CRISPR loci-PCR as Tool for Tracking Azospirillum sp. Strain B510
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Used in this Study
2.2. Primer Design
2.3. DNA Extraction and PCR Reaction
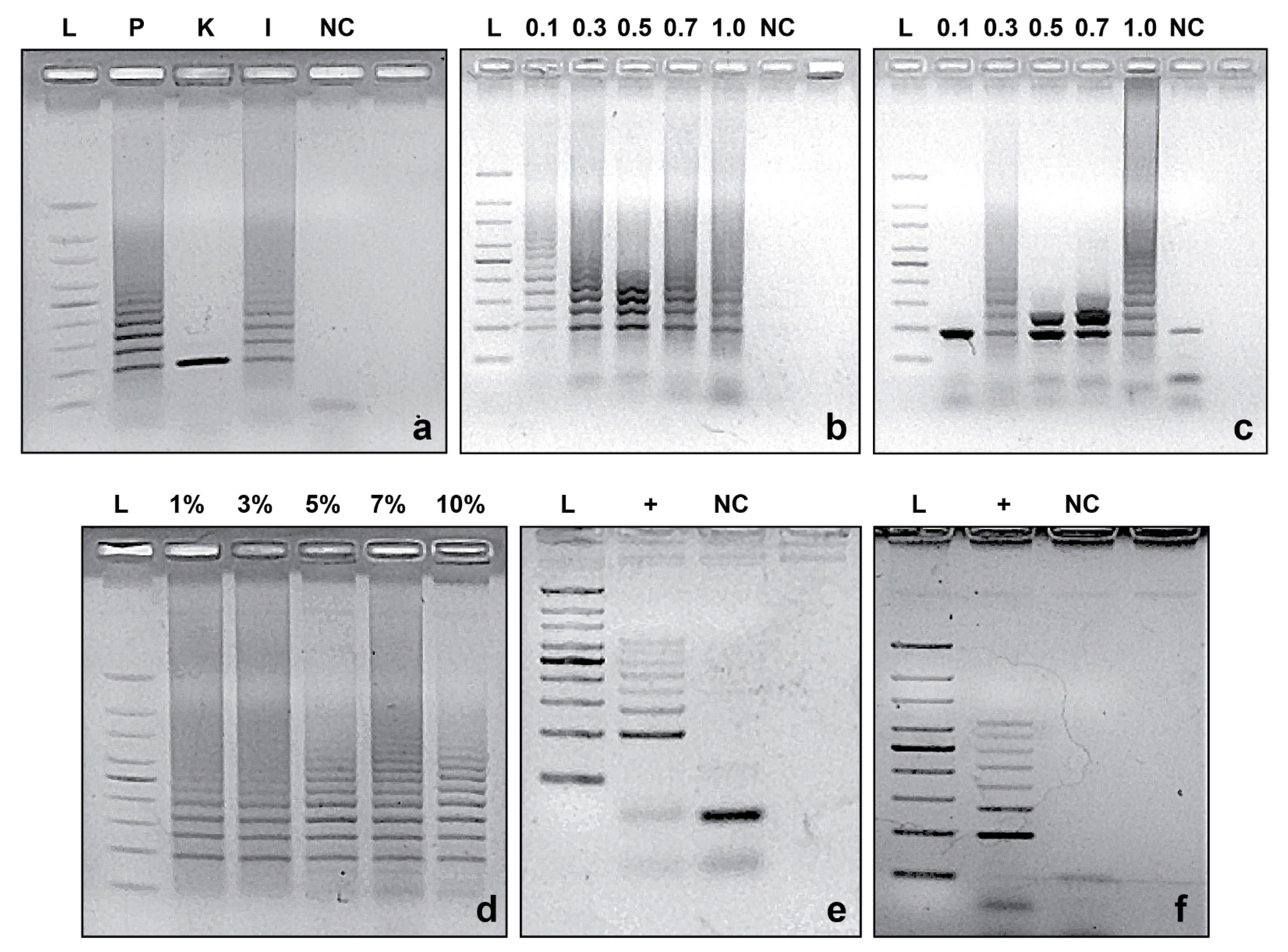
2.4. PCR Reaction Standardization
2.5. Specificity of CRISPRloci-PCR
2.6. Detection Limit of CRISPRloci-PCR
2.7. Validation of CRISPRloci-PCR
3. Results
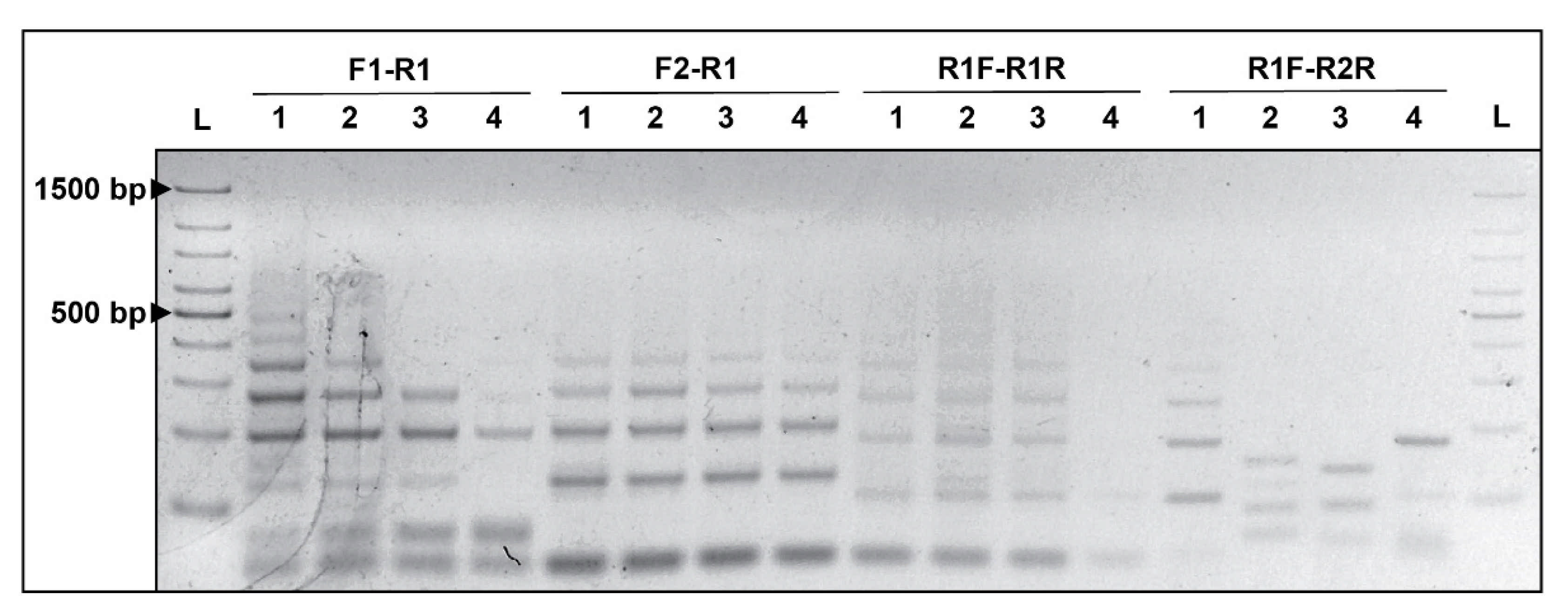
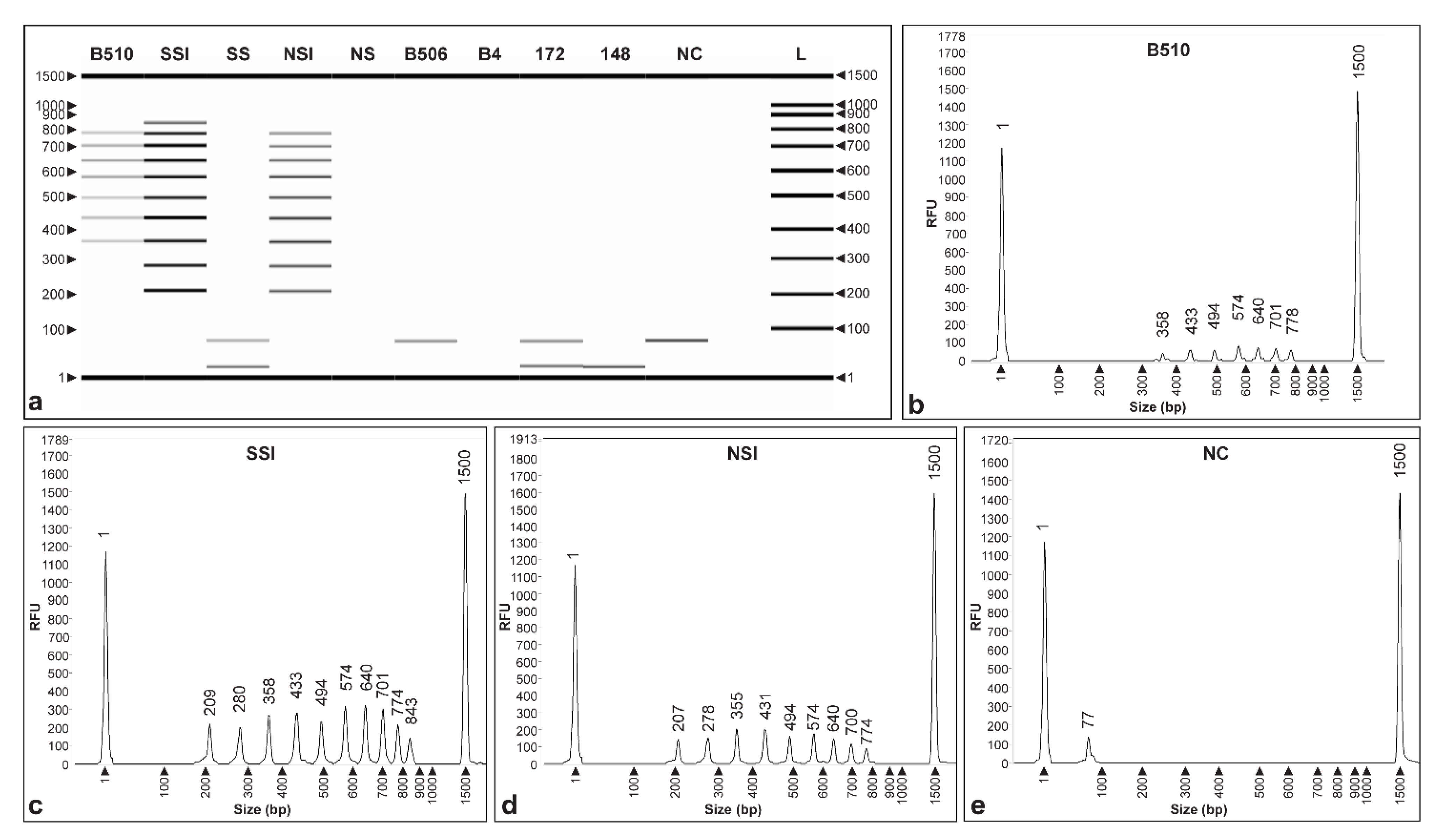
3.1. Primer Design
3.2. CRISPRloci-PCR Standardization
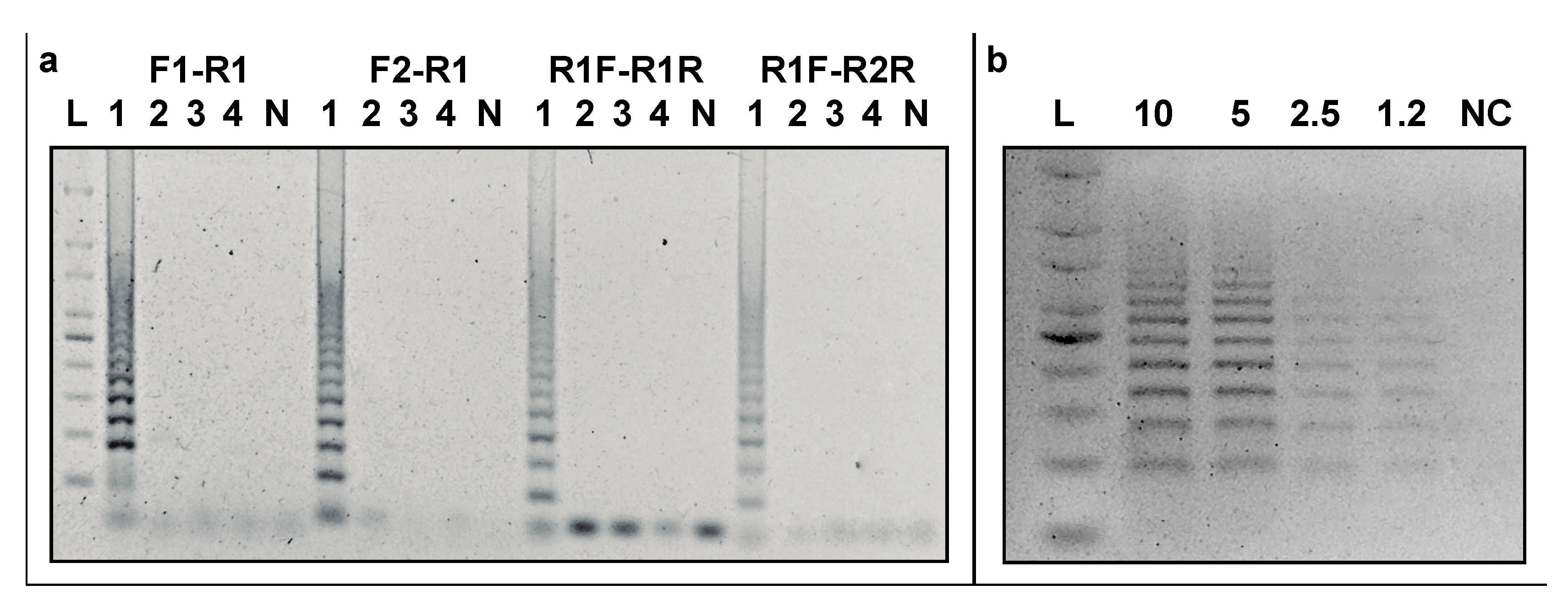
3.3. Specificity of CRISPRloci-PCR
3.4. Detection Limit
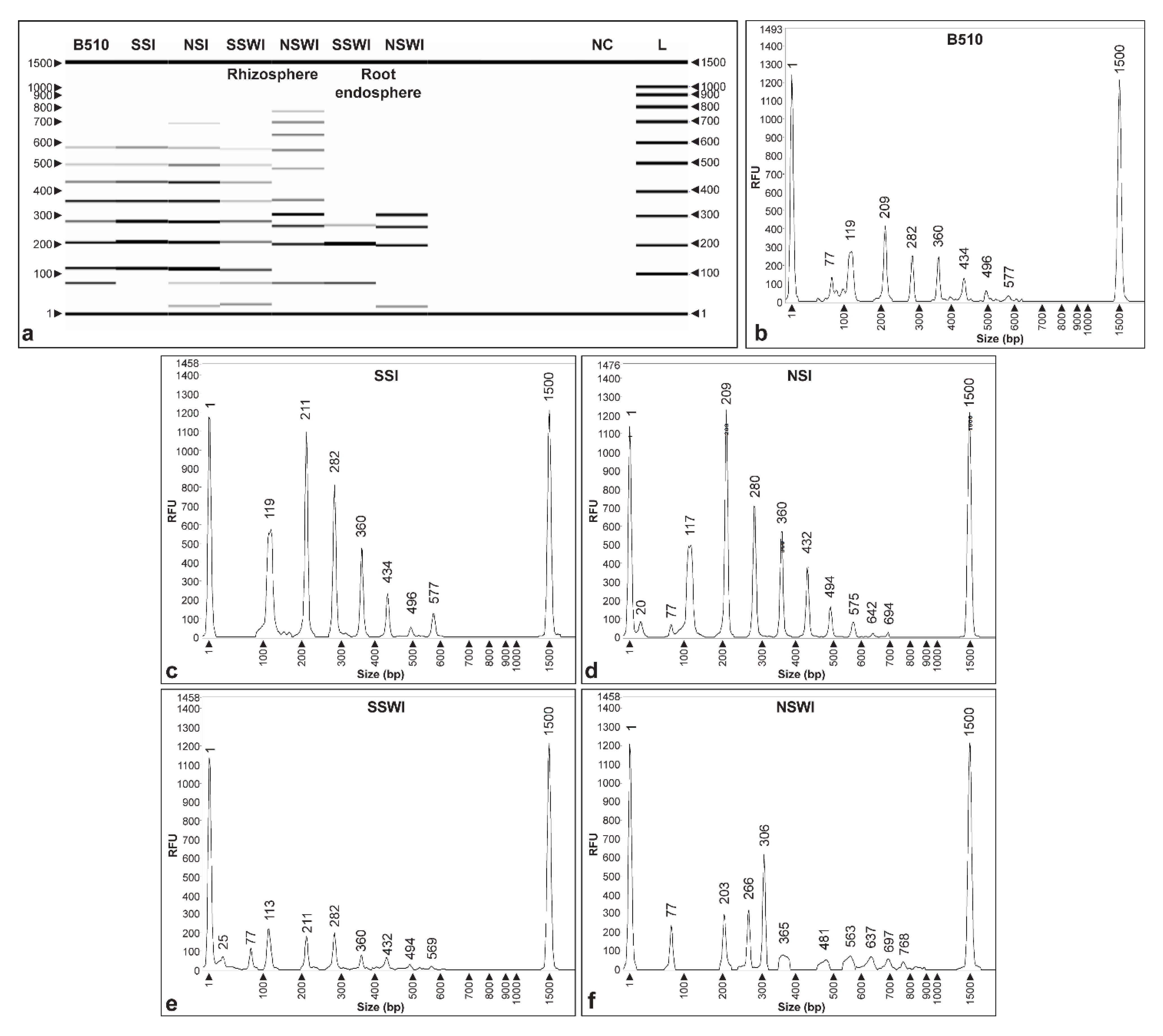
3.5. Method Validation Using Plant Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peter, A.J.; Amalraj, E.L.D.; Talluri, V.R. Commercial Aspects of Biofertilizers and Biostimulants Development Utilizing Rhizosphere Microbes: Global and Indian Scenario. In Rhizosphere Microbes. Microorganisms for Sustainability; Springer: Singapore, 2021; Volume 23, pp. 655–682. ISBN 978-981-15-9154-9. [Google Scholar]
- Rossi, F.A.; Medeot, D.B.; Liaudat, J.P.; Pistorio, M.; Jofré, E. In Azospirillum Brasilense, mutations in flmA or flmB genes affect polar flagellum assembly, surface polysaccharides, and attachment to maize roots. Microbiol. Res. 2016, 190, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Greer-Phillips, S.E.; Stephens, B.B.; Alexandre, G. An Energy Taxis Transducer Promotes Root Colonization by Azospirillum brasilense. J. Bacteriol. 