Discovery and Heterologous Production of New Cyclic Depsibosamycins
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Procedures
2.2. Metabolite Extraction and Analysis by Mass Spectrometry (MS)
2.3. Nuclear Magnetic Resonance (NMR) Spectroscopy and Optical Rotation (OD)
2.4. Isolation of Bosamycin B–D and Depsibosamycin C
2.4.1. Bosamycin B
2.4.2. Bosamycin C
2.4.3. Bosamycin D
2.4.4. Depsibosamycin C
2.5. Marfey’s Analysis
2.5.1. DL-Erythro-Hydroxyaspartic Acid
2.5.2. DL-Threo-Hydroxyaspartic Acid
2.6. Isolation and Manipulation of DNA
2.7. Genome Mining and Bioinformatics Analysis
3. Results and Discussion
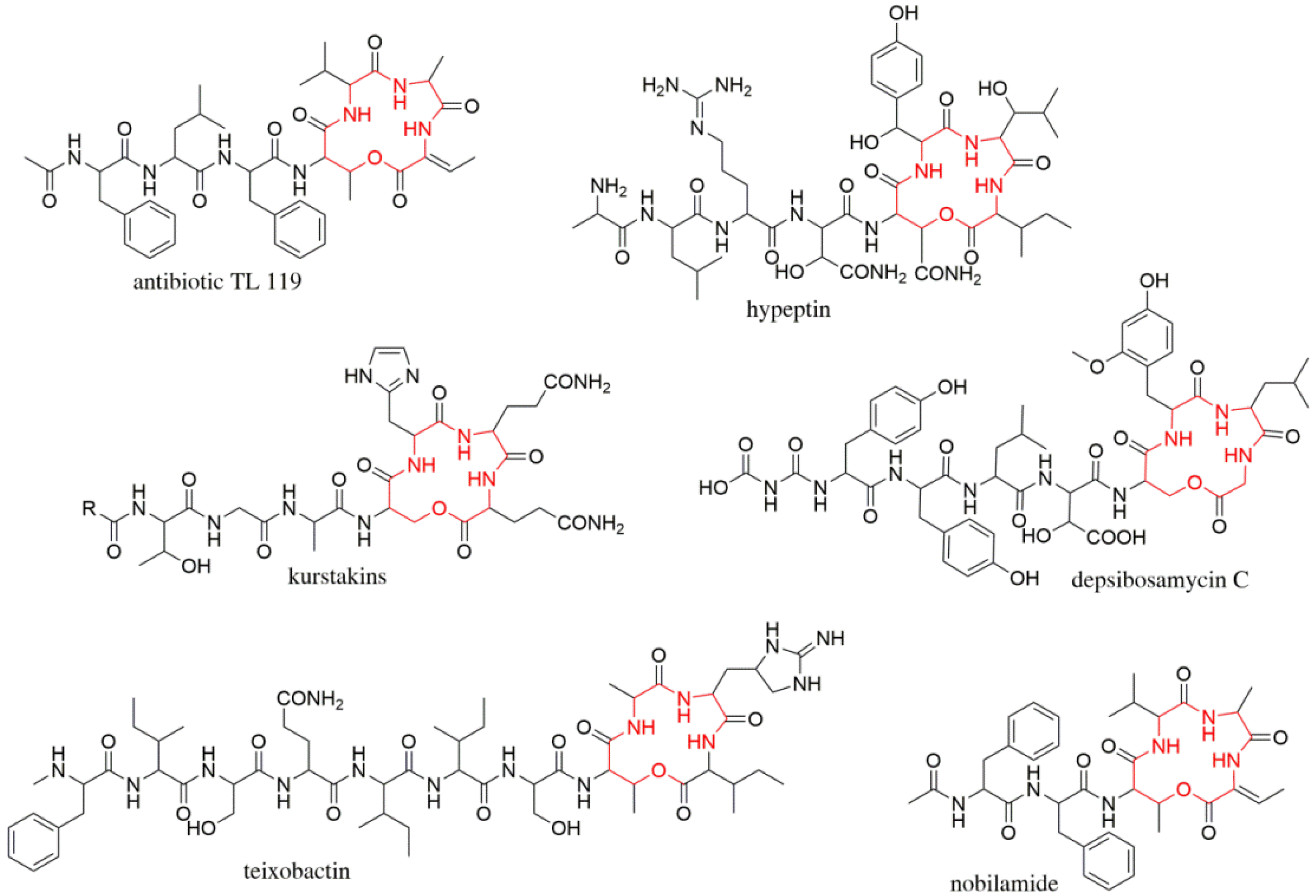
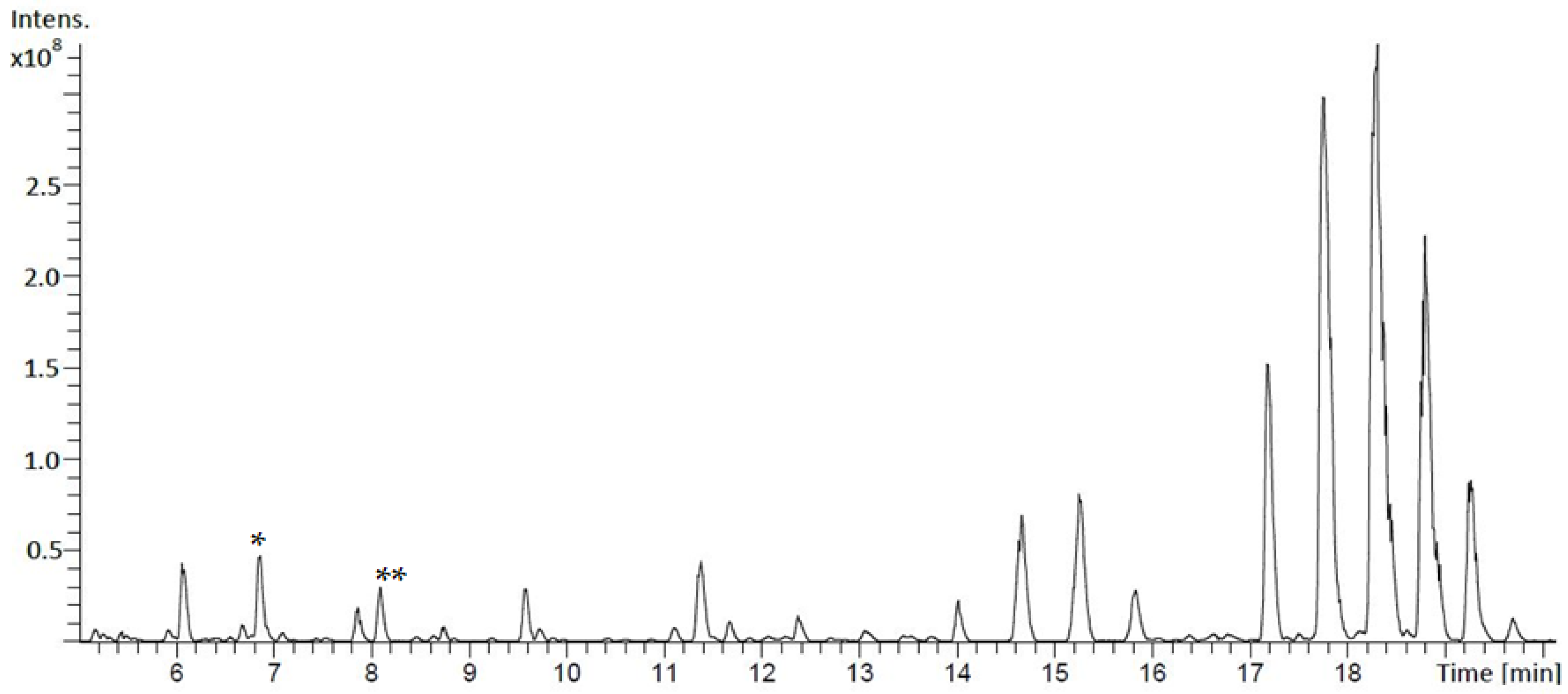
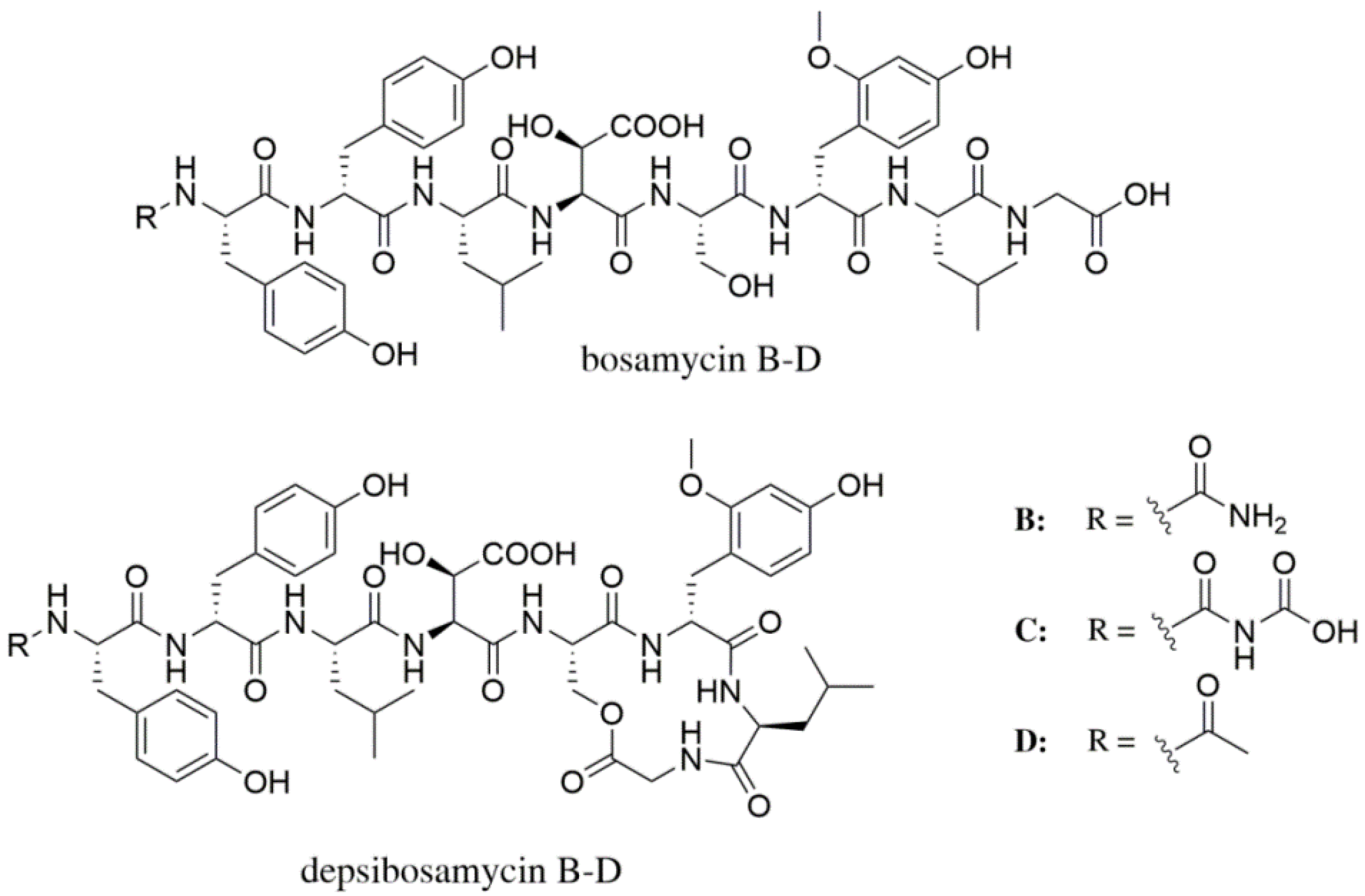
3.1. Identification and Structural Elucidation of Depsibosamycins B, C, and D
3.2. Heterologous Expression of the Depsibosamycin (dbm) Gene Cluster
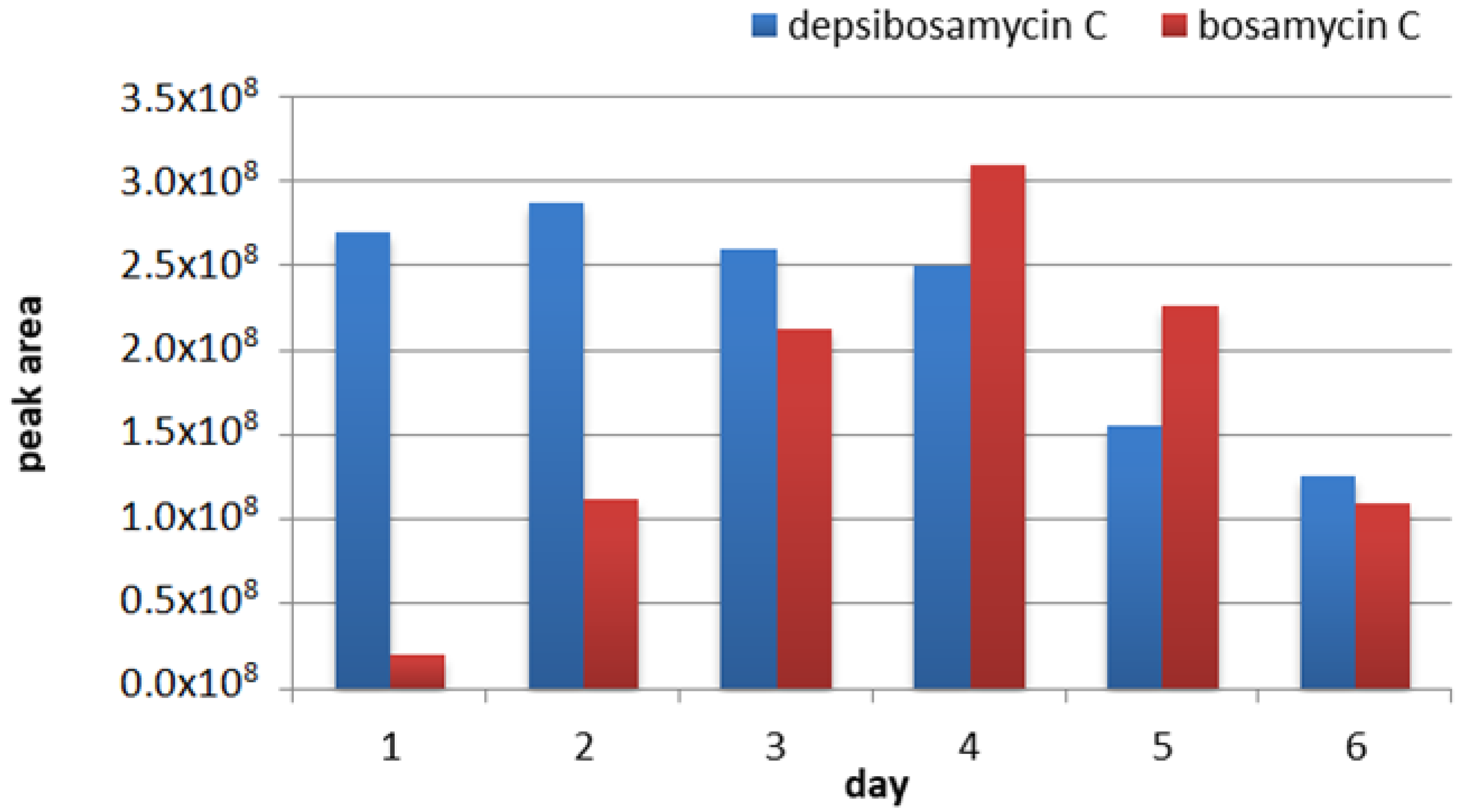
3.3. Determination of the Direct Biosynthetic Product of the dbm Gene Cluster
