Effects of the Denitrification Inhibitor “Procyanidins” on the Diversity, Interactions, and Potential Functions of Rhizosphere-Associated Microbiome
Abstract
:1. Introduction
2. Materials and Methods
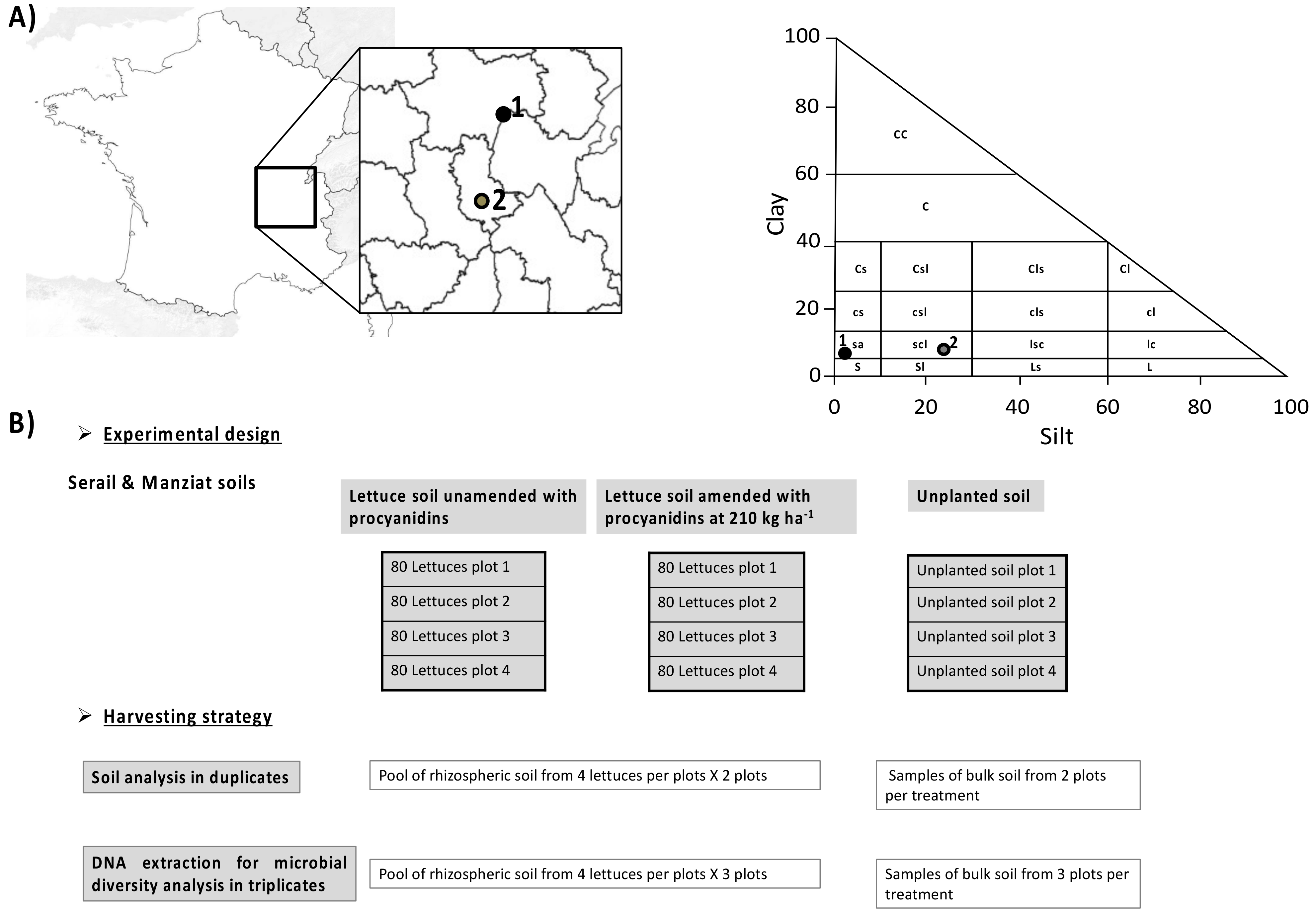
2.1. Plant Growth, Experimental Design, and Harvesting
2.2. Measurement of Environmental Variables
2.3. DNA Extraction
2.4. Amplicon Libraries and Illumina Sequencing
2.5. Bioinformatic Analysis
2.6. Statistical Analyses
2.7. Functional Inference
2.8. Microbial Networks
3. Results
3.1. Soil Type: Predominant Factor Affecting Microbiome Richness and Diversity
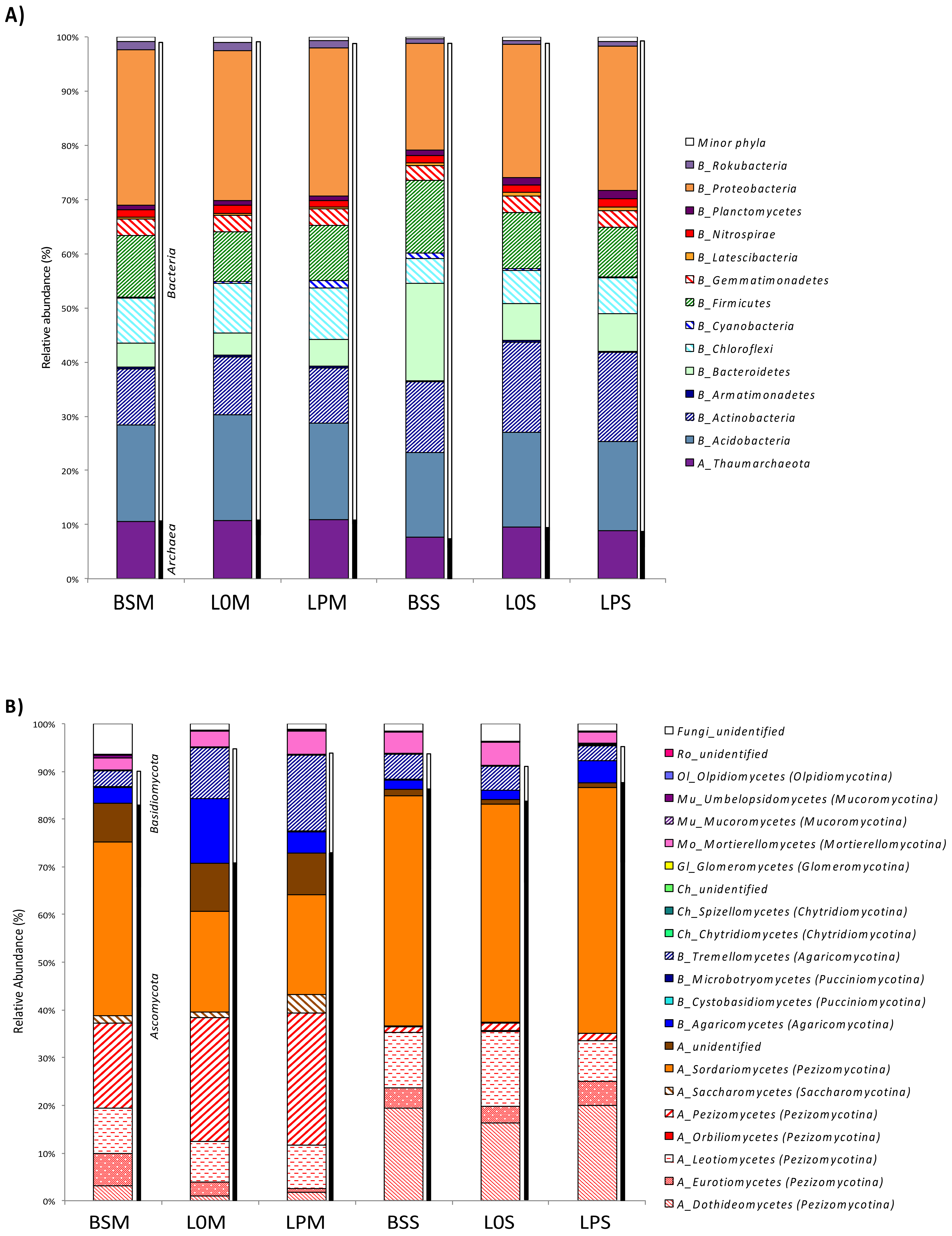
3.2. Microbiome Composition Among Treatments
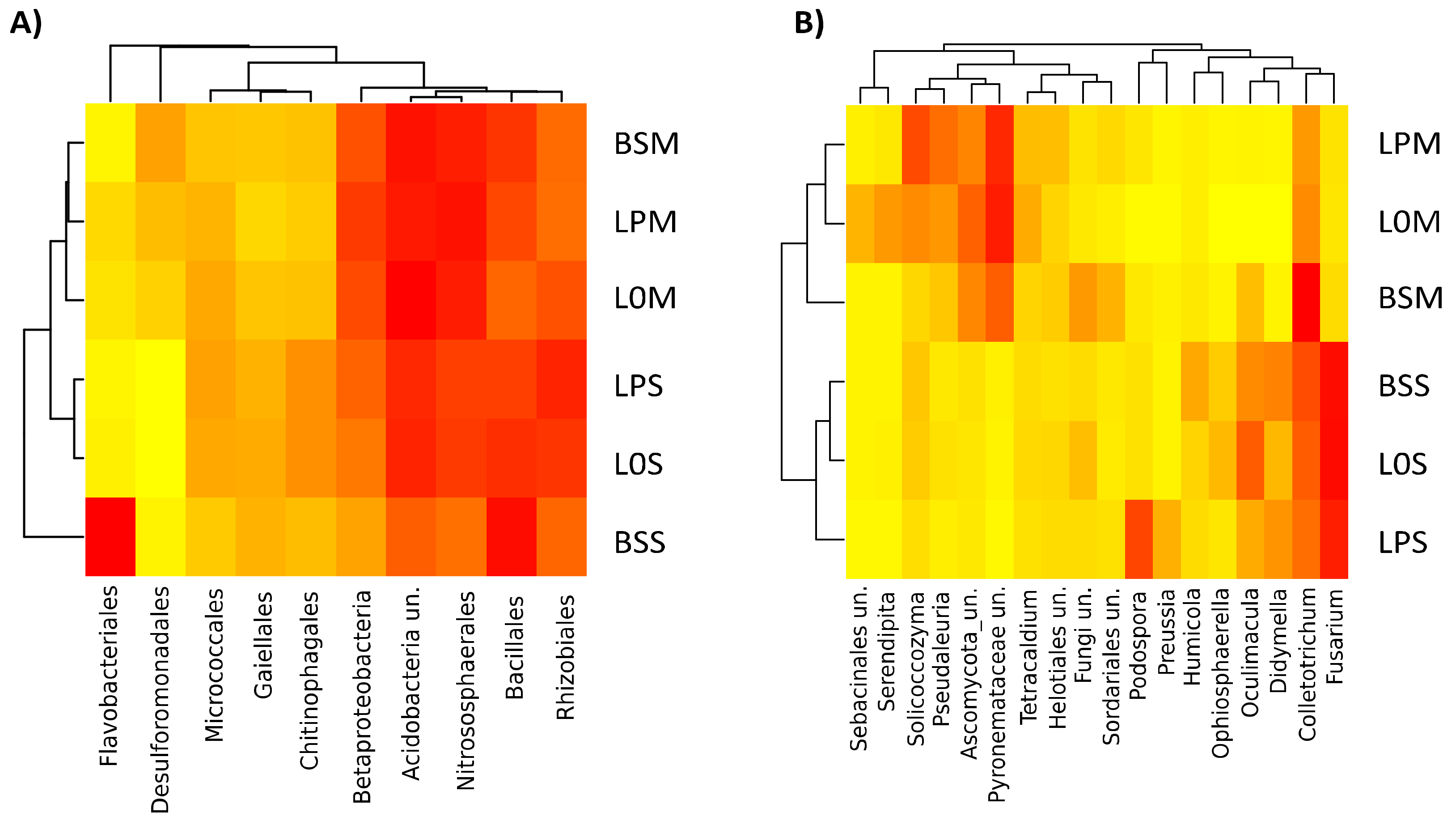
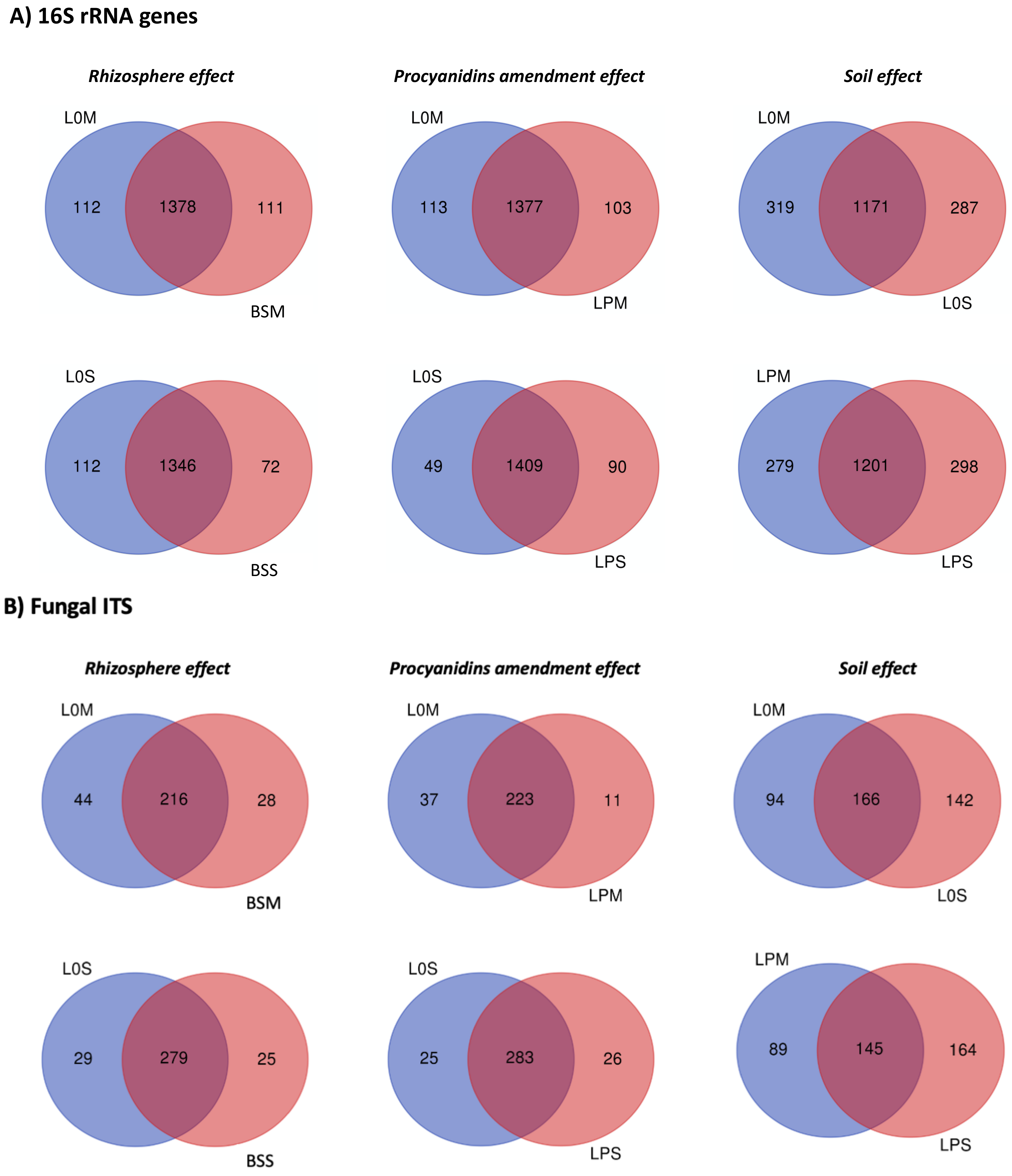
3.3. Core Microbiome and Differential Microbial Community Composition among Amended and Unamended Treatments
3.4. Relationship between Microbiome Diversity and Edaphic Variables
3.5. Prediction of Fungal and Prokaryotic Ecological Functions
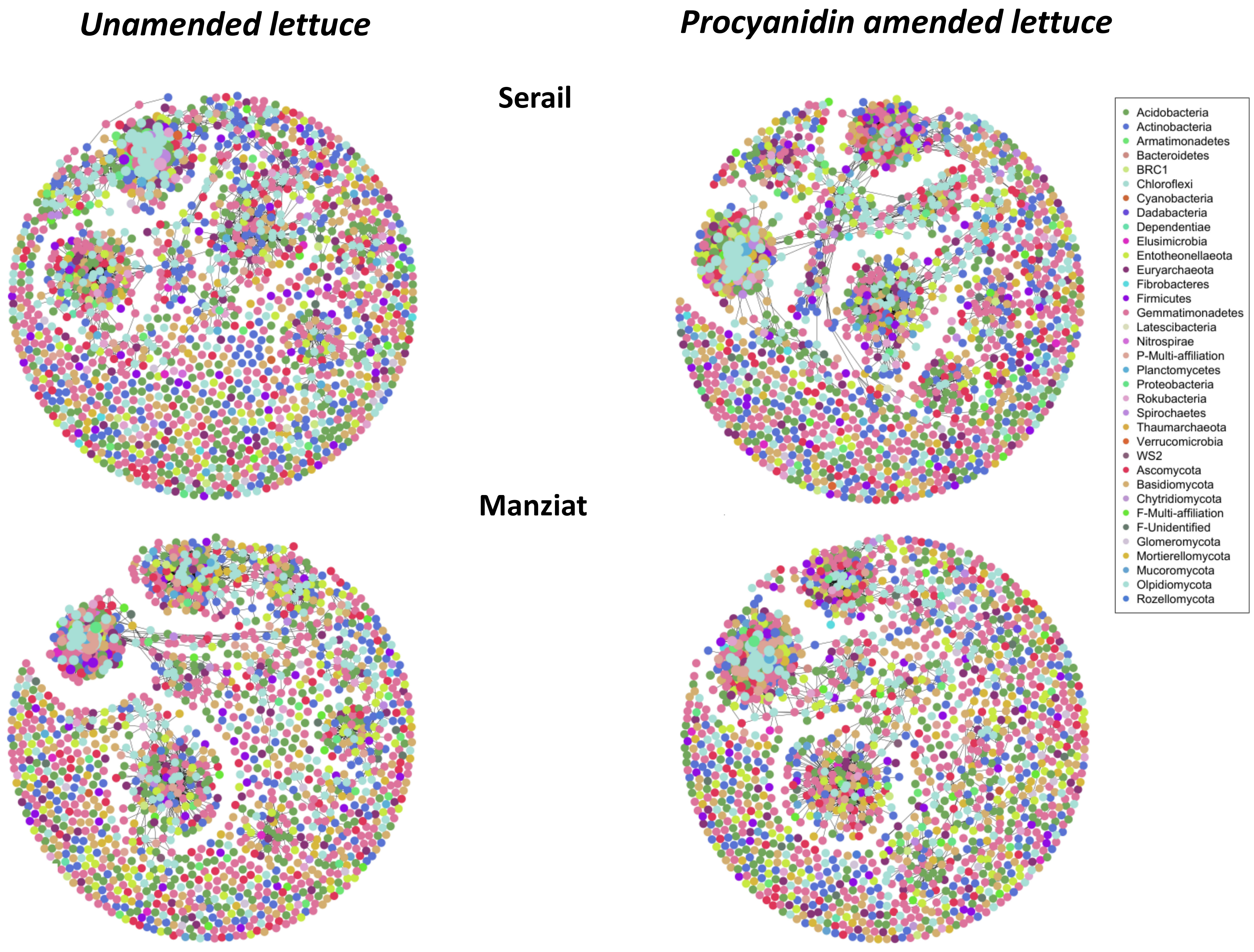
3.6. Microbial Network Description
4. Discussion
4.1. Soil Type and Edaphic Variables as Driver of Bacterial and Fungal Diversity
4.2. Procyanidin Amendment Does Not Affect the Diversity and Potential Activity of the Plant Microbiome
4.3. Procyanidin Amendment Modifies Microbial Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boserup, E. The Conditions of Agricultural Growth: The Economics of Agrarian Change under Population Pressure; Routledge: London, UK, 2017. [Google Scholar]
- Tilman, D.; Cassman, K.G.; Matson, P.A.; Naylor, R.; Polasky, S. Agricultural sustainability and intensive production practices. Nature 2002, 418, 671–677. [Google Scholar] [CrossRef]
- Bloom, A.J. The increasing importance of distinguishing among plant nitrogen sources. Curr. Opin. Plant Biol. 2015, 25, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Ronen, E. Micro-elements in agriculture. Pract. Hydroponics Greenh. 2016, 35. [Google Scholar]