2004, 186, 6595–6604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naher, K.; Miwa, H.; Okazaki, S.; Yasuda, M. Effects of Different Sources of Nitrogen on Endophytic Colonization of Rice Plants by Azospirillum sp. B510. Microbes Environ. 2018, 33, 301–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, J.E.; Kent, A.D.; Brisson, V.L.; Gaudin, A.C.M. Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 2019, 7, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Rilling, J.I.; Acuña, J.J.; Nannipieri, P.; Cassan, F.; Maruyama, F.; Jorquera, M.A. Current opinion and perspectives on the methods for tracking and monitoring plant growth‒promoting bacteria. Soil Biol. Biochem. 2019, 130, 205–219. [Google Scholar] [CrossRef]
- Jansen, R.; van Embden, J.; Gaastra, W.; Schouls, L. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Elbeltagy, A.; Nishioka, K.; Sato, T.; Suzuki, H.; Ye, B.; Hamada, T.; Isawa, T.; Mitsui, H.; Minamisawa, K. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 2001, 67, 5285–5293. [Google Scholar] [CrossRef] [Green Version]
- Elbeltagy, A.; Nishioka, K.; Suzuki, H.; Sato, T.; Sato, Y.-I.; Morisaki, H.; Mitsui, H.; Minamisawa, K. Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci. Plant Nutr. 2000, 46, 617–629. [Google Scholar] [CrossRef]