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Ntai, I.; Ju, K.-S.; Unger, M.; Zamdborg, L.; Robinson, S.J.; Doroghazi, J.R.; Labeda, D.P.; Metcalf, W.W.; Kelleher, N.L. A Proteomic Survey of Nonribosomal Peptide and Polyketide Biosynthesis in Actinobacteria. J. Proteome Res. 2011, 11, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Běhal, V. Bioactive products from streptomyces. Adv. Virus Res. 2000, 47, 113–156. [Google Scholar] [CrossRef]
- Strieker, M.; Tanović, A.; Marahiel, M.A. Nonribosomal peptide synthetases: Structures and dynamics. Curr. Opin. Struct. Biol. 2010, 20, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Bozhüyük, K.A.J.; Linck, A.; Tietze, A.; Kranz, J.; Wesche, F.; Nowak, S.; Fleischhacker, F.; Shi, Y.-N.; Grün, P.; Bode, H.B. Modification and de novo design of non-ribosomal peptide synthetases using specific assembly points within condensation domains. Nat. Chem. 2019, 11, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Süssmuth, R.D.; Mainz, A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew. Chem. Int. Ed. 2017, 56, 3770–3821. [Google Scholar] [CrossRef] [PubMed]
- Finking, R.; Marahiel, M.A. Biosynthesis of Nonribosomal Peptides. Annu. Rev. Microbiol. 2004, 58, 453–488. [Google Scholar] [CrossRef]
