Can Potato Plants Be Colonized with the Fungi Metarhizium and Beauveria under Their Natural Load in Agrosystems?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Locations and Soil Properties
- (1)
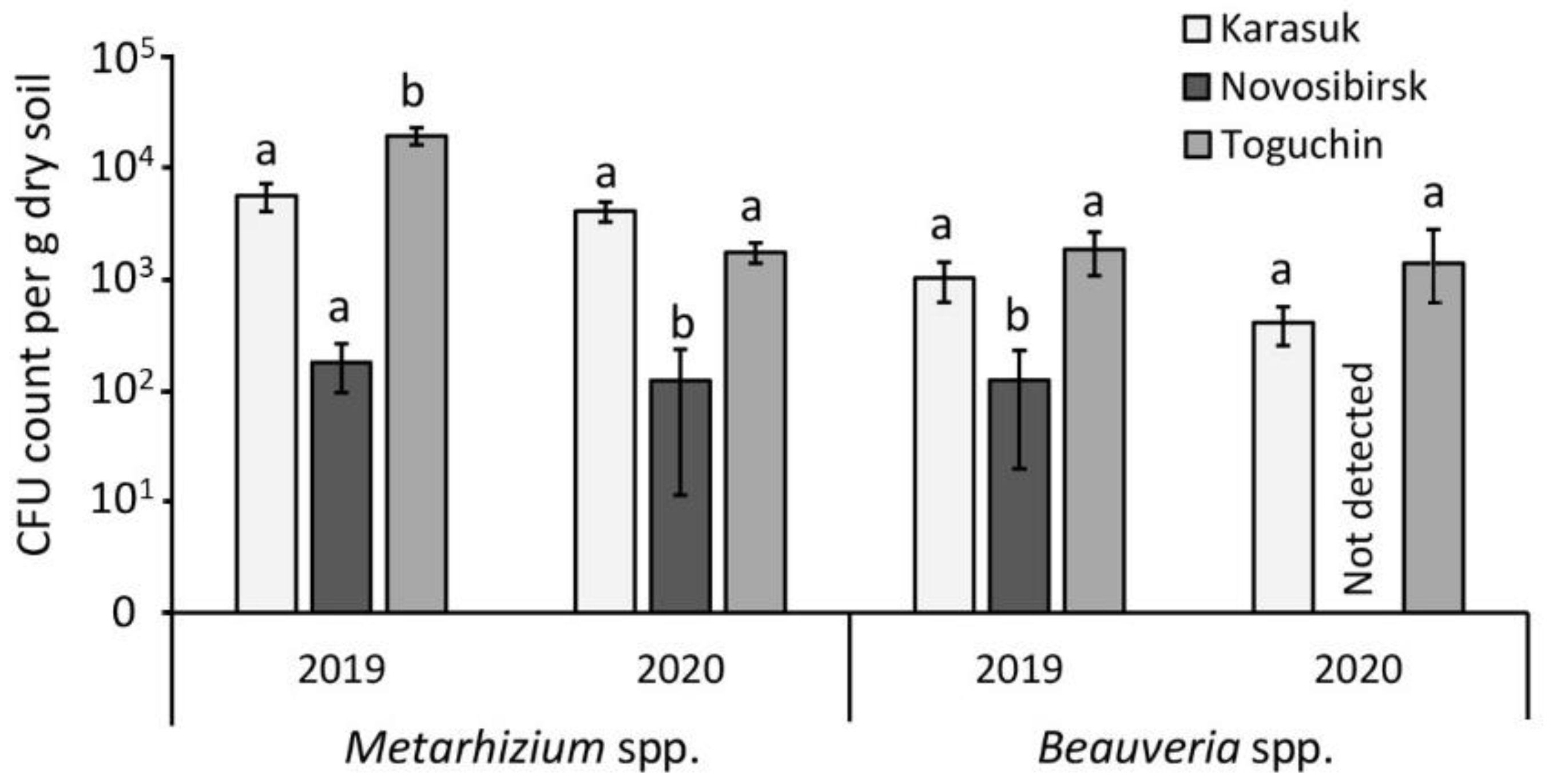
- The neighborhood of Karasuk town (53°729096′ N, 77°650617′ E) in the steppe zone. This is a more arid area than the other locations [22] (Figure S2). The agrosystem consists of kitchen gardens with continuous potato cultivation (more than 10 years). The agricultural practices are domestic, with only 1 mechanized tillage per year before potato planting. The soil is sandy clay that is acidic, with low nitrogen but high phosphorus contents (Tables S1 and S2, Figure S3). There was a high density of the Colorado potato beetle (up to 100 individuals per plant) over 10 years. Irregular applications of insecticides are conducted.