- Acuña, J.J.; Marileo, L.G.; Araya, M.A.; Rilling, J.I.; Larama, G.A.; Mora, M.L.; Epstein, S.; Jorquera, M.A. In Situ Cultivation Approach to Increase the Culturable Bacterial Diversity in the Rhizobiome of Plants. J. Soil Sci. Plant Nutr. 2020. [Google Scholar] [CrossRef]
- Rilling, J.I.; Acuña, J.J.; Sadowsky, M.J.; Jorquera, M.A. Putative Nitrogen-Fixing Bacteria Associated with the Rhizosphere and Root Endosphere of Wheat Plants Grown in an Andisol from Southern Chile. Front. Microbiol. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Reasoner, D.J.; Geldreich, E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Wilson, K. Preparation of Genomic DNA from Bacteria. In Current Protocols in Molecular Biology; Wiley and Sons: Hoboken, NJ, USA, 1997; Volume 529, pp. 2.4.1–2.4.5. ISBN 9780124186873. [Google Scholar]
- Ahmad, I.; Ahmad, F.; Pichtel, J. Rhizosphere and Root Colonization by Bacterial Inoculants and Their Monitoring Methods: A critical area in PGPR Research. In Microbes and Microbial Technology: Agricultural and Environmental Applications; Springer: New York, NY, USA, 2011; pp. 363–391. ISBN 9781441979308. [Google Scholar]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRfinder: A website to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. PILER-CR: Fast and accurate identification of CRISPR repeats. BMC Bioinform. 2007, 8, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [Green Version]
