One Pathway, Two Cyclic Non-Ribosomal Pentapeptides: Heterologous Expression of BE-18257 Antibiotics and Pentaminomycins from Streptomyces cacaoi CA-170360
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. Growth and Culture Conditions
2.3. Identification of cpp Cluster from Strain CA-170360 Whole Genome Sequence
2.4. Cloning and Heterologous Expression of the cpp Gene Cluster
2.5. Extraction and Detection of BE-18257 Antibiotics and Pentaminomycins
3. Results and Discussion
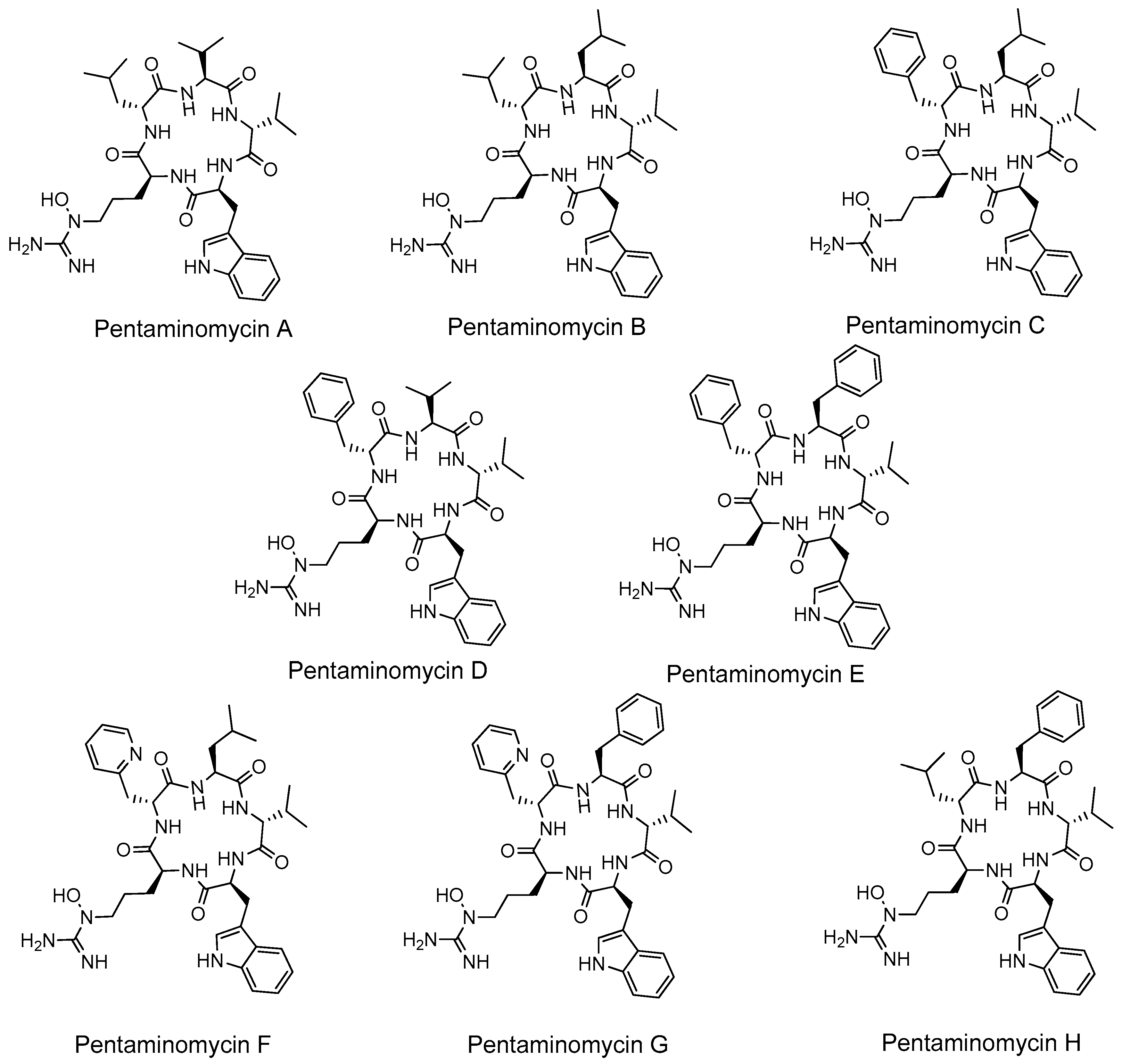
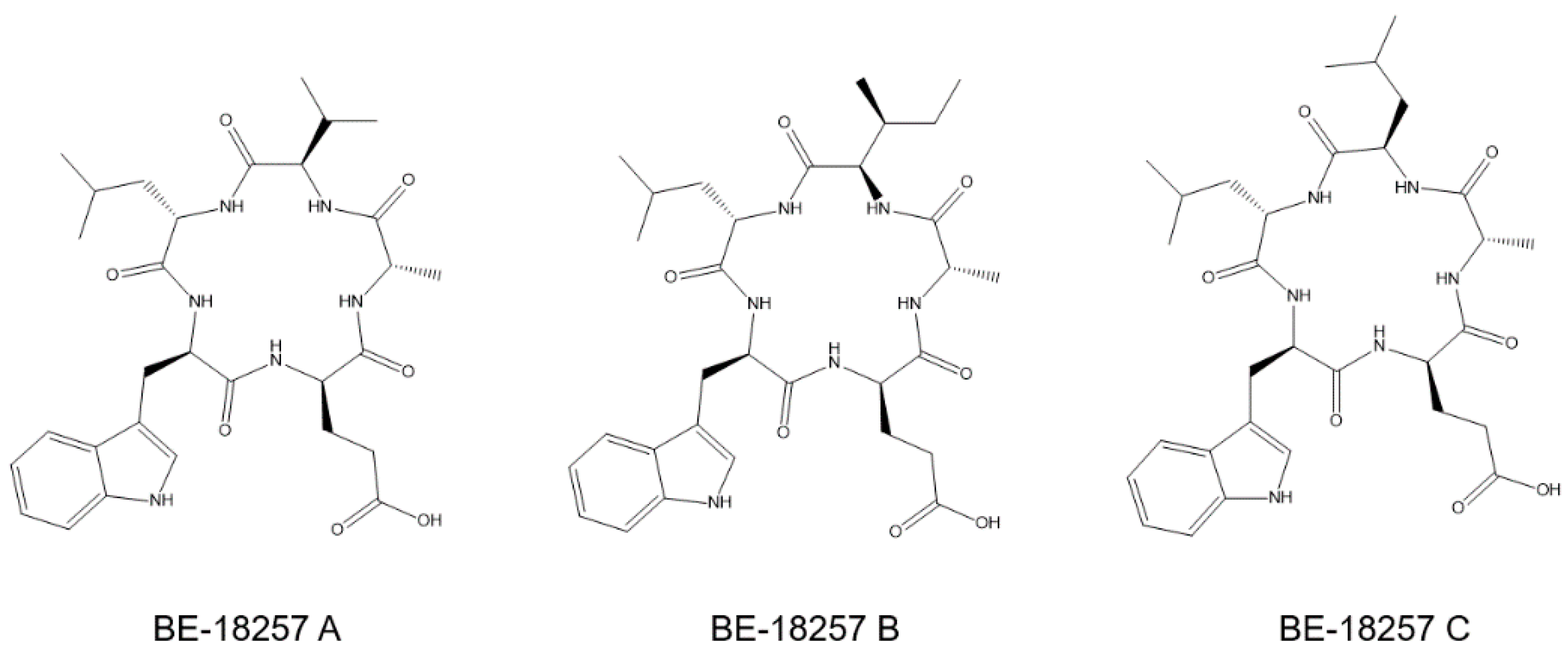
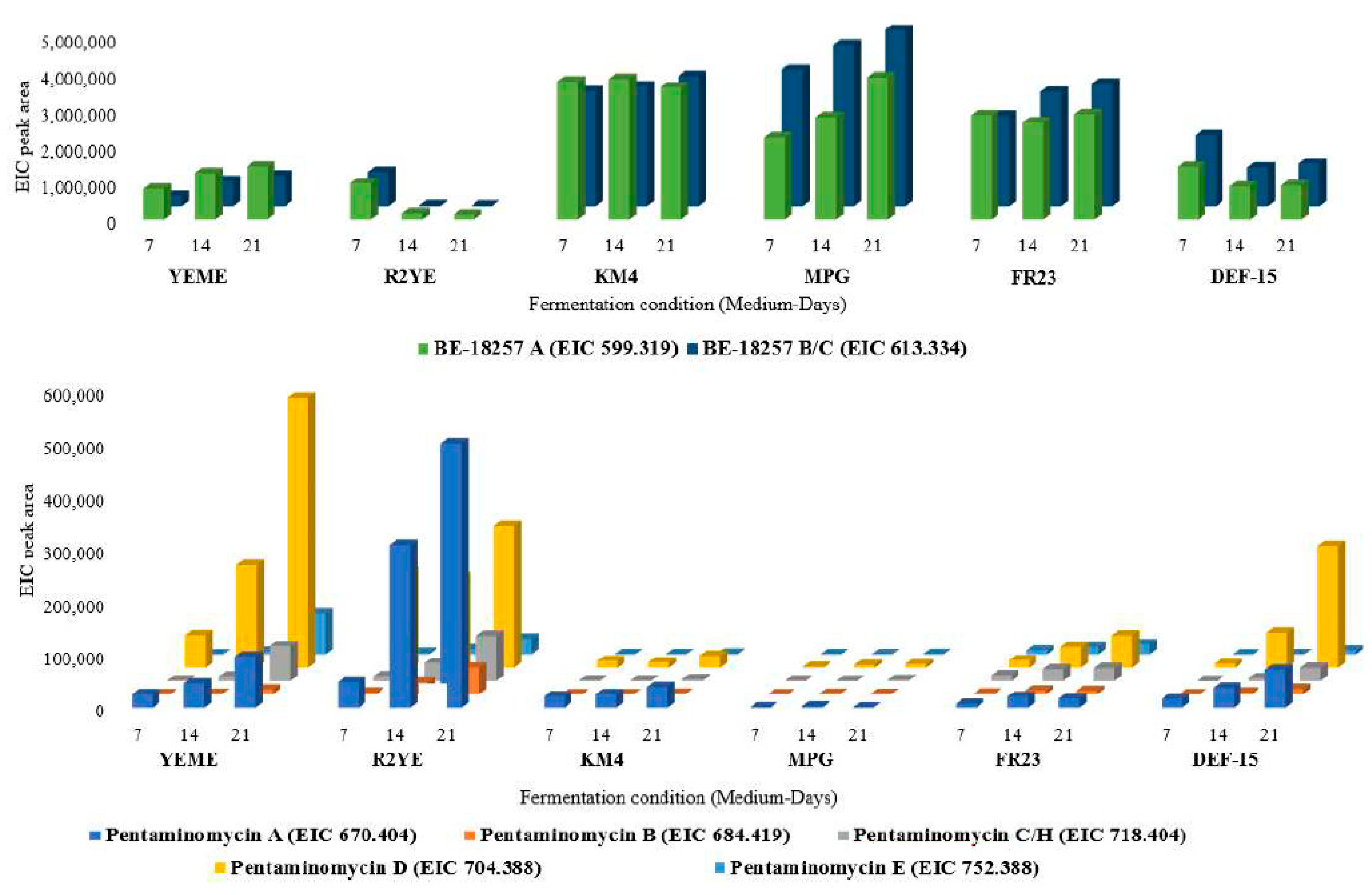
3.1. Production of Cyclic Pentapeptides by Strain CA-170360
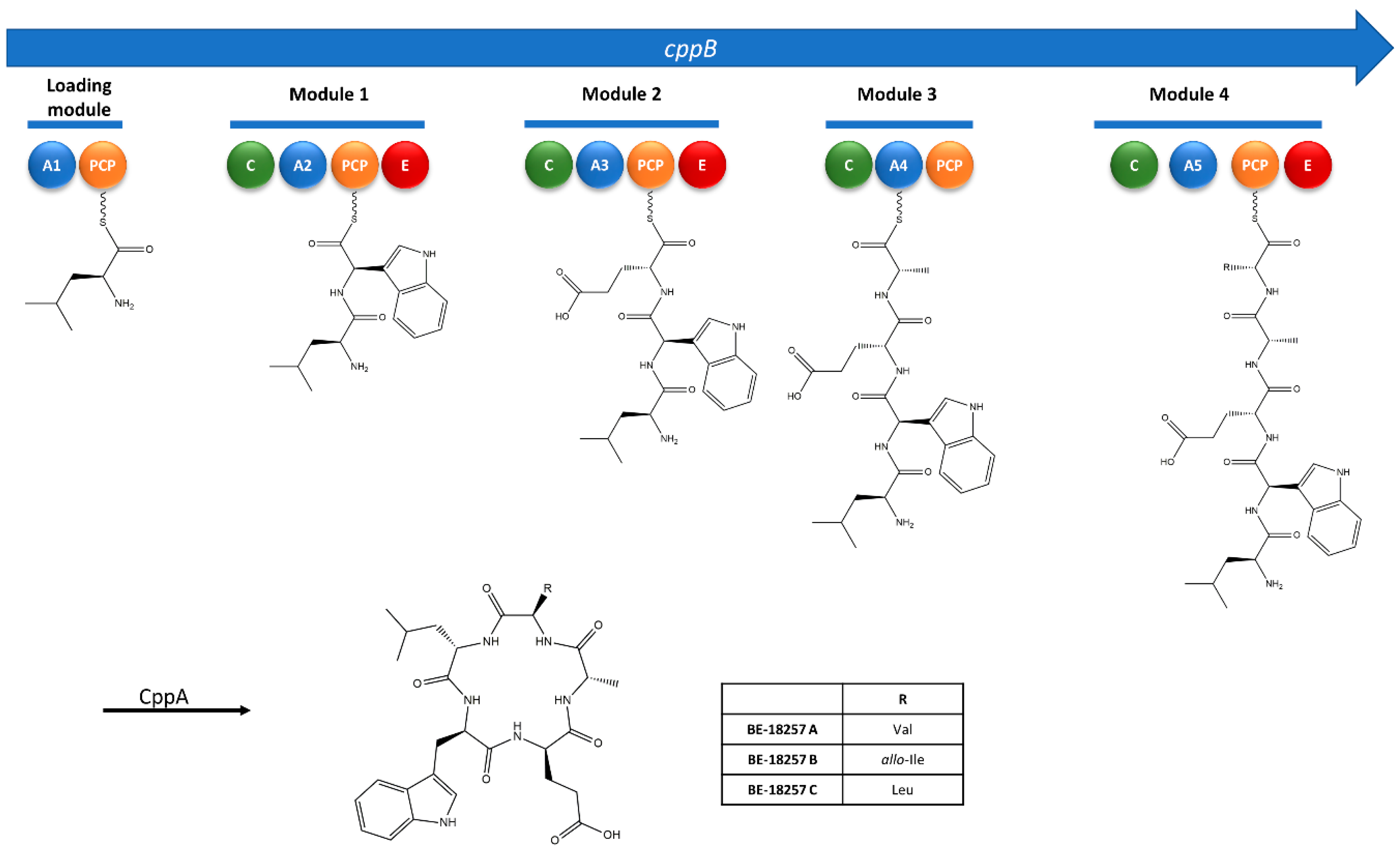
3.2. Identification of the cpp Gene Cluster from the Whole Genome Sequence of Strain CA-170360
3.3. Cloning and Heterologous Expression of the cpp Gene Cluster
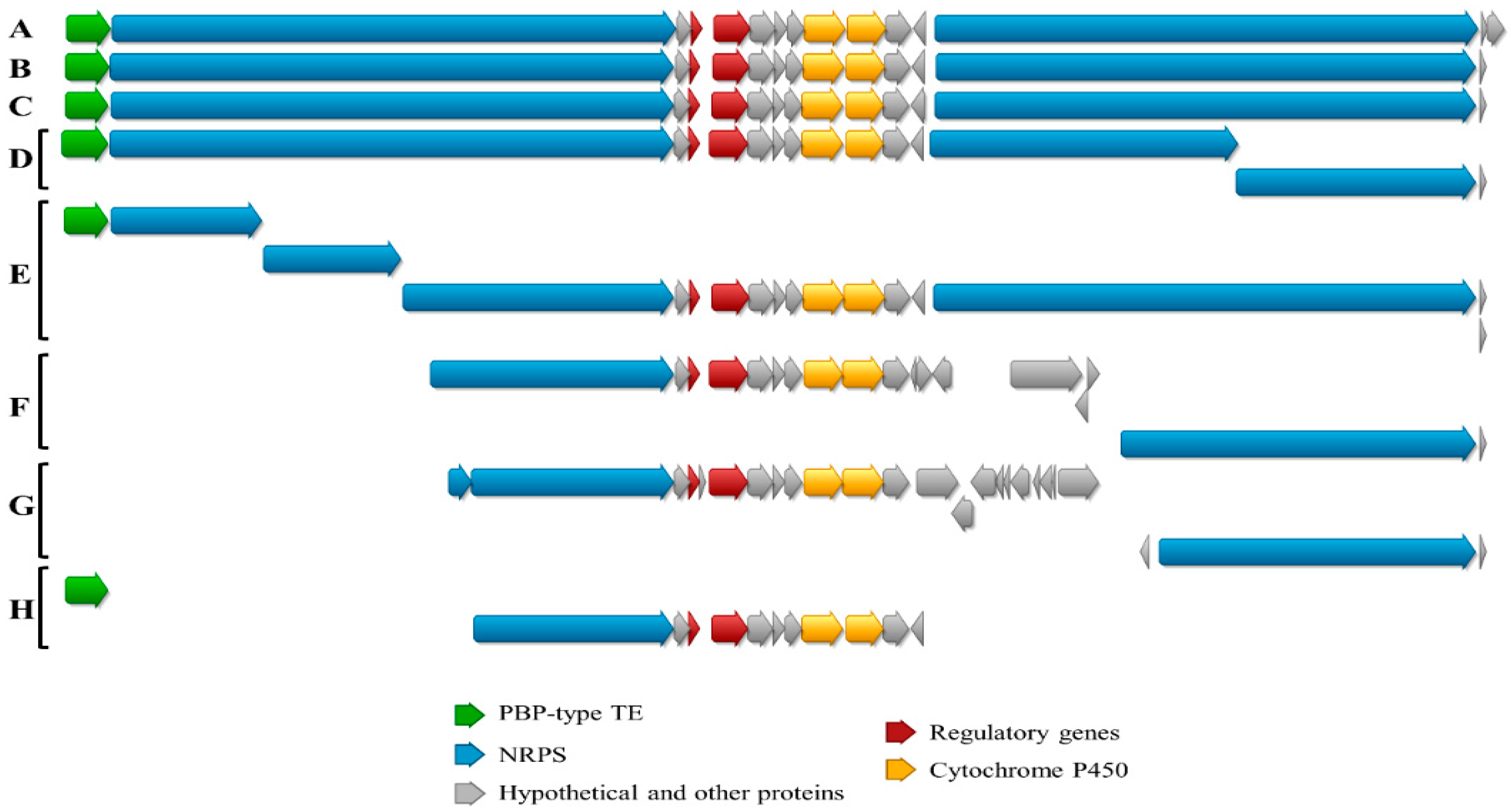
3.4. Genome Mining of cpp-like BGCs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdalla, M.A.; McGaw, L.J. Natural Cyclic Peptides as an Attractive Modality for Therapeutics: A Mini Review. Molecules 2018, 23, 2080. [Google Scholar] [CrossRef]
- Jing, X.; Jin, K. A gold mine for drug discovery: Strategies to develop cyclic peptides into therapies. Med. Res. Rev. 2020, 40, 753–810. [Google Scholar] [CrossRef] [PubMed]
- Mogi, T.; Kita, K. Gramicidin S and polymyxins: The revival of cationic cyclic peptide antibiotics. Cell. Mol. Life Sci. 2009, 66, 3821–3826. [Google Scholar] [CrossRef] [PubMed]