- (2)
- The neighborhood of Novosibirsk city (55°063772′ N, 82°760315′ E) in the forest–steppe zone. This is a conventional agrosystem with crop rotation and intensive farming (more than 40 years). Potatoes are cultivated using Dutch technology [23]. Autumn tillage is also performed. The soil is a sandy clay loam with an alkalescent pH and the lowest nitrogen content compared with the other locations (Tables S1 and S2, Figure S3). Insecticides, herbicides, and fungicides are regularly applied. The density of pest insects is extremely low.
- (3)
- The neighborhood of Toguchin town (55°043662′ N, 84°803181′ E) in the forest–steppe zone. This is the most humid location [22] (Figure S2). The agrosystem is made up of kitchen gardens under continuous potato cultivation (at least 10 years). Potatoes are cultivated in the same way as in the Karasuk location. The soil is silty clay with a neutral pH, with the highest nitrogen and microelement contents compared with the other locations (Tables S1 and S2, Figure S3). The density of Colorado potato beetles is 0–5 larvae per plant.
2.2. Sample Collection
2.3. Metarhizium and Beauveria CFU Count in Soils
2.4. Microbiological Analysis of Plant Colonization
2.5. Phylogenetic Analysis of Metarhizium Fungi Isolated from Plants and Soils
2.6. ITS Metagenomics
2.7. Statistical Analyses
3. Results
3.1. CFU Count in Soils
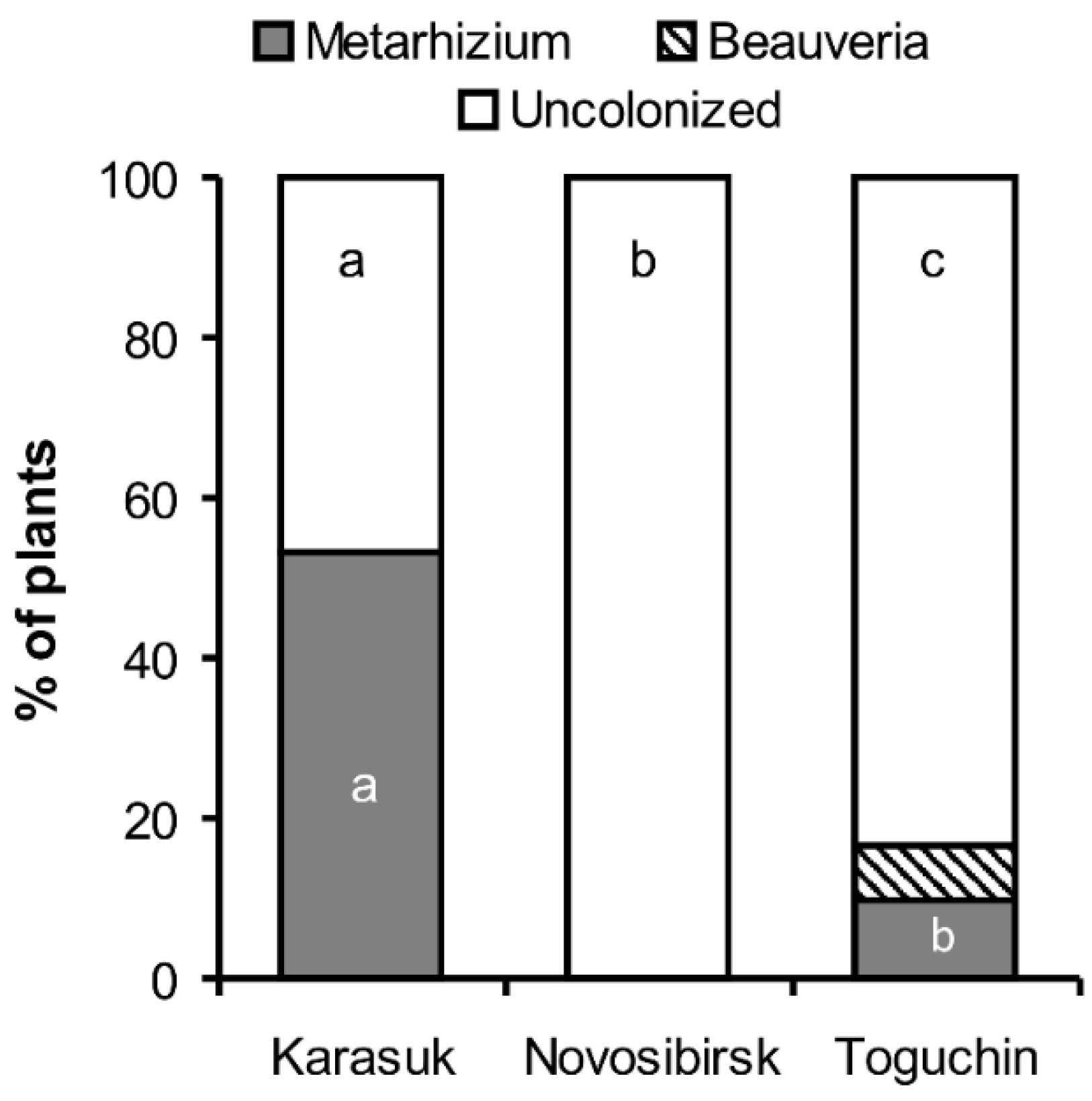
3.2. Frequency of Endophytic Colonization of Potato Plants
3.3. Frequency of Isolation from Non-Sterilized Roots
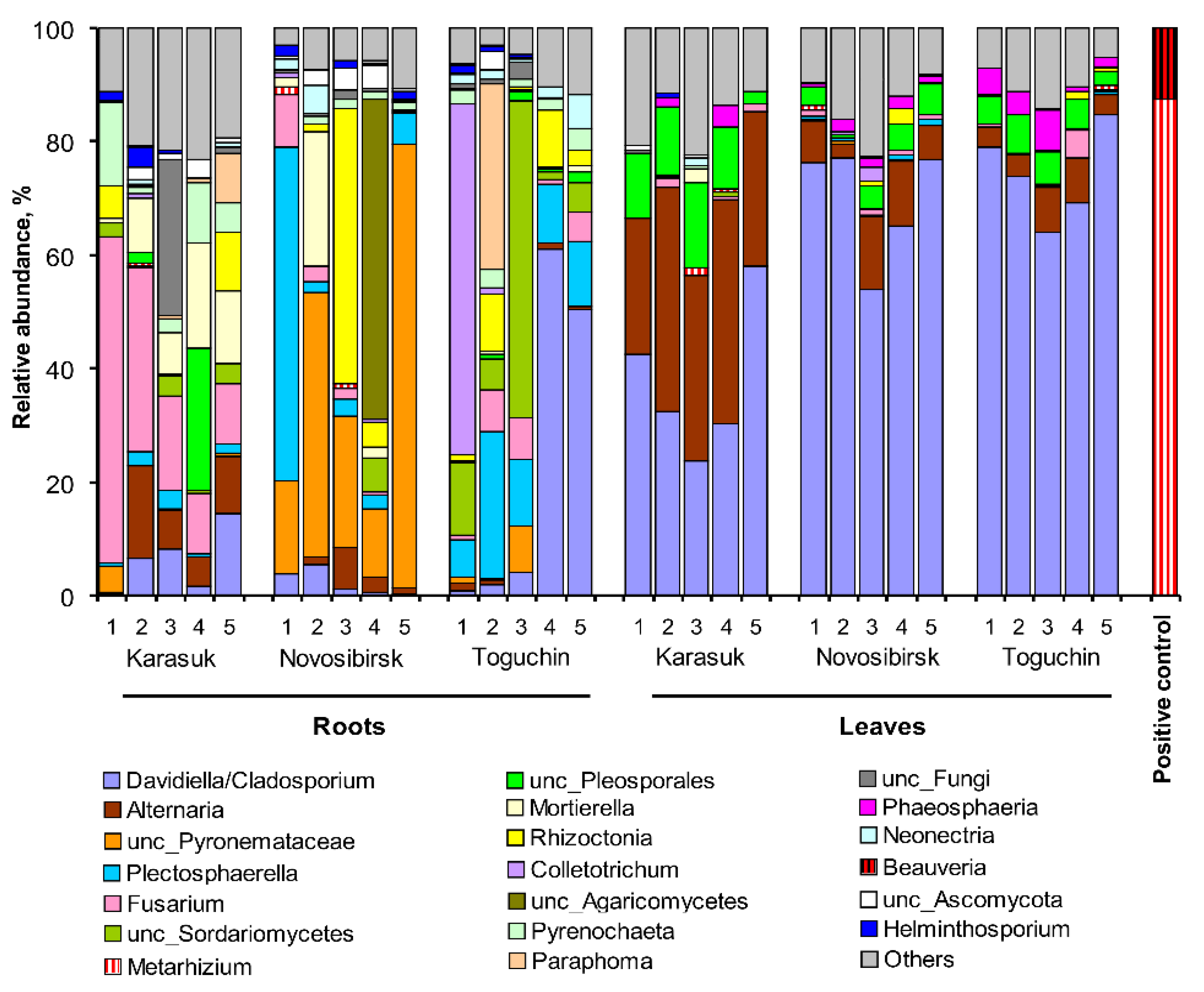
3.4. Analysis of Plant Fungal Communities by ITS Metagenomics
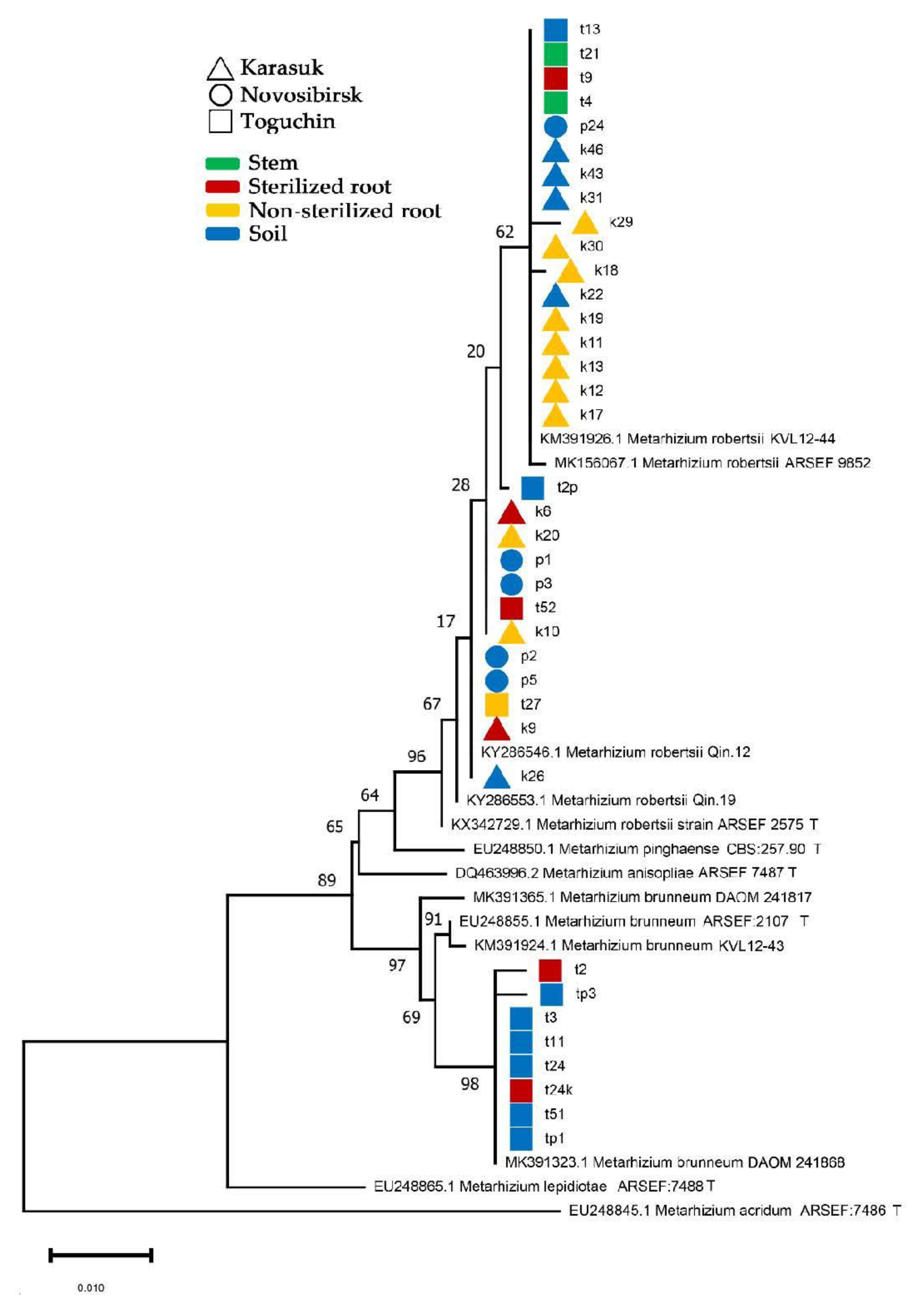
3.5. Identification of Metarhizium Species Isolated from Soils and Plants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lacey, L.; Grzywacz, D.; Shapiro-Ilan, D.; Frutos, R.; Brownbridge, M.; Goettel, M. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef] [Green Version]
- Moonjely, S.; Barelli, L.; Bidochka, M. Insect Pathogenic Fungi as Endophytes. Fungal Genomics 2016, 94, 107–135. [Google Scholar] [CrossRef]
- Vega, F. The use of fungal entomopathogens as endophytes in biological control: A review. Mycologia 2018, 110, 4–30. [Google Scholar] [CrossRef]
