Identification of Resistance Determinants for a Promising Antileishmanial Oxaborole Series
Abstract
1. Introduction
2. Materials and Methods
2.1. Parasite Cultures
2.2. Animals
2.3. Test Substances and Formulations
2.4. Intracellular Amastigote Susceptibility Assay
2.5. Promastigote Susceptibility Assay
2.6. In Vivo Resistance Selection
2.7. In Vitro Intracellular Amastigote Resistance Selection
2.8. In Vitro Extracellular Promastigote Resistance Selection
2.9. Intracellular Efflux Susceptibility Assay
2.10. Extracellular Efflux Susceptibility Assay
2.11. Cosmid Library Primary Screen (L. infantum)
2.12. Cosmid Library Secondary Screen (L. donovani)
2.13. Cosmid Transfection and Impact on Drug Susceptibility
2.14. Statistical Analysis
3. Results
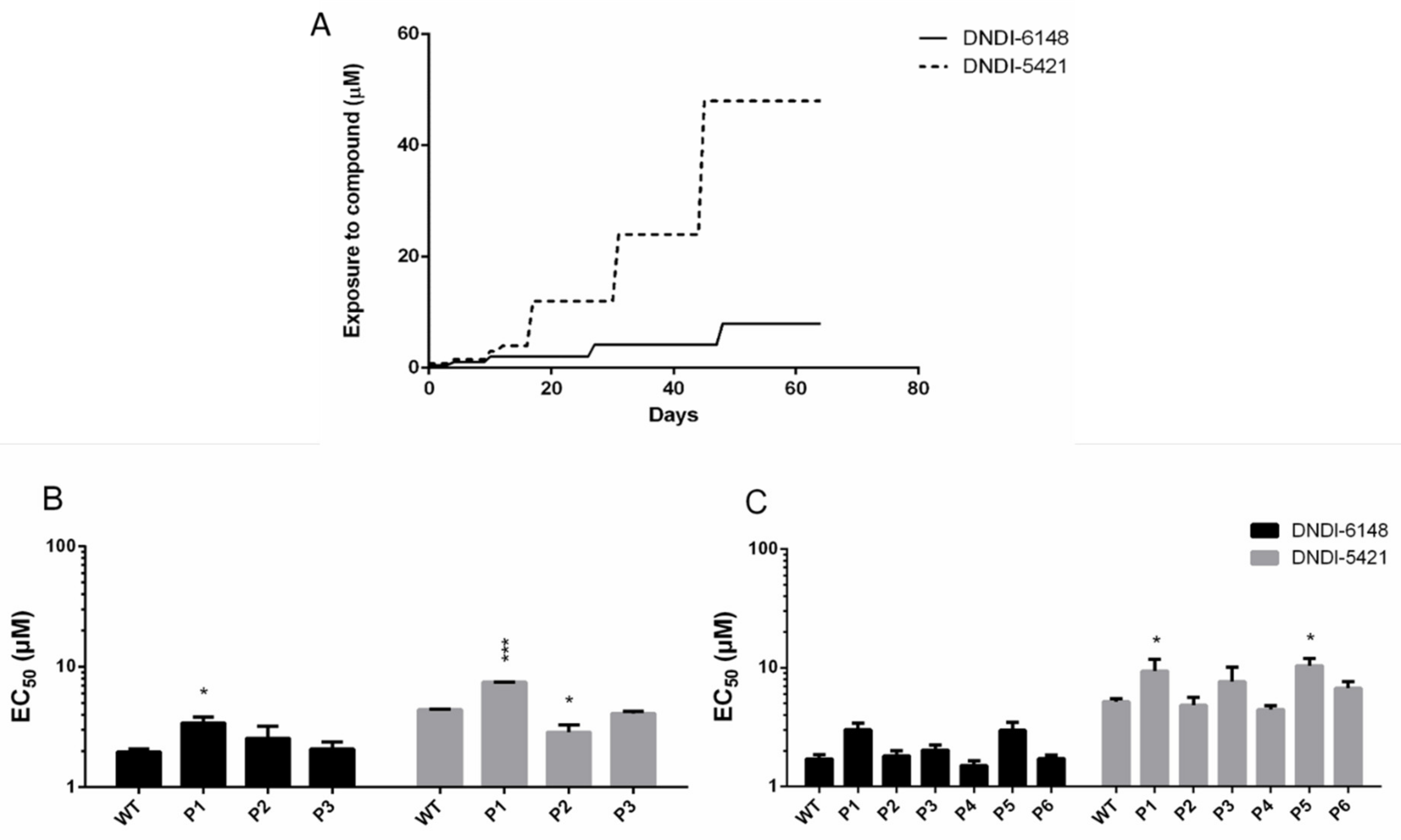
3.1. In Vivo and In Vitro Intracellular Amastigote Resistance Selection
3.2. DNDI-6148 Resistance Selection in Promastigotes
3.3. The Involvement of Efflux Pumps
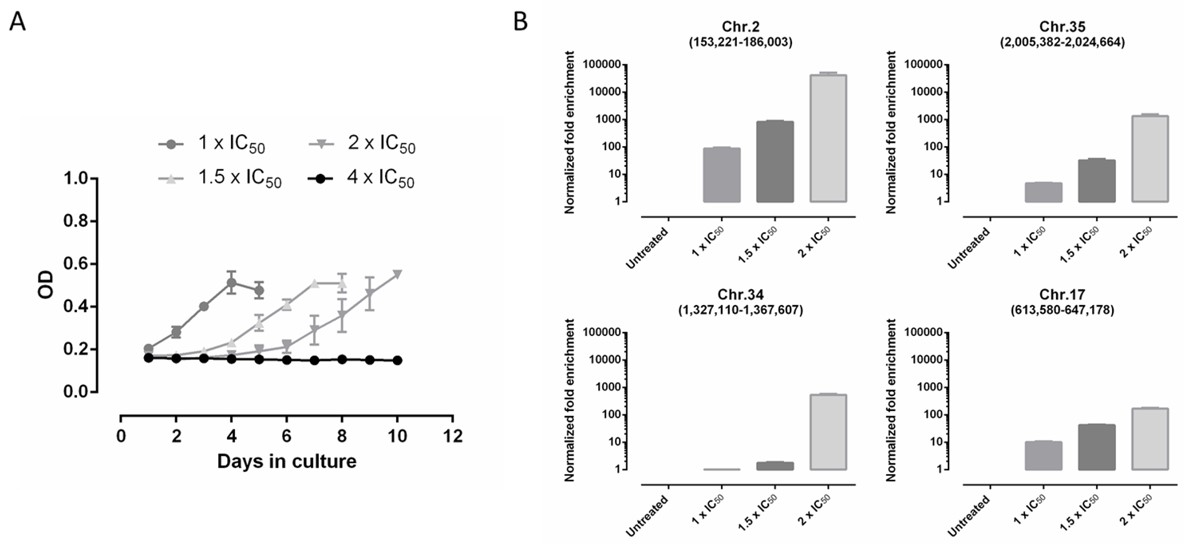
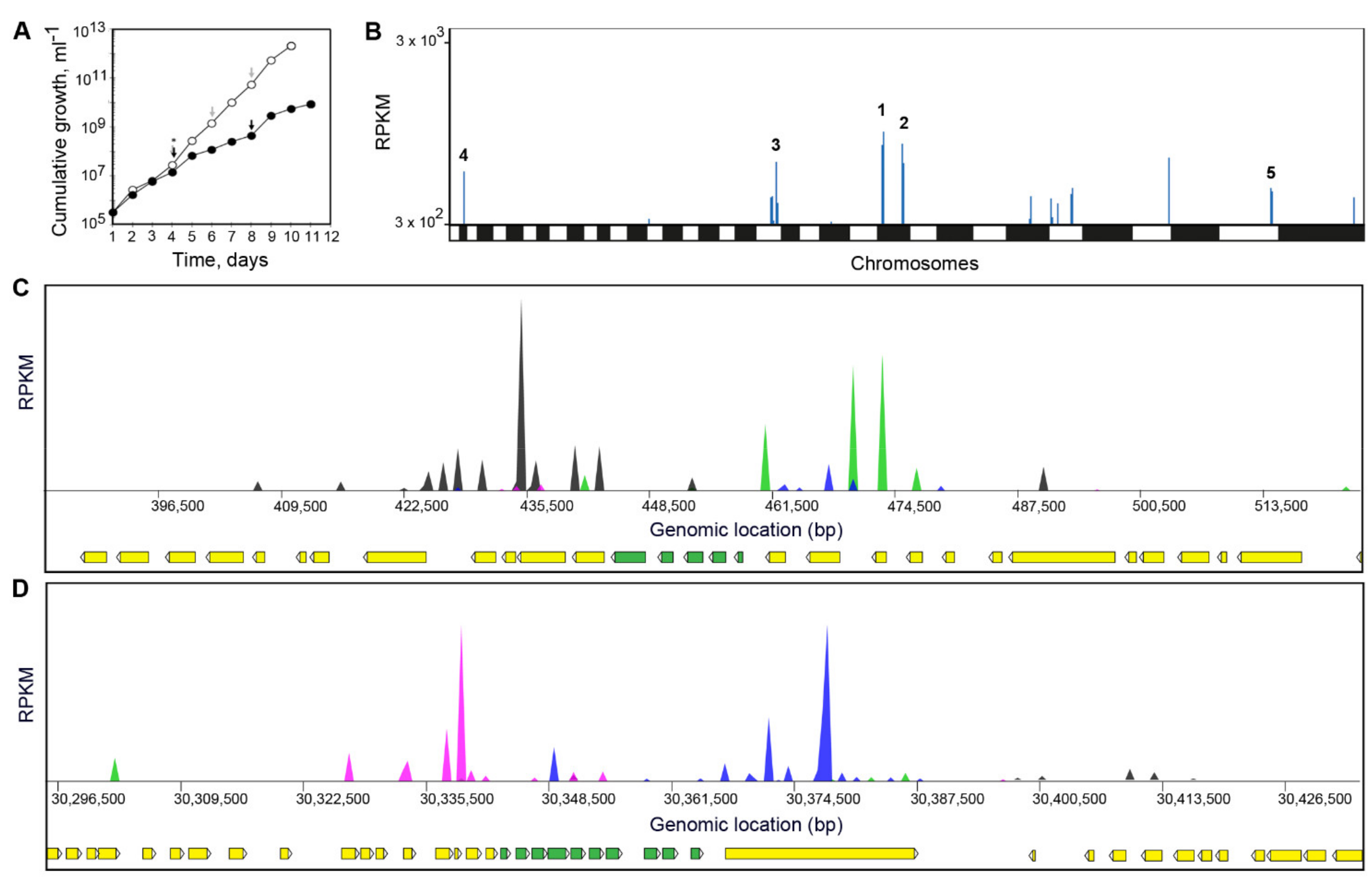
3.4. Screening of DNDI-6148 against a L. infantum Cosmid-Based Overexpression Library
3.5. Screening of DNDI-6148 against a L. Donovani Cosmid-Based Overexpression Library
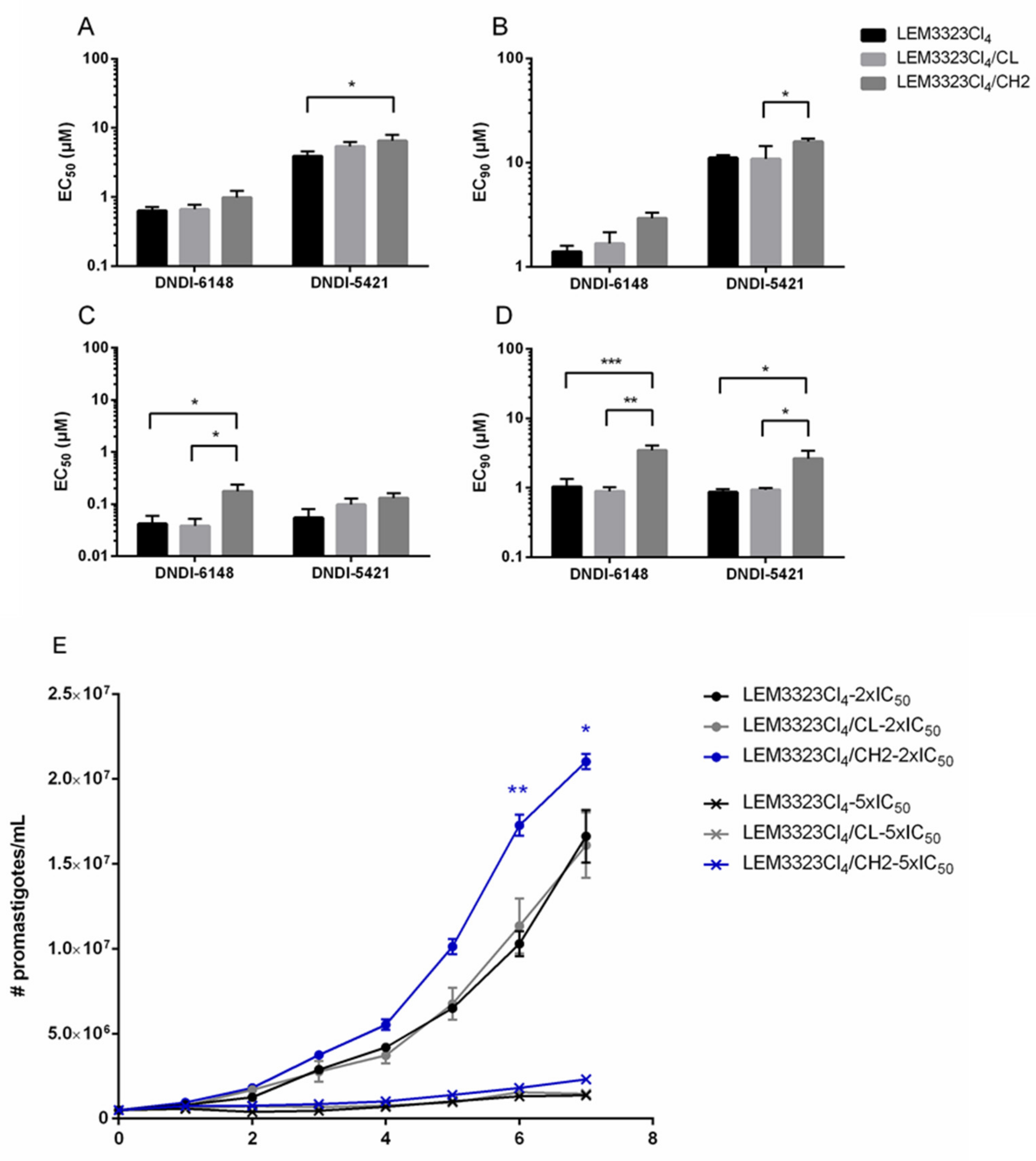
3.6. Validation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pace, D. Leishmaniasis. J. Infect. 2014, 69 (Suppl. 1), S10–S18. [Google Scholar] [CrossRef]
- Maroli, M.; Feliciangeli, M.D.; Bichaud, L.; Charrel, R.N.; Gradoni, L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med. Vet. Entomol. 2013, 27, 123–147. [Google Scholar] [CrossRef]
- Sacks, D.; Kamhawi, S. Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu. Rev. Microbiol. 2001, 55, 453–483. [Google Scholar] [CrossRef] [PubMed]
- Hommel, M. Visceral leishmaniasis: Biology of the parasite. J. Infect. 1999, 39, 101–111. [Google Scholar] [CrossRef]
- Wilson, M.E.; Sandor, M.; Blum, A.M.; Young, B.M.; Metwali, A.; Elliott, D.; Lynch, R.G.; Weinstock, J.V. Local suppression of IFN-gamma in hepatic granulomas correlates with tissue-specific replication of Leishmania chagasi. J. Immunol. 1996, 156, 2231–2239. [Google Scholar]
- Nagle, A.S.; Khare, S.; Kumar, A.B.; Supek, F.; Buchynskyy, A.; Mathison, C.J.; Chennamaneni, N.K.; Pendem, N.; Buckner, F.S.; Gelb, M.H.; et al. Recent developments in drug discovery for leishmaniasis and human African trypanosomiasis. Chem. Rev. 2014, 114, 11305–11347. [Google Scholar] [CrossRef]
- Chatelain, E.; Ioset, J.R. Drug discovery and development for neglected diseases: The DNDi model. Drug Des. Dev. Ther. 2011, 5, 175–181. [Google Scholar] [CrossRef]