- Levine, D.P. Vancomycin: A History. Clin. Infect. Dis. 2006, 42, S5–S12. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, K.L.; Rybak, M.J. Daptomycin. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2004, 24, 41–57. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, M.; Sawa, R.; Kinoshita, N.; Hashizume, H.; Nakagawa, N.; Homma, Y.; Nishimura, Y.; Akamatsu, Y.; Igarashi, R.S.M. Pargamicin A, a Novel Cyclic Peptide Antibiotic from Amycolatopsis sp. J. Antibiot. 2008, 61, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, H.; Sawa, R.; Yamashita, K.; Nishimura, Y.; Igarashi, M. Structure and antibacterial activities of new cyclic peptide antibiotics, pargamicins B, C and D, from Amycolatopsis sp. ML1-hF4. J. Antibiot. 2017, 70, 699–704. [Google Scholar] [CrossRef]
- Abdalla, M.A. Medicinal significance of naturally occurring cyclotetrapeptides. J. Nat. Med. 2016, 70, 708–720. [Google Scholar] [CrossRef]
- Miyata, S.; Hashimoto, M.; Masui, Y.; Ezaki, M.; Takase, S.; Nishikawa, M.; Kiyoto, S.; Okuhara, M.; Kohsaka, M. WS-7338, new endothelin receptor antagonists isolated from Streptomyces sp. No. 7338. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 1992, 45, 74–82. [Google Scholar] [CrossRef]