- Bamisile, B.S.; Dash, C.K.; Akutse, K.S.; Keppanan, R.; Afolabi, O.G.; Hussain, M.; Qasim, M.; Wang, L. Prospects of endophytic fungal entomopathogens as biocontrol and plant growth promoting agents: An insight on how artificial inoculation methods affect endophytic colonization of host plants. Microbiol. Res. 2018, 217, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Márquez, S.; Bills, G.F.; Zabalgogeazcoa, I. The endophytic mycobiota of the grass Dactylis glomerata. Fungal Divers 2007, 27, 171–195. [Google Scholar]
- Pimentel, I.C.; Gabardo, J.; Poitevin, C.G.; Stuart, A.K.D.C.; Azevedo, J.L. Incidence of endophytic fungi and occurrence of Beauveria and Paecilomyces in maize (Zea mays L.) under field and greenhouse conditions. Asian J. Microbiol. Biotechnol. Environ. Sc. 2016, 18, 47–53. [Google Scholar]
- Reay, S.; Brownbridge, M.; Gicquel, B.; Cummings, N.; Nelson, T. Isolation and characterization of endophytic Beauveria spp. (Ascomycota: Hypocreales) from Pinus radiata in New Zealand forests. Biol. Control. 2010, 54, 52–60. [Google Scholar] [CrossRef]
- FAOSTAT. 2016. Available online: www.fao.org/faostat/en/#home (accessed on 20 June 2021).