- Alves, F.; Bilbe, G.; Blesson, S.; Goyal, V.; Monnerat, S.; Mowbray, C.; Muthoni Ouattara, G.; Pecoul, B.; Rijal, S.; Rode, J.; et al. Recent Development of Visceral Leishmaniasis Treatments: Successes, Pitfalls, and Perspectives. Clin. Microbiol. Rev. 2018, 31. [Google Scholar] [CrossRef] [PubMed]
- Van den Kerkhof, M.; Mabille, D.; Chatelain, E.; Mowbray, C.E.; Braillard, S.; Hendrickx, S.; Maes, L.; Caljon, G. In vitro and in vivo pharmacodynamics of three novel antileishmanial lead series. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 81–86. [Google Scholar] [CrossRef] [PubMed]
- DNDi. R&D Portofolio Visceral Leishmaniasis. Available online: https://dndi.org/research-development/portfolio/dndi-6148/ (accessed on 12 August 2020).
- Liu, C.T.; Tomsho, J.W.; Benkovic, S.J. The unique chemistry of benzoxaboroles: Current and emerging applications in biotechnology and therapeutic treatments. Bioorgan. Med. Chem. 2014, 22, 4462–4473. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Zhang, Y.K.; Liu, Y.; Ding, C.Z.; Zhou, Y.; Plattner, J.J.; Baker, S.J.; Bu, W.; Liu, L.; et al. Synthesis and SAR of acyclic HCV NS3 protease inhibitors with novel P4-benzoxaborole moieties. Bioorgan. Med. Chem. Lett. 2011, 21, 2048–2054. [Google Scholar] [CrossRef] [PubMed]
- Rock, F.L.; Mao, W.; Yaremchuk, A.; Tukalo, M.; Crepin, T.; Zhou, H.; Zhang, Y.K.; Hernandez, V.; Akama, T.; Baker, S.J.; et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 2007, 316, 1759–1761. [Google Scholar] [CrossRef]
- Hernandez, V.; Crepin, T.; Palencia, A.; Cusack, S.; Akama, T.; Baker, S.J.; Bu, W.; Feng, L.; Freund, Y.R.; Liu, L.; et al. Discovery of a novel class of boron-based antibacterials with activity against gram-negative bacteria. Antimicrob. Agents Chemother. 2013, 57, 1394–1403. [Google Scholar] [CrossRef]
- Xia, Y.; Cao, K.; Zhou, Y.; Alley, M.R.; Rock, F.; Mohan, M.; Meewan, M.; Baker, S.J.; Lux, S.; Ding, C.Z.; et al. Synthesis and SAR of novel benzoxaboroles as a new class of beta-lactamase inhibitors. Bioorgan. Med. Chem. Lett. 2011, 21, 2533–2536. [Google Scholar] [CrossRef]
- Hu, Q.H.; Liu, R.J.; Fang, Z.P.; Zhang, J.; Ding, Y.Y.; Tan, M.; Wang, M.; Pan, W.; Zhou, H.C.; Wang, E.D. Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase. Sci. Rep. 2013, 3, 2475. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.B.; Gao, N.; Hajec, L.; McKinney, D.C. Time-dependent, reversible, oxaborole inhibition of Escherichia coli leucyl-tRNA synthetase measured with a continuous fluorescence assay. Anal. Biochem. 2012, 431, 48–53. [Google Scholar] [CrossRef]