- Miyata, S.; Hashimoto, M.; Fujie, K.; Nishikawa, M.; Kiyoto, S.; Okuhara, M.; Kohsaka, M. WS-7338, new endothelin receptor antagonists isolated from Streptomyces sp. No. 7338. II. Biological characterization and pharmacological characterization of WS-7338 B. J. Antibiot. 1992, 45, 83–87. [Google Scholar] [CrossRef][Green Version]
- Jang, J.-P.; Hwang, G.J.; Kwon, M.C.; Ryoo, I.-J.; Jang, M.; Takahashi, S.; Ko, S.-K.; Osada, H.; Jang, J.-H.; Ahn, J. Pentaminomycins A and B, Hydroxyarginine-Containing Cyclic Pentapeptides from Streptomyces sp. RK88-1441. J. Nat. Prod. 2018, 81, 806–810. [Google Scholar] [CrossRef] [PubMed]
- Kaweewan, I.; Hemmi, H.; Komaki, H.; Kodani, S. Isolation and structure determination of a new antibacterial peptide pentaminomycin C from Streptomyces cacaoi subsp. cacaoi. J. Antibiot. 2020, 73, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Le, L.T.H.L.; Shin, J.; Shin, J.; Lee, M.J.; Oh, D.-C. Pentaminomycins C–E: Cyclic Pentapeptides as Autophagy Inducers from a Mealworm Beetle Gut Bacterium. Microorganisms 2020, 8, 1390. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Kobayashi, M.; Kuranaga, T.; Takada, K.; Ikeda, H.; Matsunaga, S.; Wakimoto, T. SurE is a trans-acting thioesterase cyclizing two distinct non-ribosomal peptides. Org. Biomol. Chem. 2019, 17, 1058–1061. [Google Scholar] [CrossRef]
- Matsuda, K.; Kuranaga, T.; Wakimoto, T. A New Cyclase Family Catalyzing Head-to-Tail Macrolactamization of Non-ribosomal Peptides. J. Synth. Org. Chem. Jpn. 2019, 77, 1106–1115. [Google Scholar] [CrossRef]