- Myronovskyi, M.; Luzhetskyy, A. Heterologous production of small molecules in the optimized Streptomyces hosts. Nat. Prod. Rep. 2019, 36, 1281–1294. [Google Scholar] [CrossRef]
- Hill, R.A.; Sutherland, A. Hot off the Press. Nat. Prod. Rep. 2020, 37, 1294–1299. [Google Scholar] [CrossRef]
- Green, J.M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012. [Google Scholar]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics. A Laboratory Manual; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Robertson, A.W.; McCarville, N.G.; MacIntyre, L.W.; Correa, H.; Haltli, B.; Marchbank, D.H.; Kerr, R.G. Isolation of Imaqobactin, an Amphiphilic Siderophore from the Arctic Marine Bacterium Variovorax Species RKJM285. J. Nat. Prod. 2018, 81, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Rebets, Y.; Kormanec, J.; Luzhetskyy, A.; Bernaerts, K.; Anné, J. Metagenomics: Methods and Protocols; Streit, W., Daniel, R., Eds.; Springer: New York, NY, USA, 2017; pp. 99–144. [Google Scholar]
- Flett, F.; Mersinias, V.; Smith, C.P. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997, 155, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.; Weber, T.; Takano, E.; Breitling, R. Antismash: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Whittle, M.; Willett, P.; Klaffke, W.; van Noort, P. Evaluation of Similarity Measures for Searching the Dictionary of Natural Products Database. J. Chem. Inf. Comput. Sci. 2003, 43, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.L. An additional source of macrotetrolide antibiotics. J. Antibiot. 1975, 28, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.; Ravel, J.; Townsend, C.A. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 2000, 7, 211–224. [Google Scholar] [CrossRef] [Green Version]
- Prieto, C.; García-Estrada, C.; Lorenzana, D.; Martín, J.F. NRPSsp: Non-ribosomal peptide synthase substrate predictor. Bioinformatics 2012, 28, 426–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rausch, C. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 2005, 33, 5799–5808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Röttig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2—A web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011, 