- Gupta, A.K.; Versteeg, S.G. Tavaborole—A treatment for onychomycosis of the toenails. Expert Rev. Clin. Pharmacol. 2016, 9, 1145–1152. [Google Scholar] [CrossRef]
- Baker, S.J.; Tomsho, J.W.; Benkovic, S.J. Boron-containing inhibitors of synthetases. Chem. Soc. Rev. 2011, 40, 4279–4285. [Google Scholar] [CrossRef] [PubMed]
- Jinna, S.; Finch, J. Spotlight on tavaborole for the treatment of onychomycosis. Drug Des. Devel. Ther. 2015, 9, 6185–6190. [Google Scholar] [CrossRef]
- Sonoiki, E.; Ng, C.L.; Lee, M.C.; Guo, D.; Zhang, Y.K.; Zhou, Y.; Alley, M.R.; Ahyong, V.; Sanz, L.M.; Lafuente-Monasterio, M.J.; et al. A potent antimalarial benzoxaborole targets a Plasmodium falciparum cleavage and polyadenylation specificity factor homologue. Nat. Commun. 2017, 8, 14574. [Google Scholar] [CrossRef] [PubMed]
- Palencia, A.; Bougdour, A.; Brenier-Pinchart, M.P.; Touquet, B.; Bertini, R.L.; Sensi, C.; Gay, G.; Vollaire, J.; Josserand, V.; Easom, E.; et al. Targeting Toxoplasma gondii CPSF3 as a new approach to control toxoplasmosis. EMBO Mol. Med. 2017, 9, 385–394. [Google Scholar] [CrossRef]
- Begolo, D.; Vincent, I.M.; Giordani, F.; Pohner, I.; Witty, M.J.; Rowan, T.G.; Bengaly, Z.; Gillingwater, K.; Freund, Y.; Wade, R.C.; et al. The trypanocidal benzoxaborole AN7973 inhibits trypanosome mRNA processing. PLoS Pathog. 2018, 14, e1007315. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.C.; Foth, B.J.; Urbaniak, M.D.; Patterson, S.; Ong, H.B.; Berriman, M.; Fairlamb, A.H. Genomic and Proteomic Studies on the Mode of Action of Oxaboroles against the African Trypanosome. PLoS Negl. Trop. Dis. 2015, 9, e0004299. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wall, R.J.; Rico, E.; Lukac, I.; Zuccotto, F.; Elg, S.; Gilbert, I.H.; Freund, Y.; Alley, M.R.K.; Field, M.C.; Wyllie, S.; et al. Clinical and veterinary trypanocidal benzoxaboroles target CPSF3. Proc. Natl. Acad. Sci. USA 2018, 115, 9616–9621. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zoltner, M.; Leung, K.F.; Scullion, P.; Hutchinson, S.; Del Pino, R.C.; Vincent, I.M.; Zhang, Y.K.; Freund, Y.R.; Alley, M.R.K.; et al. Host-parasite co-metabolic activation of antitrypanosomal aminomethyl-benzoxaboroles. PLoS Pathog. 2018, 14, e1006850. [Google Scholar] [CrossRef]
- Giordani, F.; Paape, D.; Vincent, I.M.; Pountain, A.W.; Fernandez-Cortes, F.; Rico, E.; Zhang, N.; Morrison, L.J.; Freund, Y.; Witty, M.J.; et al. Veterinary trypanocidal benzoxaboroles are peptidase-activated prodrugs. PLoS Pathog. 2020, 16, e1008932. [Google Scholar] [CrossRef]