- Zhou, Y.; Lin, X.; Xu, C.; Shen, Y.; Wang, S.-P.; Liao, H.; Li, L.; Deng, H.; Yang, F. Investigation of Penicillin Binding Protein (PBP)-like Peptide Cyclase and Hydrolase in Surugamide Non-ribosomal Peptide Biosynthesis. Cell Chem. Biol. 2019, 26, 737–744. [Google Scholar] [CrossRef]
- Carretero-Molina, D.; Ortiz-López, F.J.; Martín, J.; González, I.; Sánchez-Hidalgo, M.; Román-Hurtado, F.; Díaz, C.; de la Cruz, M.; Genilloud, O.; Reyes, F. Pentaminomycins F and G, first non-ribosomal peptides containing 2-pyridylalanine. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Ortiz--López, F.J.; Carretero--Molina, D.; Sánchez--Hidalgo, M.; Martín, J.; González, I.; Román--Hurtado, F.; De La Cruz, M.; García--Fernández, S.; Reyes, F.; Deisinger, J.P.; et al. Cacaoidin, First Member of the New Lanthidin RiPP Family. Angew. Chem. Int. Ed. 2020, 59, 12654–12658. [Google Scholar] [CrossRef]
- Román-Hurtado, F.; Sánchez-Hidalgo, M.; Martín, J.; Ortiz-López, F.J.; Genilloud, O. Biosynthesis and heterologous expression of cacaoidin, the first member of the lanthidin family of RiPPs. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, J.J.; Yamanaka, K.; Tang, X.; Moore, B.S. Direct cloning and heterologous expression of natural product biosynthetic gene clusters by transformation-associated recombination. Methods Enzym. 2019, 621, 87–110. [Google Scholar] [CrossRef]