- Reddypriya, P.; Soumare, A.; Balachandar, D. Multiplex and quantitative PCR targeting SCAR markers for strain-level detection and quantification of biofertilizers. J. Basic Microbiol. 2019, 59, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Holmberg, A.; Melin, P.; Levenfors, J.P.; Sundh, I. Development and evaluation of SCAR markers for a Pseudomonas brassicacearum strain used in biological control of snow mould. Biol. Control 2009, 48, 181–187. [Google Scholar] [CrossRef]
- Couillerot, O.; Poirier, M.A.; Prigent-Combaret, C.; Mavingui, P.; Caballero-Mellado, J.; Moënne-Loccoz, Y. Assessment of SCAR markers to design real-time PCR primers for rhizosphere quantification of Azospirillum brasilense phytostimulatory inoculants of maize. J. Appl. Microbiol. 2010, 109, 528–538. [Google Scholar] [CrossRef]
- Wnuk, E.; Waśko, A.; Walkiewicz, A.; Bartmiński, P.; Bejger, R.; Mielnik, L.; Bieganowski, A. The effects of humic substances on DNA isolation from soils. PeerJ 2020, 8, 1–15. [Google Scholar] [CrossRef]
- Hinsinger, P.; Bengough, A.G.; Vetterlein, D.; Young, I.M. Rhizosphere: Biophysics, biogeochemistry and ecological relevance. Plant Soil 2009, 321, 117–152. [Google Scholar] [CrossRef]
- Simmons, T.; Caddell, D.F.; Deng, S.; Coleman-Derr, D. Exploring the root microbiome: Extracting bacterial community data from the soil, rhizosphere, and root endosphere. J. Vis. Exp. 2018, 2018, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Fernández-González, A.J.; Villadas, P.J.; Gómez-Lama Cabanás, C.; Valverde-Corredor, A.; Belaj, A.; Mercado-Blanco, J.; Fernández-López, M. Defining the root endosphere and rhizosphere microbiomes from the World Olive Germplasm Collection. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- McGhee, G.C.; Sundin, G.W. Erwinia amylovora CRISPR elements provide new tools for evaluating strain diversity and for microbial source tracking. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
| Strain | Plant Niche Source | Country | 16S Accession No. a | Genome Accession No. b | Reference |
|---|---|---|---|---|---|
| Target bacterium | |||||
| Azospirillum sp. B510 | Rice stems endosphere | Japan | AB049111 | AP010946.1 | Elbetagy et al. 2001 |
| Azospirillum control strains | |||||
| Azospirillum sp. B506 | Rice stems endosphere | Japan | AB049110 | BADK01000001:BADK01001143 | Elbetagy et al. 2001 |
| Azospirillum sp. B4 | Rice stems endosphere | Japan | AB027690 | BACU01000001:BACU01000744 | Elbetagy et al. 2000 |
| Azospirillum sp. I53 | Wheat rhizosphere | Chile | MK216930 | n.s. | Acuña et al. 2020 |
| Azospirillum sp. I84 | Wheat rhizosphere | Chile | MK216960 | n.s. | |
| Azospirillum sp. I118 | Wheat rhizosphere | Chile | MK216988 | n.s. | |
| Azospirillum sp. I146 | Wheat rhizosphere | Chile | MK217013 | n.s. | |
| Azospirillum sp. I148 | Wheat rhizosphere | Chile | MK217015 | n.s. | |
| Azospirillum sp. I171 | Wheat rhizosphere | Chile | MK217036 | n.s. | |