- Kirchmann, H.; Johnston, A.J.; Bergström, L.F. Possibilities for reducing nitrate leaching from agricultural land. AMBIO J. Hum. Environ. 2002, 31, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, N.; Saad, J.K.; Van Laethem, C.; Machet, J.M.; Maucorps, J.; Mary, B. Nitrate leaching in intensive agriculture in Northern France: Effect of farming practices, soils and crop rotations. Agric. Ecosyst. Environ. 2005, 111, 292–310. [Google Scholar] [CrossRef]
- Cassman, K.G.; Dobermann, A.; Walters, D.T. Agroecosystems, nitrogen-use efficiency, and nitrogen management. AMBIO J. Hum. Environ. 2002, 31, 132–141. [Google Scholar] [CrossRef]
- Di, H.J.; Cameron, K.C. Nitrate leaching in temperate agroecosystems: Sources, factors and mitigating strategies. Nutr. Cycl. Agroecosyst. 2002, 64, 237–256. [Google Scholar] [CrossRef]
- Bouwman, A.F.; Boumans, L.J.M.; Batjes, N.H. Estimation of global NH3 volatilization loss from synthetic fertilizers and animal manure applied to arable lands and grasslands. Glob. Biogeochem. Cycles 2002, 16, 8-1–8-14. [Google Scholar] [CrossRef]
- Strebel, O.; Duynisveld, W.H.M.; Böttcher, J. Nitrate pollution of groundwater in Western Europe. Agric. Ecosyst. Environ. 1989, 26, 189–214. [Google Scholar] [CrossRef]
- Lewis, W.M., Jr.; Wurtsbaugh, W.A.; Paerl, H.W. Rationale for control of anthropogenic nitrogen and phosphorus to reduce eutrophication of inland waters. Environ. Sci. Technol. 2011, 45, 10300–10305. [Google Scholar] [CrossRef]
- Kononen, K. Eutrophication, harmful algal blooms and species diversity in phytoplankton communities: Examples from the Baltic Sea. AMBIO J. Hum. Environ. 2001, 30, 184–190. [Google Scholar] [CrossRef]
- Yang, X.; Wu, X.; Hao, H.; He, Z. Mechanisms and assessment of water eutrophication. J. Zhejiang Univ. Sci. B 2008, 9, 197–209. [Google Scholar] [CrossRef]
- Zumft, W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar] [CrossRef]
- Oenema, O.; Witzke, H.P.; Klimont, Z.; Lesschen, J.P.; Velthof, G.L. Integrated assessment of promising measures to decrease nitrogen losses from agriculture in EU-27. Agric. Ecosyst. Environ. 2009, 133, 280–288. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [Green Version]
- Ravishankara, A.R.; Daniel, J.S.; Portmann, R.W. Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 2009, 326, 123–125. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, H.; Yan, X.; Yagi, K. Evaluation of effectiveness of enhanced-efficiency fertilizers as mitigation options for N2O and NO emissions from agricultural soils: Meta-analysis. Glob. Change Biol. 2010, 16, 1837–1846. [Google Scholar] [CrossRef]