- Chater, K.F.; Wilde, L.C. Streptomyces albus G Mutants Defective in the SalGI Restriction-Modification System. J. Gen. Microbiol. 1980, 116, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhao, X.; Gabrieli, T.; Lou, C.; Ebenstein, Y.; Zhu, T.F. Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat. Commun. 2015, 6, 8101. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Robertsen, H.L.; Blin, K.; Weber, T.; Lee, S.Y. CRISPR-Cas9 Toolkit for Actinomycete Genome Editing. Methods Mol. Biol. 2018, 1671, 163–184. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhu, T.F. Targeted isolation and cloning of 100-kb microbial genomic sequences by Cas9-assisted targeting of chromosome segments. Nat. Protoc. 2016, 11, 960–975. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Kuranaga, T.; Matsuda, K.; Sano, A.; Kobayashi, M.; Ninomiya, A.; Takada, K.; Matsunaga, S.; Wakimoto, T. Total Synthesis of the Nonribosomal Peptide Surugamide B and Identification of a New Offloading Cyclase Family. Angew. Chem. Int. Ed. 2018, 57, 9447–9451. [Google Scholar] [CrossRef]
- Fazal, A.; Webb, M.E.; Seipke, R.F. The Desotamide Family of Antibiotics. Antibiot. 2020, 9, 452. [Google Scholar] [CrossRef]
- Son, S.; Hong, Y.-S.; Jang, M.; Heo, K.T.; Lee, B.; Jang, J.-P.; Kim, J.-W.; Ryoo, I.-J.; Kim, W.-G.; Ko, S.-K.; et al. Genomics-Driven Discovery of Chlorinated Cyclic Hexapeptides Ulleungmycins A and B from a Streptomyces Species. J. Nat. Prod. 2017, 80, 3025–3031. [Google Scholar] [CrossRef]