- Bajan, C.; Kmitova, K. Contribution of entomopathogenic fungi to natural winter reduction of Colorado beetle adults. Pol. Ecol. Stud. 1977, 3, 107–114. [Google Scholar]
- Wraight, S.P.; Sporleder, M.; Poprawski, T.J.; Lacey, A.L. Chapter VII-1. Application and evaluation of entomopathogens in potato. In Field Manual of Techniques in Invertebrate Pathology. Application and Evaluation of Pathogens for Control of Insects and Other Invertebrate Pests; Lacey, L.A., Kaya, H.K., Eds.; Springer: London, UK, 2007; pp. 329–359. [Google Scholar]
- Ríos-Moreno, A.; Garrido-Jurado, I.; Resquín-Romero, G.; Arroyo-Manzanares, N.; Arce, L.; Quesada-Moraga, E. Destruxin A production byMetarhizium brunneumstrains during transient endophytic colonisation ofSolanum tuberosum. Biocontrol Sci. Technol. 2016, 26, 1574–1585. [Google Scholar] [CrossRef]
- Krell, V.; Unger, S.; Jakobs-Schoenwandt, D.; Patel, A.V. Importance of phosphorus supply through endophytic Metarhizium brunneum for root:shoot allocation and root architecture in potato plants. Plant Soil 2018, 430, 87–97. [Google Scholar] [CrossRef]
- Krell, V.; Unger, S.; Jakobs-Schoenwandt, D.; Patel, A.V. Endophytic Metarhizium brunneum mitigates nutrient deficits in potato and improves plant productivity and vitality. Fungal Ecol. 2018, 34, 43–49. [Google Scholar] [CrossRef]
- Tomilova, O.G.; Shaldyaeva, E.M.; Kryukova, N.A.; Pilipova, Y.V.; Schmidt, N.S.; Danilov, V.P.; Kryukov, V.Y.; Glupov, V.V. Entomopathogenic fungi decrease Rhizoctonia disease in potato in field conditions. PeerJ 2020, 8, e9895. [Google Scholar] [CrossRef]
- Scheepmaker, J.; Butt, T. Natural and released inoculum levels of entomopathogenic fungal biocontrol agents in soil in relation to risk assessment and in accordance with EU regulations. Biocontrol Sci. Technol. 2010, 20, 503–552. [Google Scholar] [CrossRef]
- Jaronski, S.T. Soil ecology of the entomopathogenic ascomycetes: A critical examination of what we (think) we know. In Use of Entomopathogenic Fungi in Biological Pest Management; Ekesi, S., Maniania, N.K., Eds.; Research Signpost: Kerala, India, 2007; pp. 91–144. [Google Scholar]
- Jaros-Su, J.; Groden, E.; Zhang, J. Effects of selected fungicides and timing of fungicide application on Beauveria bassiana in-duced mortality of the Colorado potato beetle (Coleoptera: Chrysomelidae). Biol. Control 1999, 15, 259–269. [Google Scholar] [CrossRef]
- Bruck, D.J. Impact of fungicides on Metarhizium anisopliae in the rhizosphere, bulk soil and in vitro. BioControl 2009, 54, 597–606. [Google Scholar] [CrossRef]
- Clifton, E.H.; Jaronski, S.T.; Hodgson, E.W.; Gassmann, A.J. Abundance of Soil-Borne Entomopathogenic Fungi in Organic and Conventional Fields in the Midwestern USA with an Emphasis on the Effect of Herbicides and Fungicides on Fungal Persistence. PLoS ONE 2015, 10, e0133613. [Google Scholar] [CrossRef]
- Sosa-Gómez, D.R.; Moscardi, F. Effect of Till and No-Till Soybean Cultivation on Dynamics of Entomopathogenic Fungi in the Soil. Fla. Èntomol. 1994, 77, 284. [Google Scholar] [CrossRef]
- Klingen, I.; Eilenberg, J.; Meadow, R. Effects of farming system, field margins and bait insect on the occurrence of insect pathogenic fungi in soils. Agric. Ecosyst. Environ. 2002, 91, 191–198. [Google Scholar] [CrossRef]
- Kravtsov, V.M.; Donukalova, R.P. Geography of Novosibirsk Region; INFOLIO: Novosibirsk, Russia, 1999; 208p. (In Russian) [Google Scholar]
- Dutch Potato Growing Technology. Available online: https://en.blabto.com/1565-dutch-potato-growing-technology.html (accessed on 20 June 2021).