- Santamaria, P.; Elia, A.; Serio, F.; Gonnella, M.; Parente, A. Comparison between nitrate and ammonium nutrition in fennel, celery, and Swiss chard. J. Plant Nutr. 1999, 22, 1091–1106. [Google Scholar] [CrossRef]
- Santamaria, P. Nitrate in vegetables: Toxicity, content, intake and EC regulation. J. Sci. Food Agric. 2006, 86, 10–17. [Google Scholar] [CrossRef]
- Green, L.C.; De Luzuriaga, K.R.; Wagner, D.A.; Rand, W.; Istfan, N.; Young, V.R.; Tannenbaum, S.R. Nitrate biosynthesis in man. Proc. Natl. Acad. Sci. USA 1981, 78, 7764–7768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Velzen, A.G.; Sips, A.J.; Schothorst, R.C.; Lambers, A.C.; Meulenbelt, J. The oral bioavailability of nitrate from nitrate-rich vegetables in humans. Toxicol. Lett. 2008, 181, 177–181. [Google Scholar] [CrossRef]
- Du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Gilsanz, C.; Báez, D.; Misselbrook, T.H.; Dhanoa, M.S.; Cárdenas, L.M. Development of emission factors and efficiency of two nitrification inhibitors, DCD and DMPP. Agric. Ecosyst. Environ. 2016, 216, 1–8. [Google Scholar] [CrossRef]
- Liu, C.; Wang, K.; Zheng, X. Effects of nitrification inhibitors (DCD and DMPP) on nitrous oxide emission, crop yield and nitrogen uptake in a wheat-maize cropping system. Biogeosciences 2013, 10, 2427–2437. [Google Scholar] [CrossRef] [Green Version]
- Nardi, P.; Laanbroek, H.J.; Nicol, G.W.; Renella, G.; Cardinale, M.; Pietramellara, G.; Weckwerth, W.; Trinchera, A.; Ghatak, A.; Nannipieri, P. Biological nitrification inhibition in the rhizosphere: Determining interactions and impact on microbially mediated processes and potential applications. FEMS Microbiol. Rev. 2020, 44, 874–908. [Google Scholar] [CrossRef] [PubMed]
- Abalos, D.; Jeffery, S.; Sanz-Cobena, A.; Guardia, G.; Vallejo, A. Meta-analysis of the effect of urease and nitrification inhibitors on crop productivity and nitrogen use efficiency. Agric. Ecosyst. Environ. 2014, 189, 136–144. [Google Scholar] [CrossRef]
