Antimicrobial Resistance Mechanisms and Virulence of Colistin- and Carbapenem-Resistant Acinetobacter baumannii Isolated from a Teaching Hospital in Taiwan
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Clinical Isolates, Species Identification, and Antimicrobial Susceptibility Testing
2.2. Whole-Genome Sequencing and Molecular Characterization
2.3. Sequence Analysis of Genes Related to Colistin Resistance and Virulence
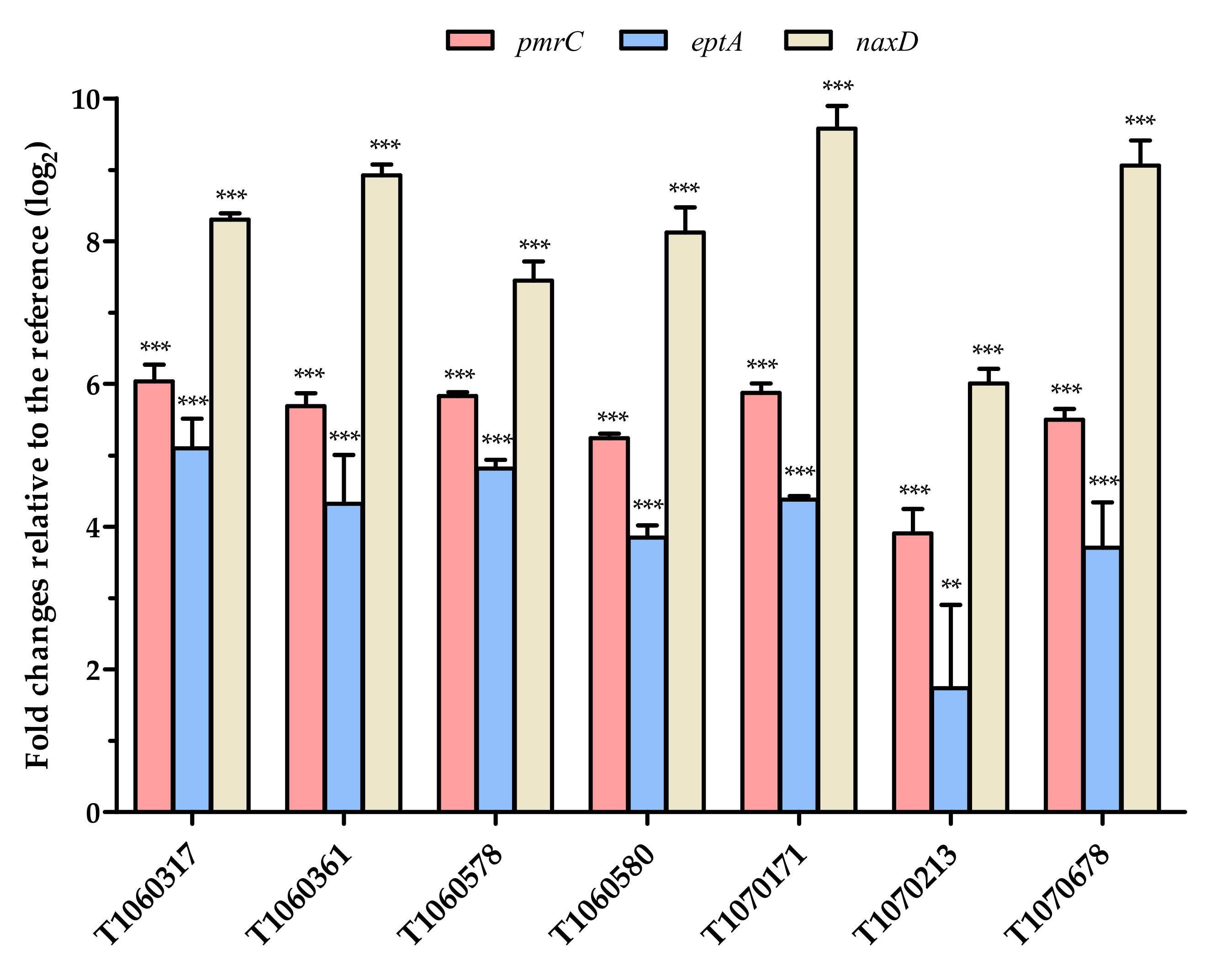
2.4. Detection of Gene Expression Levels
