias, M.; Quintas, C. Evaluation of Microbial Quality and Yeast Diversity in Fresh-Cut Apple. Food Microbiol. 2015, 51, 179–185. [Google Scholar] [CrossRef]
- Albertin, W.; Setati, M.E.; Miot-Sertier, C.; Mostert, T.T.; Colonna-Ceccaldi, B.; Coulon, J.; Girard, P.; Moine, V.; Pillet, M.; Salin, F.; et al. Hanseniaspora uvarum from Winemaking Environments Show Spatial and Temporal Genetic Clustering. Front. Microbiol. 2015, 6, 1569. [Google Scholar] [CrossRef]
- Cai, Z.; Yang, R.; Xiao, H.; Qin, X.; Si, L. Effect of Preharvest Application of Hanseniaspora uvarum on Postharvest Diseases in Strawberries. Postharvest Biol. Technol. 2015, 100, 52–58. [Google Scholar] [CrossRef]
- Al-qaysi, S. Killer Activity of Hanseniaspora uvarum Isolated from Dates Vinegar: Partially Purification and Characterization of Killer Toxin. Baghdad Sci. J. 2019, 16, 140–150. [Google Scholar] [CrossRef]
- Raspor, P.; Milek, D.M.; Polanc, J.; Mozina, S.S.; Cadez, N. Yeasts Isolated from Three Varieties of Grapes Cultivated in Different Locations of the Dolenjska Vine-Growing Region, Slovenia. Int. J. Food Microbiol. 2006, 109, 97–102. [Google Scholar] [CrossRef]
- Pelliccia, C.; Antonielli, L.; Corte, L.; Bagnetti, A.; Fatichenti, F.; Cardinali, G. Preliminary Prospection of the Yeast Biodiversity on Apple and Pear Surfaces from Northern Italy Orchards. Ann. Microbiol. 2011, 61, 965–972. [Google Scholar] [CrossRef]
- Diskin, S.; Feygenberg, O.; Feygenberg, D.; Droby, S.; Prusky, D.; Alkan, N. Microbiome Alterations Are Correlated with Occurrence of Postharvest Stem-End Rot in Mango Fruit. Phytobiomes 2017, 1. [Google Scholar] [CrossRef]
- Firrincieli, A.; Otillar, R.; Salamov, A.; Schmutz, J.; Khan, Z.; Redman, R.S.; Fleck, N.D.; Lindquist, E.; Grigoriev, I.V.; Doty, S.L. Genome Sequence of the Plant Growth Promoting Endophytic Yeast Rhodotorula graminis WP1. Front. Microbiol. 2015, 6, 978. [Google Scholar] [CrossRef]
- Sen, S.; Borah, S.N.; Bora, A.; Deka, S. Production, Characterization, and Antifungal Activity of a Biosurfactant Produced by Rhodotorula babjevae YS3. Microb. Cell Fact. 2017, 16, 95. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Paul, K.; Saha, C.; Mukherjee, G.; Nag, M.; Ghosh, S.; Das, A.; Seal, A.; Tripathy, S. A Unique Life-Strategy of an Endophytic Yeast Rhodotorula Mucilaginosa JGTA-S1-a Comparative Genomics Viewpoint. DNA Res. 2019, 26, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Janisiewicz, W. Control of Storage Decay of Apples with Sporobolomyces roseus. Plant Dis. 1994, 78, 466. [Google Scholar] [CrossRef]
- Gramisci, B.R.; Lutz, M.C.; Lopes, C.A.; Sangorrín, M.P. Enhancing the Efficacy of Yeast Biocontrol Agents against Postharvest Pathogens through Nutrient Profiling and the Use of Other Additives. Biol. Control 2018, 121, 151–158. [Google Scholar] [CrossRef]