- Pathak, H.; Bhatia, A.; Prasad, S.; Singh, S.; Kumar, S.; Jain, M.C.; Kumar, U. Emission of nitrous oxide from rice-wheat systems of Indo-Gangetic plains of India. Environ. Monit. Assess. 2002, 77, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Meijide, A.; Díez, J.A.; Sánchez-Martín, L.; López-Fernández, S.; Vallejo, A. Nitrogen oxide emissions from an irrigated maize crop amended with treated pig slurries and composts in a Mediterranean climate. Agric. Ecosyst. Environ. 2007, 121, 383–394. [Google Scholar] [CrossRef] [Green Version]
- Di, H.J.; Cameron, K.C. How does the application of different nitrification inhibitors affect nitrous oxide emissions and nitrate leaching from cow urine in grazed pastures? Soil Use Manag. 2012, 28, 54–61. [Google Scholar] [CrossRef]
- Tedeschi, A.; De Marco, A.; Polimeno, F.; Di Tommasi, P.; Maglione, G.; Ottaiano, L.; Arena, C.; Magliulo, V.; Vitale, L. Effects of the fertilizer added with DMPP on soil nitrous oxide emissions and microbial functional diversity. Agriculture 2021, 11, 12. [Google Scholar] [CrossRef]
- Bardon, C.; Poly, F.; Piola, F.; Pancton, M.; Comte, G.; Meiffren, G.; el Zahar Haichar, F. Mechanism of biological denitrification inhibition: Procyanidins induce an allosteric transition of the membrane-bound nitrate reductase through membrane alteration. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [Green Version]
- Bardon, C.; Poly, F.; el Zahar Haichar, F.; Le Roux, X.; Simon, L.; Meiffren, G.; Comte, G.; Rouifed, S.; Piola, F. Biological denitrification inhibition (BDI) with procyanidins induces modification of root traits, growth and N status in Fallopia x bohemica. Soil Biol. Biochem. 2017, 107, 41–49. [Google Scholar] [CrossRef]
- Galland, W.; Piola, F.; Mathieu, C.; Bouladra, L.; Simon, L.; el Zahar Haichar, F. Does biological denitrification inhibition (BDI) in the field induce an increase in plant growth and nutrition in Apium graveolens L. grown for a long period? Microorganisms 2020, 8, 1204. [Google Scholar] [CrossRef]
- Galland, W.; Piola, F.; Burlet, A.; Mathieu, C.; Nardy, M.; Poussineau, S.; Blazère, L.; Gervaix, J.; Puijalon, S.; Simon, L.; et al. Biological denitrification inhibition (BDI) in the field: A strategy to improve plant nutrition and growth. Soil Biol. Biochem. 2019, 136, 107513. [Google Scholar] [CrossRef]