2.5. Lipopolysaccharide Analysis by SDS-PAGE
2.6. Measurement of Generation Time
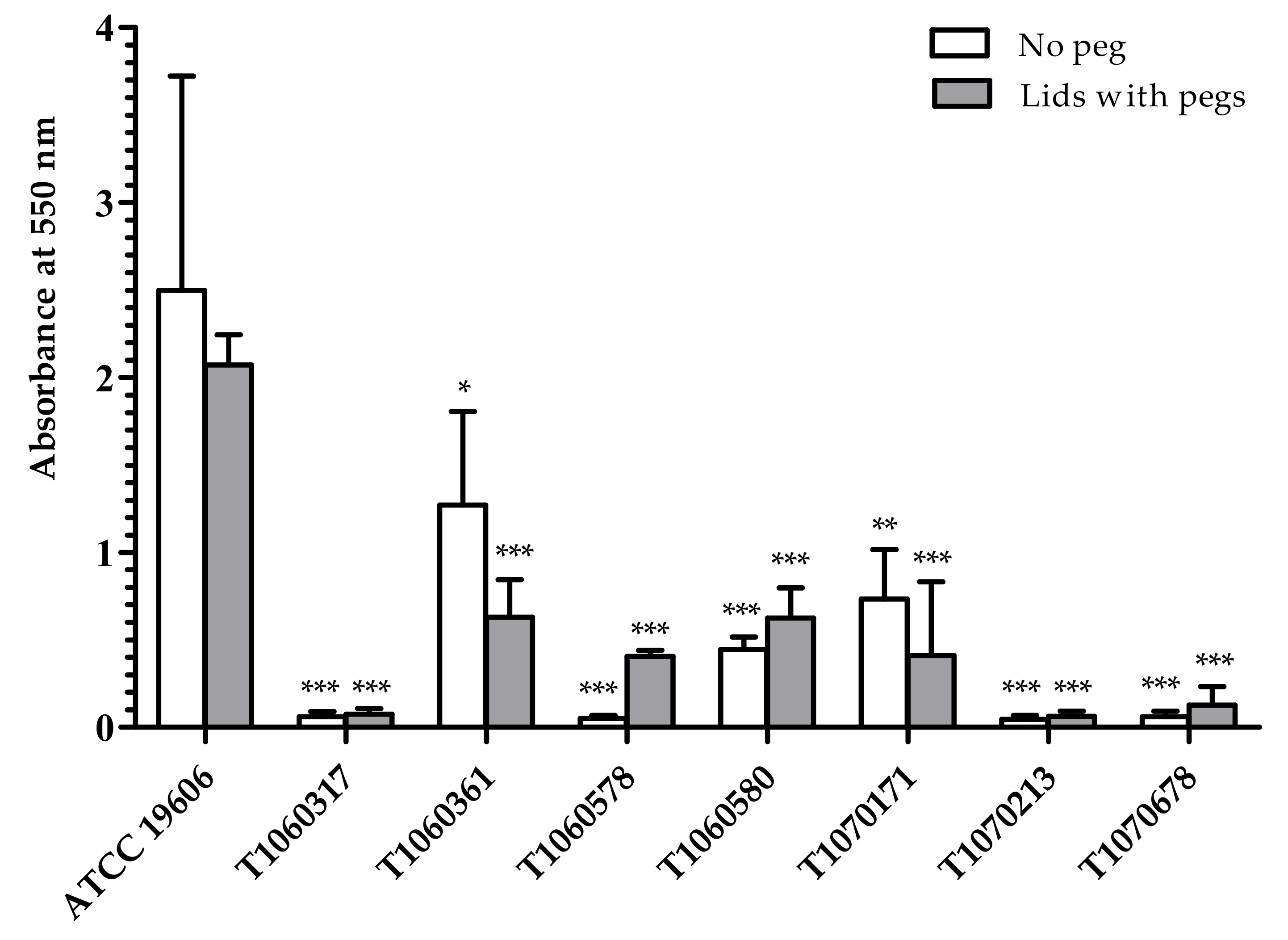
2.7. Biofilm Formation Ability
2.8. Determination of Colistin Heteroresistance
2.9. Virulence Analysis in Galleria Mellonella Infection Model
3. Results and Discussion
3.1. Epidemiology of Colistin- and Carbapenem-Resistant A. baumannii (CCR-AB)
3.2. Molecular Characterizations of CCR-AB Isolates
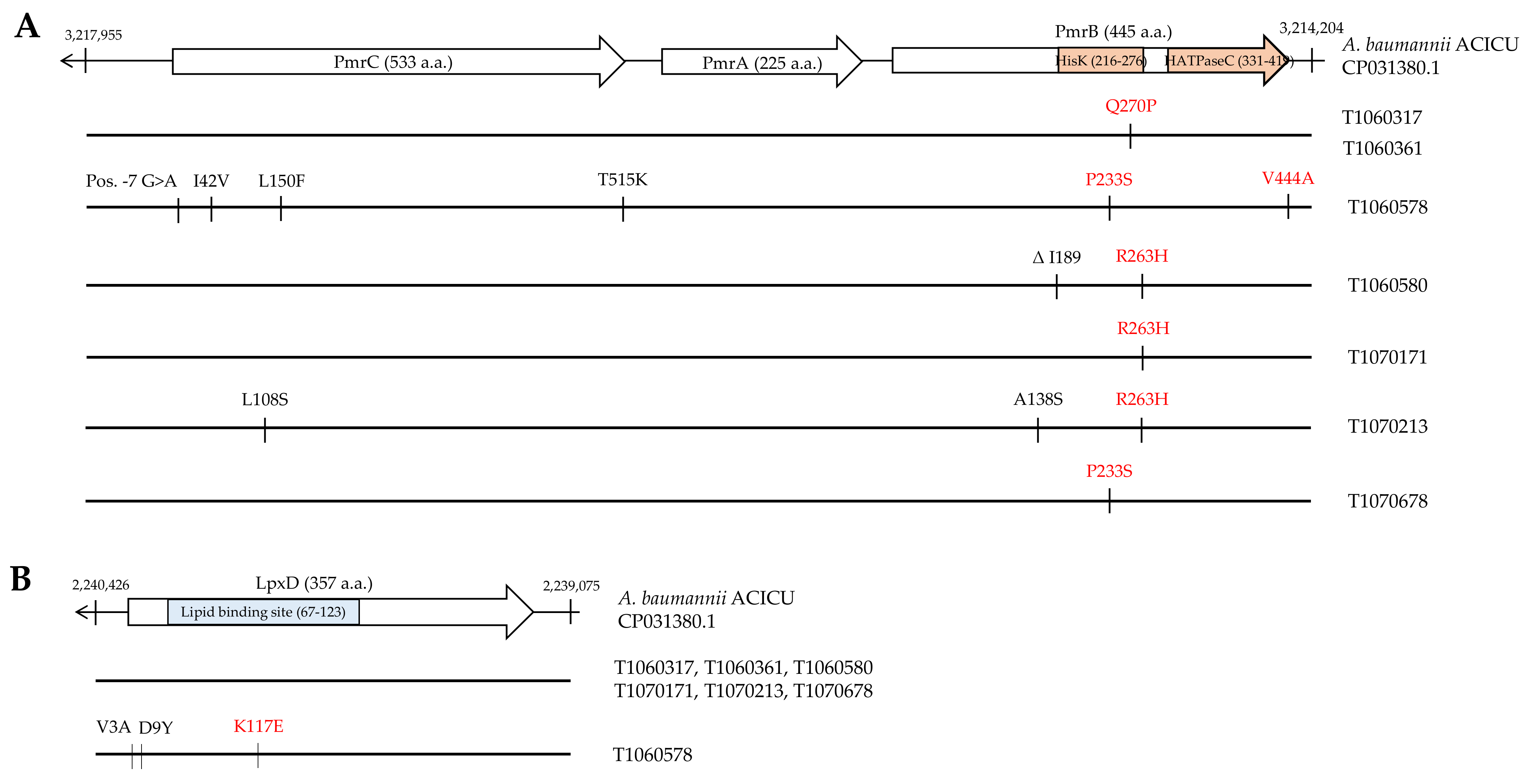
3.3. Mutation Analysis of Genes Conferring Colistin Resistance
3.4. Effects of PmrB and LpxD Mutations
3.5. Fitness, Biofilm Formation, and Heteroresistance to Colistin
3.6. Virulence to Wax Moth
3.7. In Silico Analysis of Virulence Factors
3.8. Potentially Alternative Therapies against MDR Bacteria
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. In Review on Antimicrobial Resistance; Wellcome Trust: London, UK, 2014; pp. 1–16. Available online: http://www.jpiamr.eu/wp-content/uploads/2014/12/AMR-Review-Paper-Tackling-a-crisis-for-the-health-and-wealth-of-nations_1-2.pdf (accessed on 31 March 2021).