- Janisiewicz, W.J.; Kurtzman, C.P.; Buyer, J.S. Yeasts Associated with Nectarines and Their Potential for Biological Control of Brown Rot. Yeast 2010, 27, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Hashem, M.; Alamri, S.A.; Hesham, A.E.-L.; Al-Qahtani, F.M.H.; Kilany, M. Biocontrol of Apple Blue Mould by New Yeast Strains: Cryptococcus albidus KKUY0017 and Wickerhamomyces anomalus KKUY0051 and Their Mode of Action. Biocontrol Sci. Technol. 2014, 24, 1137–1152. [Google Scholar] [CrossRef]
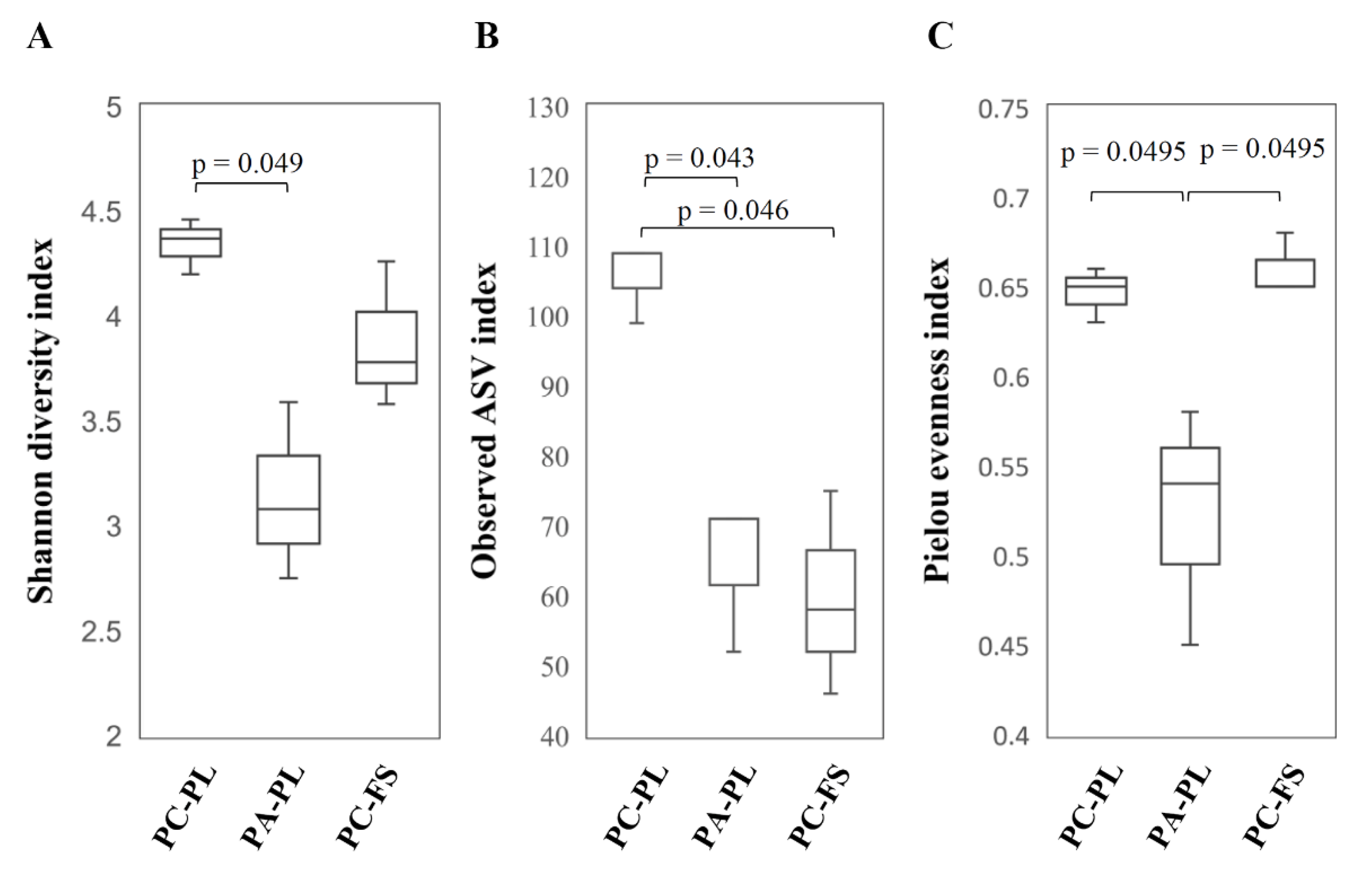


| Reads Obtained | HQ Reads | Observed ASVs | Pielou Evenness | Shannon Diversity | Simpson Index | |
|---|---|---|---|---|---|---|
| PC-PL1 | 26,895 | 17,126 | 99 | 0.63 | 4.2 | 0.86 |
| PC-PL2 | 28,665 | 19,082 | 109 | 0.66 | 4.46 | 0.91 |
| PC-PL3 | 30,732 | 21,140 | 109 | 0.65 | 4.37 | 0.92 |
| PA-PL1 | 27,571 | 20,325 | 71 | 0.45 | 2.75 | 0.72 |
| PA-PL2 | 28,473 | 17,509 | 52 | 0.54 | 3.08 | 0.78 |
| PA-PL3 | 29,021 | 19,901 | 71 | 0.58 | 3.59 | 0.86 |
| PC-FS1 | 27,825 | 17,104 | 46 | 0.65 | 3.58 | 0.85 |
| PC-FS2 | 30,369 | 19,242 | 58 | 0.65 | 3.78 | 0.87 |
| PC-FS3 | 28,433 | 17,504 | 75 | 0.68 | 4.26 | 0.92 |
| Total: | 257,984 | 168,933 | 690 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanevičienė, R.; Lukša, J.; Strazdaitė-Žielienė, Ž.; Ravoitytė, B.; Losinska-Sičiūnienė, R.; Mozūraitis, R.; Servienė, E. Mycobiota in the Carposphere of Sour and Sweet Cherries and Antagonistic Features of Potential Biocontrol Yeasts. Microorganisms 2021, 9, 1423. https://doi.org/10.3390/microorganisms9071423
Stanevičienė R, Lukša J, Strazdaitė-Žielienė Ž, Ravoitytė B, Losinska-Sičiūnienė R, Mozūraitis R, Servienė E. Mycobiota in the Carposphere of Sour and Sweet Cherries and Antagonistic Features of Potential Biocontrol Yeasts. Microorganisms. 2021; 9(7):1423. https://doi.org/10.3390/microorganisms9071423
Chicago/Turabian StyleStanevičienė, Ramunė, Juliana Lukša, Živilė Strazdaitė-Žielienė, Bazilė Ravoitytė, Regina Losinska-Sičiūnienė, Raimondas Mozūraitis, and Elena Servienė. 2021. "Mycobiota in the Carposphere of Sour and Sweet Cherries and Antagonistic Features of Potential Biocontrol Yeasts" Microorganisms 9, no. 7: 1423. https://doi.org/10.3390/microorganisms9071423
APA StyleStanevičienė, R., Lukša, J., Strazdaitė-Žielienė, Ž., Ravoitytė, B., Losinska-Sičiūnienė, R., Mozūraitis, R., & Servienė, E. (2021). Mycobiota in the Carposphere of Sour and Sweet Cherries and Antagonistic Features of Potential Biocontrol Yeasts. Microorganisms, 9(7), 1423. https://doi.org/10.3390/microorganisms9071423