| Azospirillum sp. I51 | Wheat rhizosphere | Chile | MK216928 | n.s. | |
| Control strains | |||||
| Microbacterium sp. 62CR | Wheat rhizosphere | Chile | MG835569 | n.s. | Rilling et al. 2018 |
| Bacillus sp. 72CR | Wheat rhizosphere | Chile | MG835570 | n.s. | |
| Bacillus sp. 102CR | Wheat rhizosphere | Chile | MG835571 | n.s. | |
| Bacillus sp. 112BR | Wheat rhizosphere | Chile | MG835572 | n.s. | |
| Bacillus sp. 154AR | Wheat rhizosphere | Chile | MG835573 | n.s. | |
| Bacillus sp. 173CR | Wheat rhizosphere | Chile | MG835574 | n.s. | |
| Microbacterium sp. 184AR | Wheat rhizosphere | Chile | MG835575 | n.s. | |
| Bacillus sp. 184AR-1 | Wheat rhizosphere | Chile | MG835576 | n.s. | |
| Bacillus sp. 214AR | Wheat rhizosphere | Chile | MG835577 | n.s. | |
| Bacillus sp. 214AR-1 | Wheat rhizosphere | Chile | MG835578 | n.s. | |
| Bacillus sp. 222BR | Wheat rhizosphere | Chile | MG835579 | n.s. | |
| Chitinophaga sp. 623EA | Wheat rhizosphere | Chile | MG835588 | n.s. | |
| Arthrobacter sp. 243AR | Wheat rhizosphere | Chile | MG835580 | n.s. | |
| Bacillus sp. 322CR | Wheat rhizosphere | Chile | MG835583 | n.s. | |
| Bacillus sp. 342CR | Wheat rhizosphere | Chile | MG835584 | n.s. | |
| Microbacterium sp. 354AR | Wheat rhizosphere | Chile | MG835585 | n.s. | |
| Bacillus sp. 354AR-1 | Wheat rhizosphere | Chile | MG835586 | n.s. | |
| Bacillus sp. 372EC | Wheat rhizosphere | Chile | MG835587 | n.s. | |
| Bacillus sp. 503CR | Wheat rhizosphere | Chile | MG835581 | n.s. | |