- Van den Kerkhof, M.; Sterckx, Y.G.; Leprohon, P.; Maes, L.; Caljon, G. Experimental Strategies to Explore Drug Action and Resistance in Kinetoplastid Parasites. Microorganisms 2020, 8, 950. [Google Scholar] [CrossRef]
- Gazanion, E.; Fernandez-Prada, C.; Papadopoulou, B.; Leprohon, P.; Ouellette, M. Cos-Seq for high-throughput identification of drug target and resistance mechanisms in the protozoan parasite Leishmania. Proc. Natl. Acad. Sci. USA 2016, 113, E3012–E3021. [Google Scholar] [CrossRef]
- Corpas-Lopez, V.; Moniz, S.; Thomas, M.; Wall, R.J.; Torrie, L.S.; Zander-Dinse, D.; Tinti, M.; Brand, S.; Stojanovski, L.; Manthri, S.; et al. Pharmacological Validation of N-Myristoyltransferase as a Drug Target in Leishmania donovani. ACS Infect. Dis. 2019, 5, 111–122. [Google Scholar] [CrossRef]
- Berg, M.; Mannaert, A.; Vanaerschot, M.; Van Der Auwera, G.; Dujardin, J.C. (Post-) Genomic approaches to tackle drug resistance in Leishmania. Parasitology 2013, 140, 1492–1505. [Google Scholar] [CrossRef]
- Leprohon, P.; Fernandez-Prada, C.; Gazanion, E.; Monte-Neto, R.; Ouellette, M. Drug resistance analysis by next generation sequencing in Leishmania. Int. J. Parasitol. Drugs Drug Resist. 2015, 5, 26–35. [Google Scholar] [CrossRef]
- Hendrickx, S.; Boulet, G.; Mondelaers, A.; Dujardin, J.C.; Rijal, S.; Lachaud, L.; Cos, P.; Delputte, P.; Maes, L. Experimental selection of paromomycin and miltefosine resistance in intracellular amastigotes of Leishmania donovani and L. infantum. Parasitol. Res. 2014, 113, 1875–1881. [Google Scholar] [CrossRef]
- Hendrickx, S.; Mondelaers, A.; Eberhardt, E.; Delputte, P.; Cos, P.; Maes, L. In Vivo Selection of Paromomycin and Miltefosine Resistance in Leishmania donovani and L. infantum in a Syrian Hamster Model. Antimicrob. Agents Chemother. 2015, 59, 4714–4718. [Google Scholar] [CrossRef]
- Hendrickx, S.; Mondelaers, A.; Eberhardt, E.; Lachaud, L.; Delputte, P.; Cos, P.; Maes, L. Intracellular amastigote replication may not be required for successful in vitro selection of miltefosine resistance in Leishmania infantum. Parasitol. Res. 2015, 114, 2561–2565. [Google Scholar] [CrossRef] [PubMed]
- Perry, M.R.; Prajapati, V.K.; Menten, J.; Raab, A.; Feldmann, J.; Chakraborti, D.; Sundar, S.; Fairlamb, A.H.; Boelaert, M.; Picado, A. Arsenic exposure and outcomes of antimonial treatment in visceral leishmaniasis patients in Bihar, India: A retrospective cohort study. PLoS Negl. Trop. Dis. 2015, 9, e0003518. [Google Scholar] [CrossRef]
- Srivastava, S.; Mishra, J.; Gupta, A.K.; Singh, A.; Shankar, P.; Singh, S. Laboratory confirmed miltefosine resistant cases of visceral leishmaniasis from India. Parasites Vectors 2017, 10, 49. [Google Scholar] [CrossRef]