- Mudalungu, C.M.; Von Törne, W.J.; Voigt, K.; Rückert, C.; Schmitz, S.; Sekurova, O.N.; Zotchev, S.B.; Süssmuth, R.D. Noursamycins, Chlorinated Cyclohexapeptides Identified from Molecular Networking of Streptomyces noursei NTR-SR4. J. Nat. Prod. 2019, 82, 1478–1486. [Google Scholar] [CrossRef]
- Kaweewan, I.; Komaki, H.; Hemmi, H.; Kodani, S. Isolation and Structure Determination of New Antibacterial Peptide Curacomycin Based on Genome Mining. Asian J. Org. Chem. 2017, 6, 1838–1844. [Google Scholar] [CrossRef]
- Magarvey, N.A.; Haltli, B.; He, M.; Greenstein, M.; Hucul, J.A. Biosynthetic Pathway for Mannopeptimycins, Lipoglycopeptide Antibiotics Active against Drug-Resistant Gram-Positive Pathogens. Antimicrob. Agents Chemother. 2006, 50, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Reynolds, K.A.; Kersten, R.D.; Ryan, K.S.; Gonzalez, D.J.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. USA 2014, 111, 1957–1962. [Google Scholar] [CrossRef] [PubMed]
- Vicente, C.M.; Thibessard, A.; Lorenzi, J.-N.; Benhadj, M.; Hôtel, L.; Gacemi-Kirane, D.; Lespinet, O.; Leblond, P.; Aigle, B. Comparative Genomics among Closely Related Streptomyces Strains Revealed Specialized Metabolite Biosynthetic Gene Cluster Diversity. Antibiotics 2018, 7, 86. [Google Scholar] [CrossRef] [PubMed]



| ORF | Length (aa) | Closest BLAST Match (Organism) GenBank Reference | Identity (%)/Similarity (%) |
|---|---|---|---|
| cppA | 502 | Hypothetical protein DEH18_05445 (Streptomyces sp. NHF165) QHF93414.1 | 99/99 |