- Galland, W.; el Zahar Haichar, F.; Czarnes, S.; Mathieu, C.; Demorge, J.-L.; Simon, L.; Puijalon, S.; Piola, F. Biological inhibition of denitrification (BDI) in the field: Effect on plant growth in two different soils. Appl. Soil Ecol. 2021, 159, 103857. [Google Scholar] [CrossRef]
- Preece, D.A. RA fisher and experimental design: A review. Biometrics 1990, 46, 925–935. [Google Scholar] [CrossRef]
- Hénin, S. Cours de Physique du Sol: Texture-Structure-Aération; Initiations-Documentations Techniques; ORSTOM: Paris, France, 1976; p. 159. [Google Scholar]
- Despujols, J.; Station d’Experimentation et d’Information R.A.L. Control of nitrate level in autumn greenhouse lettuce. Infos CTIFL Fr. 1997. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S RRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Qian, P.-Y. Conservative fragments in bacterial 16S RRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- White, T.J.; Burns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Appl. Lab. Man. 1990, 18, 315–322. [Google Scholar]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarm: Robust and fast clustering method for amplicon-based studies. PeerJ 2014, 2, e593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of RRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [Green Version]
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, rapidly, OTUs with galaxy solution. Bioinformatics 2017, 34, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aßhauer, K.P.; Meinicke, P. On the estimation of metabolic profiles in metagenomics. In Proceedings of the German Conference on Bioinformatics 2013, Gottingen, Germany, 10–13 September 2013; pp. 1–13. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [Green Version]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Constructing and analyzing microbiome networks in R. In Microbiome Analysis; Humana Press: New York, NY, USA, 2018; Volume 1849, pp. 243–266. [Google Scholar] [CrossRef]