- Cojean, S.; Houze, S.; Haouchine, D.; Huteau, F.; Lariven, S.; Hubert, V.; Michard, F.; Bories, C.; Pratlong, F.; Le Bras, J.; et al. Leishmania resistance to miltefosine associated with genetic marker. Emerg. Infect. Dis. 2012, 18, 704–706. [Google Scholar] [CrossRef]
- Purkait, B.; Kumar, A.; Nandi, N.; Sardar, A.H.; Das, S.; Kumar, S.; Pandey, K.; Ravidas, V.; Kumar, M.; De, T.; et al. Mechanism of amphotericin B resistance in clinical isolates of Leishmania donovani. Antimicrob. Agents Chemother. 2012, 56, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, P.K.; Alam, M.N.; Roy Chowdhury, D.; Chakraborti, T. Drug Resistance in Protozoan Parasites: An Incessant Wrestle for Survival. J. Glob. Antimicrob. Resist. 2019, 18, 1–11. [Google Scholar] [CrossRef]
- Muriithi, W.; Macharia, L.W.; Heming, C.P.; Echevarria, J.L.; Nyachieo, A.; Filho, P.N.; Neto, V.M. ABC transporters and the hallmarks of cancer: Roles in cancer aggressiveness beyond multidrug resistance. Cancer Biol. Med. 2020, 17, 253–269. [Google Scholar] [CrossRef]
- Leprohon, P.; Legare, D.; Ouellette, M. Intracellular localization of the ABCC proteins of Leishmania and their role in resistance to antimonials. Antimicrob. Agents Chemother. 2009, 53, 2646–2649. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Messaritakis, I.; Christodoulou, V.; Mazeris, A.; Koutala, E.; Vlahou, A.; Papadogiorgaki, S.; Antoniou, M. Drug resistance in natural isolates of Leishmania donovani s.l. promastigotes is dependent of Pgp170 expression. PLoS ONE 2013, 8, e65467. [Google Scholar] [CrossRef] [PubMed]
- Coelho, A.C.; Boisvert, S.; Mukherjee, A.; Leprohon, P.; Corbeil, J.; Ouellette, M. Multiple mutations in heterogeneous miltefosine-resistant Leishmania major population as determined by whole genome sequencing. PLoS Negl. Trop. Dis. 2012, 6, e1512. [Google Scholar] [CrossRef]
- BoseDasgupta, S.; Ganguly, A.; Roy, A.; Mukherjee, T.; Majumder, H.K. A novel ATP-binding cassette transporter, ABCG6 is involved in chemoresistance of Leishmania. Mol. Biochem. Parasitol. 2008, 158, 176–188. [Google Scholar] [CrossRef]
- El Fadili, K.; Messier, N.; Leprohon, P.; Roy, G.; Guimond, C.; Trudel, N.; Saravia, N.G.; Papadopoulou, B.; Legare, D.; Ouellette, M. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob. Agents Chemother. 2005, 49, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.S.; Monte Neto, R.L.; Andrade, J.M.; Santi, A.M.; Reis, P.G.; Frezard, F.; Murta, S.M. Molecular characterization of the MRPA transporter and antimony uptake in four New World Leishmania spp. susceptible and resistant to antimony. Int. J. Parasitol. Drugs Drug Resist. 