- Posada, F.; Aime, M.C.; Peterson, S.W.; Rehner, S.A.; Vega, F.E. Inoculation of coffee plants with the fungal entomopathogen Beauveria bassiana (Ascomycota: Hypocreales). Mycol. Res. 2007, 111, 748–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKinnon, A.C.; Saari, S.; Moran-Diez, M.E.; Meyling, N.V.; Raad, M.; Glare, T.R. Beauveria bassiana as an endophyte: A critical review on associated methodology and biocontrol potential. BioControl 2017, 62, 1–17. [Google Scholar] [CrossRef]
- Kryukov, V.; Yaroslavtseva, O.; Tyurin, M.; Akhanaev, Y.; Elisaphenko, E.; Wen, T.-C.; Tomilova, O.; Tokarev, Y.; Glupov, V. Ecological preferences of Metarhizium spp. from Russia and neighboring territories and their activity against Colorado potato beetle larvae. J. Invertebr. Pathol. 2017, 149, 1–7. [Google Scholar] [CrossRef]
- Bischoff, J.F.; Rehner, S.A.; Humber, R.A. A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia 2009, 101, 512–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Tanabe, A.; Yamamoto, S.; Sato, H. High-Coverage ITS Primers for the DNA-Based Identification of Ascomycetes and Basidiomycetes in Environmental Samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Shinsky, J.J., White, T.J., Eds.; PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Wemheuer, F.; Berkelmann, D.; Wemheuer, B.; Daniel, R.; Vidal, S.; Daghela, H.B.B. Agroforestry Management Systems Drive the Composition, Diversity, and Function of Fungal and Bacterial Endophyte Communities in Theobroma Cacao Leaves. Microorganisms 2020, 8, 405. [Google Scholar] [CrossRef] [Green Version]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Naumova, N.; Belanov, I.; Alikina, T.; Kabilov, M. Undisturbed Soil Pedon under Birch Forest: Characterization of Microbiome in Genetic Horizons. Soil Syst. 2021, 5, 14. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C. UNCROSS2: Identification of cross-talk in 16S rRNA OTU tables. bioRxiv 2018. [Google Scholar] [CrossRef]
- Edgar, R.C. SINTAX, a Simple Non-Bayesian Taxonomy Classifier for 16S and ITS Sequences. bioRxiv 2016. [Google Scholar] [CrossRef] [Green Version]
- UNITE Community: UNITE USEARCH/UTAX Release for Fungi Version 18.11.2018. UNITE Community. Available online: https://doi.org/10.15156/BIO/786345 (accessed on 20 June 2021).