| Bacillus sp. 503CR-1 | Wheat rhizosphere | Chile | MG835582 | n.s. | |
| Mycobacterium sp. 222EC | Wheat root endosphere | Chile | MG835593 | n.s. | |
| Microbacterium sp. 382EC | Wheat root endosphere | Chile | MG835590 | n.s. | |
| Georgenia sp. 424EC | Wheat root endosphere | Chile | MG835591 | n.s. | |
| Psychrobacillus sp. 444EC | Wheat root endosphere | Chile | MG835592 | n.s. | |
| Mycobacterium sp. 491EC | Wheat root endosphere | Chile | MG835594 | n.s. | |
| Roseomonas sp. 513EC | Wheat root endosphere | Chile | MG835595 | n.s. | |
| Roseomonas sp. 523EC | Wheat root endosphere | Chile | MG835596 | n.s. | |
| Roseomonas sp. 543EC | Wheat root endosphere | Chile | MG835598 | n.s. | |
| Bacillus sp. 564EB | Wheat root endosphere | Chile | MG835599 | n.s. | |
| Bacillus sp. 584EA-1 | Wheat root endosphere | Chile | MG835600 | n.s. | |
| Bacillus sp. 592BR | Wheat root endosphere | Chile | MG835601 | n.s. | |
| Chitinophaga sp. 643EA | Wheat root endosphere | Chile | MG835602 | n.s. | |
| Bosea sp. 693EB | Wheat root endosphere | Chile | MG835603 | n.s. | |
| Bosea sp. 703EB | Wheat root endosphere | Chile | MG835604 | n.s. | |
| Leifsonia sp. 274EB | Wheat root endosphere | Chile | MG835589 | n.s. | |
| Mycobacterium sp. 222EC | Wheat root endosphere | Chile | MG835593 | n.s. | |
| Bacillus sp. 714EA | Wheat root endosphere | Chile | MG835605 | n.s. | |