| cppB | 6187 | Non-ribosomal peptide synthase/polyketide synthase (Streptomyces cacaoi) WP_158102276.1 | 99/99 |
| cppC | 172 | MULTISPECIES: DUF2975 domain-containing protein (Streptomyces) WP_030891799.1 | 100/100 |
| cppD | 102 | Helix-turn-helix domain-containing protein (Streptomyces cacaoi) WP_149564434.1 | 99/99 |
| cppE | 420 | MULTISPECIES: sensor histidine kinase (Streptomyces) WP_051857187.1 | 99/100 |
| cppF | 280 | Hypothetical protein SCA03_67000 (Streptomyces cacaoi subsp. cacaoi) GEB54149.1 | 100/100 |
| cppG | 131 | MULTISPECIES: DUF742 domain-containing protein (Streptomyces) WP_030891786.1 | 100/100 |
| cppH | 186 | MULTISPECIES: ATP/GTP-binding protein (Streptomyces) WP_030891784.1 | 100/100 |
| cppI | 451 | MULTISPECIES: cytochrome P450 (Streptomyces) WP_030891781.1 | 100/100 |
| cppJ | 431 | Cytochrome P450 (Streptomyces cacaoi) WP_086815207.1 | 99/99 |
| cppK | 271 | 3-hydroxybutyryl-CoA dehydratase (Streptomyces cacaoi subsp. cacaoi) GEB54144.1 | 99/99 |
| cppL | 141 | MULTISPECIES: hypothetical protein (Streptomyces) WP_141275837.1 | 100/100 |
| cppM | 5901 | Non-ribosomal peptide synthase/polyketide synthase (Streptomyces sp. NHF165) WP_159784853.1 | 99/99 |
| cppN | 69 | MULTISPECIES: hypothetical protein (Streptomyces) WP_030890086.1 | 100/100 |