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: An R package for rarefaction and extrapolation of species diversity (H ill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Fiers, M.; Edel-Hermann, V.; Chatot, C.; Le Hingrat, Y.; Alabouvette, C.; Steinberg, C. Potato soil-borne diseases. A review. Agron. Sustain. Dev. 2012, 32, 93–132. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Zhang, Y.Y.; Zhao, X.J.; Wang, K.; Zhao, J. First Report of Potato Wilt Caused by Plectosphaerella cucumerina in Inner Mongolia, China. Plant Dis. 2016, 100, 2523. [Google Scholar] [CrossRef]
- Cordero-Ramírez, J.D.; López-Rivera, R.L.-R.; Figueroa-López, A.M.; Álvarez, J.C.M.; Cervantes-Gámez, R.G.; Maldonado-Mendoza, I.E. Microorganismos asociados a la rizosfera de jitomate en un agroecosistema del valle de Guasave, Sinaloa, México. Rev. Mex. Biodivers. 2012, 83, 712–730. [Google Scholar] [CrossRef]
- Simmons, D.R.; Kepler, R.M.; Renner, S.A.; Groden, E. Phylogeny of Hirsutella species (Ophiocordycipitaceae) from the USA: Remedying the paucity of Hirsutella sequence data. IMA Fungus 2015, 6, 345–356. [Google Scholar] [CrossRef]
- Gaugler, R.; Costa, S.D.; Lashomb, J. Stability and Efficacy of Beauveria bassiana Soil Inoculations. Environ. Èntomol. 1989, 18, 412–417. [Google Scholar] [CrossRef]
- Medo, J.; Michalko, J.; Medová, J.; Cagáň, Ľ. Phylogenetic structure and habitat associations of Beauveria species isolated from soils in Slovakia. J. Invertebr. Pathol. 2016, 140, 46–50. [Google Scholar] [CrossRef] [Green Version]
- Inglis, G.D.; Duke, G.M.; Goettel, M.S.; Kabaluk, J.T.; Ortega-Polo, R. Biogeography and genotypic diversity of Metarhizium brunneum and Metarhizium robertsii in northwestern North American soils. Can. J. Microbiol. 2019, 65, 261–281. [Google Scholar] [CrossRef] [PubMed]
- Tkaczuk, C.; Krzyczkowski, T.; Wegensteiner, R. The occurrence of entomopathogenic fungi in soils from mid-field woodlots and adjacent small-scale arable fields. Acta Mycol. 2013, 47, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Ramos, Y.; Portal, O.; Lysøe, E.; Meyling, N.; Klingen, I. Diversity and abundance of Beauveria bassiana in soils, stink bugs and plant tissues of common bean from organic and conventional fields. J. Invertebr. Pathol. 2017, 150, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.A.; Klingeman, W.; Ownley, B.H.; Gwinn, K.D. Evidence of Endophytic Beauveria bassiana in Seed-treated Tomato Plants Acting as a Systemic Entomopathogen to Larval Helicoverpa zea (Lepidoptera: Noctuidae). J. Èntomol. Sci. 2009, 44, 391–396. [Google Scholar] [CrossRef]
- García, J.E.; Posadas, J.B.; Perticari, A.; Lecuona, R.E. Metarhizium anisopliae (Metschnikoff) Sorokin promotes growth and has endophytic activity in tomato plants. Adv. Biol. Res. 2011, 5, 22–27. [Google Scholar]
- Sánchez-Rodríguez, A.R.; del Campillo, M.C.; Quesada-Moraga, E. Beauveria bassiana: An entomopathogenic fungus alleviates Fe chlorosis symptoms in plants grown on calcareous substrates. Sci. Hortic. 2015, 197, 193–202. [Google Scholar] [CrossRef]
- Kryukov, V.Y.; Kabilov, M.R.; Smirnova, N.; Tomilova, O.G.; Tyurin, M.V.; Akhanaev, Y.B.; Polenogova, O.V.; Danilov, V.P.; Zhangissina, S.K.; Alikina, T.; et al. Bacterial decomposition of insects post-Metarhizium infection: Possible influence on plant growth. Fungal Biol. 2019, 123, 927–935. [Google Scholar] [CrossRef]
- Spiridon, M. Endophytic Colonization of Solanum Tuberosum L. (Solanales: Solanaceae) Plants Can Affect the Infestation of Serious Pests. Appl. Microbiol. Theory Technol. 2020, 1, 51–57. [Google Scholar] [CrossRef]
- Moonjely, S.; Bidochka, M.J. Generalist and specialist Metarhizium insect pathogens retain ancestral ability to colonize plant roots. Fungal Ecol. 2019, 41, 209–217. [Google Scholar] [CrossRef]
- Barelli, L.; Waller, A.S.; Behie, S.W.; Bidochka, M.J. Plant microbiome analysis after Metarhizium amendment reveals increases in abundance of plant growth-promoting organisms and maintenance of disease-suppressive soil. PLoS ONE 2020, 15, e0231150. [Google Scholar] [CrossRef]
- Stuart, A.K.D.C.; Stuart, R.M.; Pimentel, I.C. Effect of agrochemicals on endophytic fungi community associated with crops of organic and conventional soybean (Glycine max L. Merril). Agric. Nat. Resour. 2018, 52, 388–392. [Google Scholar] [CrossRef]
- Bills, G.; Polishook, J.D. Microfungi from Carpinus caroliniana. Can. J. Bot. 1991, 69, 1477–1482. [Google Scholar] [CrossRef]
- Vega, F.E.; Simpkins, A.; Aime, M.C.; Posada, F.; Peterson, S.W.; Rehner, S.A.; Infante, F.; Castillo, A.; Arnold, A.E. Fungal endophyte diversity in coffee plants from Colombia, Hawai’i, Mexico and Puerto Rico. Fungal Ecol. 2010, 3, 122–138. [Google Scholar] [CrossRef]
- Mardanova, A.; Lutfullin, M.; Hadieva, G.; Akosah, Y.; Pudova, D.; Kabanov, D.; Shagimardanova, E.; Vankov, P.; Vologin, S.; Gogoleva, N.; et al. Structure and variation of root-associated microbiomes of potato grown in alfisol. World J. Microbiol. Biotechnol. 2019, 35, 181. [Google Scholar] [CrossRef]
- European Nucleotide Archive. Run: ERR3301606. Available online: https://www.ebi.ac.uk/ena/data/view/ERR3301606 (accessed on 22 June 2021).