- Zheng, Q.; Hu, Y.; Zhang, S.; Noll, L.; Böckle, T.; Dietrich, M.; Herbold, C.W.; Eichorst, S.A.; Woebken, D.; Richter, A.; et al. Soil multifunctionality is affected by the soil environment and by microbial community composition and diversity. Soil Biol. Biochem. 2019, 136, 107521. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Burns, K.N.; Bokulich, N.A.; Cantu, D.; Greenhut, R.F.; Kluepfel, D.A.; O’Geen, A.T.; Strauss, S.L.; Steenwerth, K.L. Vineyard soil bacterial diversity and composition revealed by 16S RRNA genes: Differentiation by vineyard management. Soil Biol. Biochem. 2016, 103, 337–348. [Google Scholar] [CrossRef] [Green Version]
- Drenovsky, R.E.; Vo, D.; Graham, K.J.; Scow, K.M. Soil water content and organic carbon availability are major determinants of soil microbial community composition. Microb. Ecol. 2004, 48, 424–430. [Google Scholar] [CrossRef]
- Ng, E.-L.; Patti, A.F.; Rose, M.T.; Schefe, C.R.; Wilkinson, K.; Smernik, R.J.; Cavagnaro, T.R. Does the chemical nature of soil carbon drive the structure and functioning of soil microbial communities? Soil Biol. Biochem. 2014, 70, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Wang, C.; Zheng, M.; Jiang, L.; Luo, Y. Patterns and mechanisms of responses by soil microbial communities to nitrogen addition. Soil Biol. Biochem. 2017, 115, 433–441. [Google Scholar] [CrossRef]
- Li, Y.; Chapman, S.J.; Nicol, G.W.; Yao, H. Nitrification and nitrifiers in acidic soils. Soil Biol. Biochem. 2018, 116, 290–301. [Google Scholar] [CrossRef]
- Ling, N.; Chen, D.; Guo, H.; Wei, J.; Bai, Y.; Shen, Q.; Hu, S. Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma 2017, 292, 25–33. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Q.; Han, X. Soil bacterial communities respond to mowing and nutrient addition in a steppe ecosystem. PLoS ONE 2013, 8, e84210. [Google Scholar] [CrossRef]
- Kim, J.M.; Roh, A.-S.; Choi, S.-C.; Kim, E.-J.; Choi, M.-T.; Ahn, B.-K.; Kim, S.-K.; Lee, Y.-H.; Joa, J.-H.; Kang, S.-S.; et al. Soil PH and electrical conductivity are key edaphic factors shaping bacterial communities of greenhouse soils in Korea. J. Microbiol. 2016, 54, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Sridevi, G.; Minocha, R.; Turlapati, S.A.; Goldfarb, K.C.; Brodie, E.L.; Tisa, L.S.; Minocha, S.C. Soil bacterial communities of a calcium-supplemented and a reference watershed at the Hubbard Brook Experimental Forest (HBEF), New Hampshire, USA. FEMS Microbiol. Ecol. 2012, 79, 728–740. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.; Duan, Y.; Schramm, A.; Eriksen, J.; Petersen, S.O. 3,4-dimethylpyrazole phosphate (DMPP) reduces activity of ammonia oxidizers without adverse effects on non-target soil microorganisms and functions. Appl. Soil Ecol. 2016, 105, 67–75. [Google Scholar] [CrossRef]
- Bardon, C.; Piola, F.; Bellvert, F.; el Zahar Haichar, F.; Comte, G.; Meiffren, G.; Pommier, T.; Puijalon, S.; Tsafack, N.; Poly, F. Evidence for biological denitrification inhibition (BDI) by plant secondary metabolites. New Phytol. 2014, 204, 620–630. [Google Scholar] [CrossRef] [PubMed]

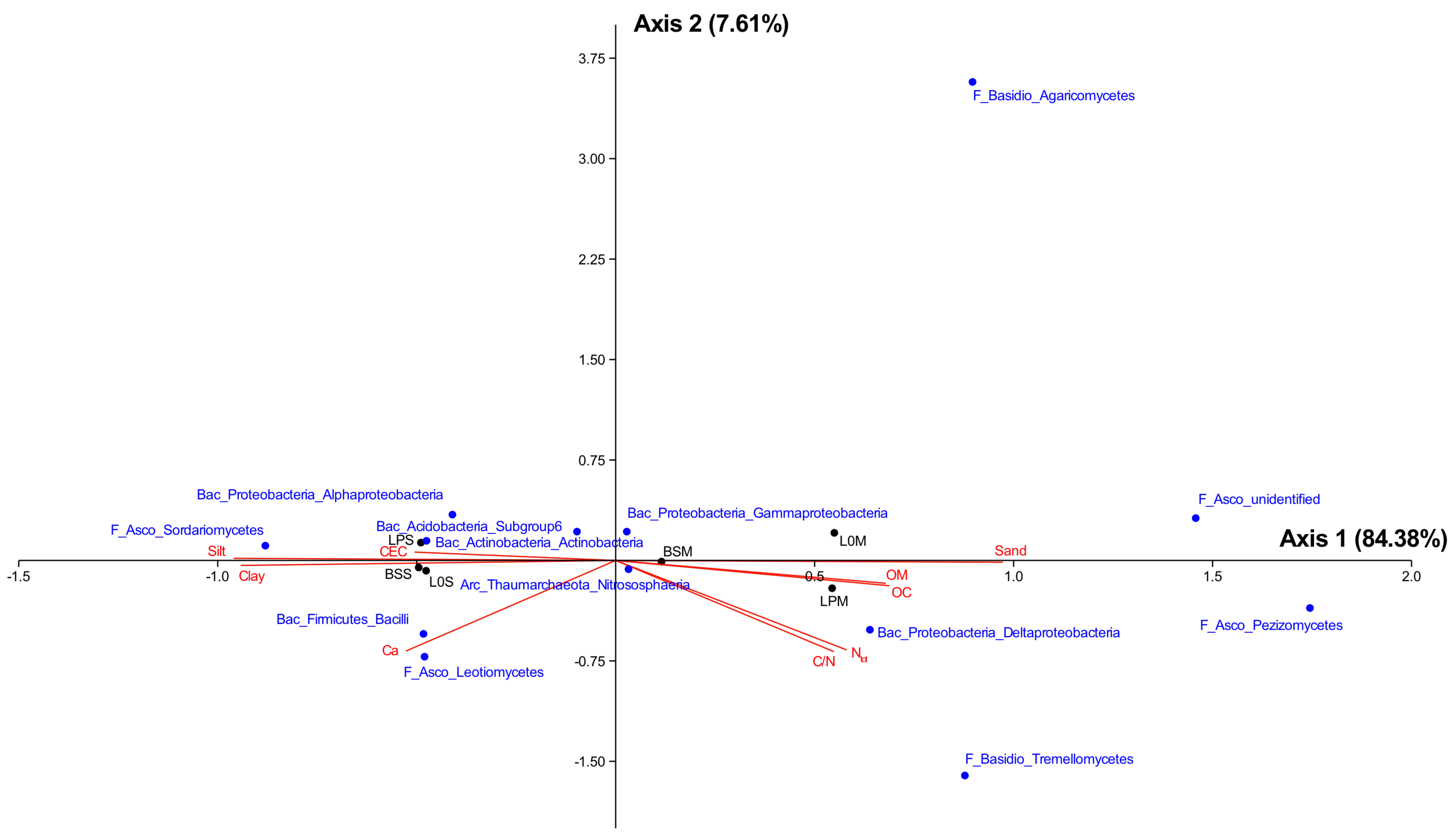
| (A) | ||||||||||||
| BSM | L0M | LPM | BSS | L0S | LPS | |||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Taxa Number | 1138.7 | 89.0 | 1162.3 | 38.0 | 1162.7 | 34.2 | 1066.3 | 80.7 | 1172.0 | 36.5 | 1183.7 | 24.1 |
| Simpson | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 |
| Shannon | 6.2 | 0.2 | 6.3 | 0.0 | 6.2 | 0.0 | 5.5 | 0.8 | 6.2 | 0.0 | 6.2 | 0.1 |
| Evenness | 0.4 | 0.1 | 0.5 | 0.0 | 0.4 | 0.0 | 0.3 | 0.2 | 0.4 | 0.0 | 0.4 | 0.1 |
| Chao-1 | 1350.0 | 74.1 | 1374.7 | 28.7 | 1380.0 | 46.5 | 1357.7 | 25.8 | 1353.3 | 33.2 | 1371.3 | 29.6 |
| (B) | ||||||||||||
| BSM | L0M | LPM | BSS | L0S | LPS | |||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Taxa Number | 193.7 | 5.1 | 202.3 | 3.5 | 186.3 | 16.8 | 245.7 | 11.9 | 249.7 | 4.9 | 242.0 | 24.9 |
| Simpson | 0.9 | 0.0 | 0.9 | 0.0 | 0.9 | 0.0 | 0.9 | 0.0 | 0.9 | 0.0 | 0.9 | 0.0 |
| Shannon | 3.4 | 0.3 | 3.4 | 0.1 | 3.2 | 0.1 | 3.6 | 0.2 | 3.6 | 0.1 | 3.6 | 0.4 |
| Evenness | 0.2 | 0.0 | 0.1 | 0.0 | 0.1 | 0.0 | 0.2 | 0.0 | 0.2 | 0.0 | 0.2 | 0.1 |
| Chao-1 | 205.2 | 7.7 | 216.9 | 6.6 | 195.7 | 18.0 | 261.9 | 13.8 | 283.4 | 14.4 | 260.7 | 10.7 |
| (A) Abundance of OTUs | ||||||||
| Prokaryotic 16S rRNA Gene | Fungal ITS | |||||||
| Df | F | p value | R2 | Df | F | p value | R2 | |
| Effect | ||||||||
| Plant | 1 | 1.40 | 0.184 | 0.048 | 1 | 1.41 | 0.138 | 0.039 |
| Treatment | 1 | 0.52 | 0.875 | 0.018 | 1 | 0.96 | 0.357 | 0.027 |