2013, 3, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Castanys-Munoz, E.; Alder-Baerens, N.; Pomorski, T.; Gamarro, F.; Castanys, S. A novel ATP-binding cassette transporter from Leishmania is involved in transport of phosphatidylcholine analogues and resistance to alkyl-phospholipids. Mol. Microbiol. 2007, 64, 1141–1153. [Google Scholar] [CrossRef]
- Castanys-Munoz, E.; Perez-Victoria, J.M.; Gamarro, F.; Castanys, S. Characterization of an ABCG-like transporter from the protozoan parasite Leishmania with a role in drug resistance and transbilayer lipid movement. Antimicrob. Agents Chemother. 2008, 52, 3573–3579. [Google Scholar] [CrossRef] [PubMed]
- Lachaud, L.; Bourgeois, N.; Plourde, M.; Leprohon, P.; Bastien, P.; Ouellette, M. Parasite susceptibility to amphotericin B in failures of treatment for visceral leishmaniasis in patients coinfected with HIV type 1 and Leishmania infantum. Clin. Infect. Dis. 2009, 48, e16–e22. [Google Scholar] [CrossRef]
- Van den Kerkhof, M.; Mabille, D.; Hendrickx, S.; Leprohon, P.; Mowbray, C.E.; Braillard, S.; Ouellette, M.; Maes, L.; Caljon, G. Antileishmanial Aminopyrazoles: Studies into Mechanisms and Stability of Experimental Drug Resistance. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Ryan, K.A.; Dasgupta, S.; Beverley, S.M. Shuttle cosmid vectors for the trypanosomatid parasite Leishmania. Gene 1993, 131, 145–150. [Google Scholar] [CrossRef]
- Potvin, J.E.; Leprohon, P.; Gazanion, E.; Sharma, M.; Fernandez-Prada, C.; Ouellette, M. Cos-Seq: A High-Throughput Gain-of-Function Screen for Drug Resistance Studies in Leishmania. Methods Mol. Biol. 2019, 1971, 141–167. [Google Scholar] [CrossRef] [PubMed]
- Steketee, P.C.; Vincent, I.M.; Achcar, F.; Giordani, F.; Kim, D.H.; Creek, D.J.; Freund, Y.; Jacobs, R.; Rattigan, K.; Horn, D.; et al. Benzoxaborole treatment perturbs S-adenosyl-L-methionine metabolism in Trypanosoma brucei. PLoS Negl. Trop. Dis. 2018, 12, e0006450. [Google Scholar] [CrossRef]
- Barrand, M.A.; Bagrij, T.; Neo, S.Y. Multidrug resistance-associated protein: A protein distinct from P-glycoprotein involved in cytotoxic drug expulsion. Gen. Pharmacol. 1997, 28, 639–645. [Google Scholar] [CrossRef]
- Chauhan, I.S.; Rao, G.S.; Singh, N. Enhancing the copy number of Ldrab6 gene in Leishmania donovani parasites mediates drug resistance through drug-thiol conjugate dependent multidrug resistance protein A (MRPA). Acta Trop. 2019, 199, 105158. [Google Scholar] [CrossRef]
- Kaur, J.; Dey, C.S. Putative P-glycoprotein expression in arsenite-resistant Leishmania donovani down-regulated by verapamil. Biochem. Biophys Res. Commun. 2000, 271, 615–619. [Google Scholar] [CrossRef]