39, W362–W367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rückert, C.; Albersmeier, A.; Busche, T.; Jaenicke, S.; Winkler, A.; Friðjónsson, Ó.H.; Hreggviðsson, G.Ó.; Lambert, C.; Badcock, D.; Bernaerts, K.; et al. Complete genome sequence of Streptomyces lividans TK24. J. Biotechnol. 2015, 199, 21–22. [Google Scholar] [CrossRef]
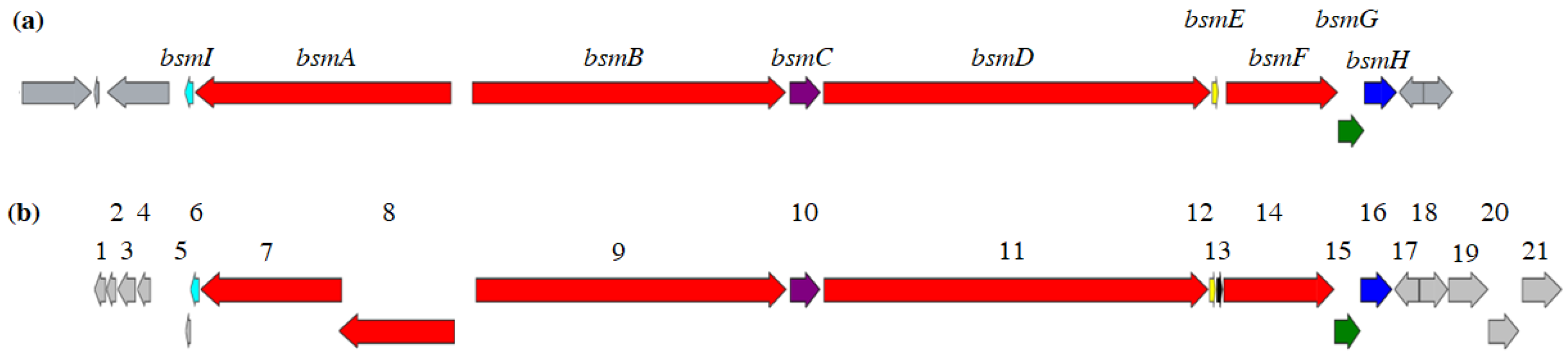
| Gene # | dbm Genes a | Putative Function | bsm Genes b |
|---|---|---|---|
| 1 | orf − 5 | Hypothetical protein | |
| 2 | orf − 4 | Hypothetical protein | |
| 3 | orf − 3 | Hypothetical protein | |
| 4 | orf − 2 | Hypothetical protein | |
| 5 | orf − 1 | Dehydrogenase | |
| 6 | dbmI | Acyl carrier protein | bsmI |
| 7 | dbmA1 | NRPS (C-A-PCP-E) | bsmA |
| 8 | dbmA2 | NRPS (C-A-PCP) | |
| 9 | dbmB | NRPS (C-A-PCP-C-A-PCP-C-A-PCP) | bsmB |
| 10 | dbmC | Dioxygenase | bsmC |
| 11 | dbmD | NRPS (C-A-PCP-E-C-A-PCP-C-A-PCP-TE) | bsmD |
| 12 | dbmE | MbtH family protein | bsmE |
| 13 | Hypothetical protein | ||
| 14 | dbmF | NRPS (P450-A-PCP) | bsmF |
| 15 | dbmG | Alpha/beta hydrolase | bsmG |
| 16 | dbmH | O-methyltransferase | bsmH |