| cppO | 175 | MULTISPECIES: hypothetical protein (unclassified Streptomyces) WP_030890089.1 | 99/100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Román-Hurtado, F.; Sánchez-Hidalgo, M.; Martín, J.; Ortiz-López, F.J.; Carretero-Molina, D.; Reyes, F.; Genilloud, O. One Pathway, Two Cyclic Non-Ribosomal Pentapeptides: Heterologous Expression of BE-18257 Antibiotics and Pentaminomycins from Streptomyces cacaoi CA-170360. Microorganisms 2021, 9, 135. https://doi.org/10.3390/microorganisms9010135
Román-Hurtado F, Sánchez-Hidalgo M, Martín J, Ortiz-López FJ, Carretero-Molina D, Reyes F, Genilloud O. One Pathway, Two Cyclic Non-Ribosomal Pentapeptides: Heterologous Expression of BE-18257 Antibiotics and Pentaminomycins from Streptomyces cacaoi CA-170360. Microorganisms. 2021; 9(1):135. https://doi.org/10.3390/microorganisms9010135
Chicago/Turabian StyleRomán-Hurtado, Fernando, Marina Sánchez-Hidalgo, Jesús Martín, Francisco Javier Ortiz-López, Daniel Carretero-Molina, Fernando Reyes, and Olga Genilloud. 2021. "One Pathway, Two Cyclic Non-Ribosomal Pentapeptides: Heterologous Expression of BE-18257 Antibiotics and Pentaminomycins from Streptomyces cacaoi CA-170360" Microorganisms 9, no. 1: 135. https://doi.org/10.3390/microorganisms9010135
APA StyleRomán-Hurtado, F., Sánchez-Hidalgo, M., Martín, J., Ortiz-López, F. J., Carretero-Molina, D., Reyes, F., & Genilloud, O. (2021). One Pathway, Two Cyclic Non-Ribosomal Pentapeptides: Heterologous Expression of BE-18257 Antibiotics and Pentaminomycins from Streptomyces cacaoi CA-170360. Microorganisms, 9(1), 135. https://doi.org/10.3390/microorganisms9010135