- Bellamy, W.T. P-glycoproteins and multidrug resistance. Annu. Rev. Pharmacol. Toxicol. 1996, 36, 161–183. [Google Scholar] [CrossRef]
- Gupta, I.; Aggarwal, S.; Singh, K.; Yadav, A.; Khan, S. Ubiquitin Proteasome pathway proteins as potential drug targets in parasite Trypanosoma cruzi. Sci. Rep. 2018, 8, 8399. [Google Scholar] [CrossRef] [PubMed]
- Bibo-Verdugo, B.; Jiang, Z.; Caffrey, C.R.; O’Donoghue, A.J. Targeting proteasomes in infectious organisms to combat disease. FEBS J. 2017, 284, 1503–1517. [Google Scholar] [CrossRef] [PubMed]
- Gannavaram, S.; Sharma, P.; Duncan, R.C.; Salotra, P.; Nakhasi, H.L. Mitochondrial associated ubiquitin fold modifier-1 mediated protein conjugation in Leishmania donovani. PLoS ONE 2011, 6, e16156. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Wang, C.C. Differentiation of Trypanosoma brucei may be stage non-specific and does not require progression of cell cycle. Mol. Microbiol. 2003, 49, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Damianou, A.; Burge, R.J.; Catta-Preta, C.M.C.; Geoghegan, V.; Nievas, Y.R.; Newling, K.; Brown, E.; Burchmore, R.; Rodenko, B.; Mottram, J.C. Essential roles for deubiquitination in Leishmania life cycle progression. PLoS Pathog. 2020, 16, e1008455. [Google Scholar] [CrossRef]
- Tan, J.; Jakob, U.; Bardwell, J.C. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 2002, 184, 2692–2698. [Google Scholar] [CrossRef]
- Bocchetta, M.; Xiong, L.; Mankin, A.S. 23S rRNA positions essential for tRNA binding in ribosomal functional sites. Proc. Natl. Acad. Sci. USA 1998, 95, 3525–3530. [Google Scholar] [CrossRef]
- Sacerdoti-Sierra, N.; Jaffe, C.L. Release of ecto-protein kinases by the protozoan parasite Leishmania major. J. Biol. Chem. 1997, 272, 30760–30765. [Google Scholar] [CrossRef]
- Bhatia, A.; Sanyal, R.; Paramchuk, W.; Gedamu, L. Isolation, characterization and disruption of the casein kinase II alpha subunit gene of Leishmania chagasi. Mol. Biochem. Parasitol. 1998, 92, 195–206. [Google Scholar] [CrossRef]
- Krober-Boncardo, C.; Lorenzen, S.; Brinker, C.; Clos, J. Casein kinase 1.2 over expression restores stress resistance to Leishmania donovani HSP23 null mutants. Sci. Rep. 2020, 10, 15969. [Google Scholar] [CrossRef] [PubMed]
- Sindhe, K.M.V.; Wu, W.; Legac, J.; Zhang, Y.K.; Easom, E.E.; Cooper, R.A.; Plattner, J.J.; Freund, Y.R.; DeRisi, J.L.; Rosenthal, P.J. Plasmodium falciparum Resistance to a Lead Benzoxaborole Due to Blocked Compound Activation and Altered Ubiquitination or Sumoylation. mBio 2020, 11. [Google Scholar] [CrossRef]