| Soil | 1 | 13.31 | 0.001 | 0.457 | 1 | 19.28 | 0.001 | 0.541 |
| Effect Interactions | ||||||||
| Plant*Soil | 1 | 1.32 | 0.231 | 0.045 | 1 | 1.15 | 0.294 | 0.032 |
| Treatment*Soil | 1 | 0.54 | 0.853 | 0.019 | 1 | 0.84 | 0.460 | 0.235 |
| Residuals | 12 | 0.412 | 12 | 0.337 | ||||
| (B) Richness of OTUs | ||||||||
| Prokaryotic 16S rRNA Gene | Fungal ITS | |||||||
| Df | F | p value | R2 | Df | F | p value | R2 | |
| Effect | ||||||||
| Plant | 1 | 1.33 | 0.156 | 0.033 | 1 | 1.88 | 0.154 | 0.024 |
| Treatment | 1 | 0.72 | 0.447 | 0.018 | 1 | 1.24 | 0.280 | 0.016 |
| Soil | 1 | 24.56 | 0.001 | 0.609 | 1 | 62.60 | 0.001 | 0.790 |
| Effect Interactions | ||||||||
| Plant*Treatment*Soil | 1 | 0.87 | 0.445 | 0.043 | 1 | 0.78 | 0.506 | 0.020 |
| Residuals | 12 | 0.198 | 12 | 0.151 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hugoni, M.; Galland, W.; Lecomte, S.; Bruto, M.; Barakat, M.; Piola, F.; Achouak, W.; Haichar, F.e.Z. Effects of the Denitrification Inhibitor “Procyanidins” on the Diversity, Interactions, and Potential Functions of Rhizosphere-Associated Microbiome. Microorganisms 2021, 9, 1406. https://doi.org/10.3390/microorganisms9071406
Hugoni M, Galland W, Lecomte S, Bruto M, Barakat M, Piola F, Achouak W, Haichar FeZ. Effects of the Denitrification Inhibitor “Procyanidins” on the Diversity, Interactions, and Potential Functions of Rhizosphere-Associated Microbiome. Microorganisms. 2021; 9(7):1406. https://doi.org/10.3390/microorganisms9071406
Chicago/Turabian StyleHugoni, Mylène, William Galland, Solène Lecomte, Maxime Bruto, Mohamed Barakat, Florence Piola, Wafa Achouak, and Feth el Zahar Haichar. 2021. "Effects of the Denitrification Inhibitor “Procyanidins” on the Diversity, Interactions, and Potential Functions of Rhizosphere-Associated Microbiome" Microorganisms 9, no. 7: 1406. https://doi.org/10.3390/microorganisms9071406
APA StyleHugoni, M., Galland, W., Lecomte, S., Bruto, M., Barakat, M., Piola, F., Achouak, W., & Haichar, F. e. Z. (2021). Effects of the Denitrification Inhibitor “Procyanidins” on the Diversity, Interactions, and Potential Functions of Rhizosphere-Associated Microbiome. Microorganisms, 9(7), 1406. https://doi.org/10.3390/microorganisms9071406