| 17 | orf + 1 | ABC transporter | |
| 18 | orf + 2 | ABC transporter | |
| 19 | orf + 3 | Hypothetical protein | |
| 20 | orf + 4 | ABC transporter | |
| 21 | orf + 5 | Hypothetical protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stierhof, M.; Myronovskyi, M.; Zapp, J.; Luzhetskyy, A. Discovery and Heterologous Production of New Cyclic Depsibosamycins. Microorganisms 2021, 9, 1396. https://doi.org/10.3390/microorganisms9071396
Stierhof M, Myronovskyi M, Zapp J, Luzhetskyy A. Discovery and Heterologous Production of New Cyclic Depsibosamycins. Microorganisms. 2021; 9(7):1396. https://doi.org/10.3390/microorganisms9071396
Chicago/Turabian StyleStierhof, Marc, Maksym Myronovskyi, Josef Zapp, and Andriy Luzhetskyy. 2021. "Discovery and Heterologous Production of New Cyclic Depsibosamycins" Microorganisms 9, no. 7: 1396. https://doi.org/10.3390/microorganisms9071396
APA StyleStierhof, M., Myronovskyi, M., Zapp, J., & Luzhetskyy, A. (2021). Discovery and Heterologous Production of New Cyclic Depsibosamycins. Microorganisms, 9(7), 1396. https://doi.org/10.3390/microorganisms9071396