| ITMAP263 | LEM4038 | |
|---|---|---|
| Wild Type | 0.24 ± 0.20 | 0.51 ± 0.11 |
| Cycle 1 | 0.61 ± 0.20 | 0.23 ± 0.08 |
| Cycle 2 | 0.44 ± 0.20 | 0.21 ± 0.04 |
| Cycle 3 | 0.48 ± 0.20 | 0.23 ± 0.06 |
| Cycle 4 | 0.31 ± 0.04 | 0.29 ± 0.05 |
| Cycle 5 | 0.44 ± 0.13 | 0.27 ± 0.06 |
| Cosmid | Gene ID | Gene Function | Max Enrichment | Rationalised L. donovani Gene ID |
|---|---|---|---|---|
| Chromo-some 2 | LINF_020008400 | EamA-like transporter family/Triose-phosphate Transporter family/UAA transporter family putative | 33,225 | |
| LINF_020008500 | Hypothetical protein conserved | 25,936 | LdBPK.02.2.000320.1 | |
| LINF_020008600 | Casein kinase II—alpha chain putative | 24,707 | LdBPK.02.2.000330.1 | |
| LINF_020008700 | Proteasome regulatory non-ATPase subunit 6 putative | 30,199 | LdBPK.02.2.000340.1 | |
| LINF_020008800 | FtsJ-like methyltransferase putative | 22,404 | LdBPK.02.2.000350.1 | |
| LINF_020008900 | Ubiquitin-conjugating enzyme E2 putative | 24,949 | LdBPK.02.2.000360.1 | |
| LINF_020009100 | Mitochondrial protein 81 | 1852 | ||
| LINF_020009200 | Hypothetical protein conserved | 44,918 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van den Kerkhof, M.; Leprohon, P.; Mabille, D.; Hendrickx, S.; Tulloch, L.B.; Wall, R.J.; Wyllie, S.; Chatelain, E.; Mowbray, C.E.; Braillard, S.; et al. Identification of Resistance Determinants for a Promising Antileishmanial Oxaborole Series. Microorganisms 2021, 9, 1408. https://doi.org/10.3390/microorganisms9071408
Van den Kerkhof M, Leprohon P, Mabille D, Hendrickx S, Tulloch LB, Wall RJ, Wyllie S, Chatelain E, Mowbray CE, Braillard S, et al. Identification of Resistance Determinants for a Promising Antileishmanial Oxaborole Series. Microorganisms. 2021; 9(7):1408. https://doi.org/10.3390/microorganisms9071408
Chicago/Turabian StyleVan den Kerkhof, Magali, Philippe Leprohon, Dorien Mabille, Sarah Hendrickx, Lindsay B. Tulloch, Richard J. Wall, Susan Wyllie, Eric Chatelain, Charles E. Mowbray, Stéphanie Braillard, and et al. 2021. "Identification of Resistance Determinants for a Promising Antileishmanial Oxaborole Series" Microorganisms 9, no. 7: 1408. https://doi.org/10.3390/microorganisms9071408
APA StyleVan den Kerkhof, M., Leprohon, P., Mabille, D., Hendrickx, S., Tulloch, L. B., Wall, R. J., Wyllie, S., Chatelain, E., Mowbray, C. E., Braillard, S., Ouellette, M., Maes, L., & Caljon, G. (2021). Identification of Resistance Determinants for a Promising Antileishmanial Oxaborole Series. Microorganisms, 9(7), 1408. https://doi.org/10.3390/microorganisms9071408