- Peters, J.C.; Lees, A.K.; Cullen, D.W.; Sullivan, L.; Stroud, G.P.; Cunnington, A.C. Characterization of Fusarium spp. responsible for causing dry rot of potato in Great Britain. Plant Pathol. 2008, 57, 262–271. [Google Scholar] [CrossRef]
- Jindo, K.; Evenhuis, A.; Kempenaar, C.; Sudré, C.P.; Zhan, X.; Teklu, M.G.; Kessel, G. Review: Holistic pest management against early blight disease towards sustainable agriculture. Pest Manag. Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Bullock, D.G. Crop rotation. Crit. Rev. Plant Sci. 1992, 11, 309–326. [Google Scholar] [CrossRef]
- Nishi, O.; Iiyama, K.; Yasunaga-Aoki, C.; Shimizu, S. Comparison of the germination rates of Metarhizium spp. conidia from Japan at high and low temperatures. Lett. Appl. Microbiol. 2013, 57, 554–560. [Google Scholar] [CrossRef]
- Russo, M.L.; Pelizza, S.A.; Cabello, M.N.; Stenglein, S.A.; Scorsetti, A.C. Endophytic colonisation of tobacco, corn, wheat and soybeans by the fungal entomopathogen Beauveria bassiana (Ascomycota, Hypocreales). Biocontrol Sci. Technol. 2014, 25, 475–480. [Google Scholar] [CrossRef]
| % of Fungus-Positive Plants (n = 30 per Location) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genus | Root | Stem | Leaf | ||||||
| K-k | N-sk | T-n | K-k | N-sk | T-n | K-k | N-sk | T-n | |
| 2019 | |||||||||
| Metarhizium | 6.7 | 3.3 | 3.3 | 0 | 0 | 3.3 | 0 | 0 | 0 |
| Beauveria | 6.7 | 0 | 0 | 3.3 | 0 | 0 | 0 | 0 | 0 |
| 2020 | |||||||||
| Metarhizium | 3.3 | 0 | 3.3 | 0 | 0 | 10.0 | 0 | 0 | 0 |
| Beauveria | 0 | 3.3 | 0 | 3.3 | 0 | 3.3 | 0 | 3.3 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyurin, M.; Kabilov, M.R.; Smirnova, N.; Tomilova, O.G.; Yaroslavtseva, O.; Alikina, T.; Glupov, V.V.; Kryukov, V.Y. Can Potato Plants Be Colonized with the Fungi Metarhizium and Beauveria under Their Natural Load in Agrosystems? Microorganisms 2021, 9, 1373. https://doi.org/10.3390/microorganisms9071373
Tyurin M, Kabilov MR, Smirnova N, Tomilova OG, Yaroslavtseva O, Alikina T, Glupov VV, Kryukov VY. Can Potato Plants Be Colonized with the Fungi Metarhizium and Beauveria under Their Natural Load in Agrosystems? Microorganisms. 2021; 9(7):1373. https://doi.org/10.3390/microorganisms9071373
Chicago/Turabian StyleTyurin, Maksim, Marsel R. Kabilov, Natalia Smirnova, Oksana G. Tomilova, Olga Yaroslavtseva, Tatyana Alikina, Viktor V. Glupov, and Vadim Yu Kryukov. 2021. "Can Potato Plants Be Colonized with the Fungi Metarhizium and Beauveria under Their Natural Load in Agrosystems?" Microorganisms 9, no. 7: 1373. https://doi.org/10.3390/microorganisms9071373
APA StyleTyurin, M., Kabilov, M. R., Smirnova, N., Tomilova, O. G., Yaroslavtseva, O., Alikina, T., Glupov, V. V., & Kryukov, V. Y. (2021). Can Potato Plants Be Colonized with the Fungi Metarhizium and Beauveria under Their Natural Load in Agrosystems? Microorganisms, 9(7), 1373. https://doi.org/10.3390/microorganisms9071373