| Georgenia sp. 734EA | Wheat root endosphere | Chile | MG835606 | n.s. |
| Primer Name | Sequence (5′-3′) | Melting Temperature (°C) | Fixed or Repeat |
|---|---|---|---|
| Forward | |||
| F1 | CGAACGCTTTCTCAAACCAC | 60.81 | Fixed |
| F2 | CCTTTTCTCTCACACACGCTAC | 59.05 | Fixed |
| R1F | ATGAGGCCCAAGCATTTCTG | 62.43 | Repeat |
| Reverse | |||
| R1 | AGAAATGCTTGGGCCTCATT | 60.96 | Repeat |
| R1R | CCACTTCGACAGCTTTCTCC | 59.99 | Fixed |
| R2R | AGGCCCAAGCATTTCTGC | 61.30 | Fixed |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rilling, J.I.; Maruyama, F.; Sadowsky, M.J.; Acuña, J.J.; Jorquera, M.A. CRISPR loci-PCR as Tool for Tracking Azospirillum sp. Strain B510. Microorganisms 2021, 9, 1351. https://doi.org/10.3390/microorganisms9071351
Rilling JI, Maruyama F, Sadowsky MJ, Acuña JJ, Jorquera MA. CRISPR loci-PCR as Tool for Tracking Azospirillum sp. Strain B510. Microorganisms. 2021; 9(7):1351. https://doi.org/10.3390/microorganisms9071351
Chicago/Turabian StyleRilling, Joaquin I., Fumito Maruyama, Michael J. Sadowsky, Jacquelinne J. Acuña, and Milko A. Jorquera. 2021. "CRISPR loci-PCR as Tool for Tracking Azospirillum sp. Strain B510" Microorganisms 9, no. 7: 1351. https://doi.org/10.3390/microorganisms9071351
APA StyleRilling, J. I., Maruyama, F., Sadowsky, M. J., Acuña, J. J., & Jorquera, M. A. (2021). CRISPR loci-PCR as Tool for Tracking Azospirillum sp. Strain B510. Microorganisms, 9(7), 1351. https://doi.org/10.3390/microorganisms9071351