- World Health Organization. Global priority list of antibiotic-resistant bacteria-To guide research, discovery, and development of new antibiotics. 2017. Available online: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed on 31 March 2021).
- Kanj, S.S.; Kanafani, Z.A. Current concepts in antimicrobial therapy against resistant Gram-negative organisms: extended-spectrum β-lactamase-producing Enterobacteriaceae, carbapenem-resistant Enterobacteriaceae, and multidrug-resistant Pseudomonas aeruginosa. Mayo Clin. Proc. 2011, 86, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Sievert, D.M.; Ricks, P.; Edwards, J.R.; Schneider, A.; Patel, J.; Srinivasan, A.; Kallen, A.; Limbago, B.; Fridkin, S. National Healthcare Safety Network, T., et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control. Hosp. Epidemiol. 2013, 34, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Kuo, S.C.; Chang, S.C.; Wang, H.Y.; Lai, J.F.; Chen, P.C.; Shiau, Y.R.; Huang, I.W.; Lauderdale, T.L. TSAR Hospitals. Emergence of extensively drug-resistant Acinetobacter baumannii complex over 10 years: nationwide data from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program. BMC Infect. Dis. 2012, 12, 200. [Google Scholar] [CrossRef]
- Taiwan Centers for Disease Control. Annual report of nosocomial infections surveillance system (2016). Taiwan Nosocomial Infections. Surveillance system. Available online: https://www.cdc.gov.tw/En/Category/Page/J63NmsvevBg2u3I2qYBenw (accessed on 31 March 2021).
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention. 2019. Available online: https://www.cdc.gov/DrugResistance/Biggest-Threats.html (accessed on 31 March 2021).
- Bakour, S.; Olaitan, A.O.; Ammari, H.; Touati, A.; Saoudi, S.; Saoudi, K.; Rolain, J.M. Emergence of colistin- and carbapenem-resistant Acinetobacter baumannii ST2 clinical isolate in Algeria: first case report. Microb. Drug Resist. 2015, 21, 279–285. [Google Scholar] [CrossRef]
- Oikonomou, O.; Sarrou, S.; Papagiannitsis, C.C.; Georgiadou, S.; Mantzarlis, K.; Zakynthinos, E.; Dalekos, G.N.; Petinaki, E. Rapid dissemination of colistin and carbapenem resistant Acinetobacter baumannii in Central Greece: mechanisms of resistance, molecular identification and epidemiological data. BMC Infect. Dis. 2015, 15, 559. [Google Scholar] [CrossRef]
- Qureshi, Z.A.; Hittle, L.E.; O’Hara, J.A.; Rivera, J.I.; Syed, A.; Shields, R.K.; Pasculle, A.W.; Ernst, R.K.; Doi, Y. Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin. Infect. Dis. 2015, 60, 1295–1303. [Google Scholar] [CrossRef]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Hameed, F.; Khan, M.A.; Muhammad, H.; Sarwar, T.; Bilal, H.; Rehman, T.U. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: first report from Pakistan. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190237. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Shen, C.; Zheng, X.; Liu, Y.; Chen, H.; Zhong, L.; Liang, Y.; Liao, K.; Xia, Y.; Tian, G.B.; et al. Identification of a novel plasmid carrying mcr-4.3 in an Acinetobacter baumannii strain in China. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Martins-Sorenson, N.; Snesrud, E.; Xavier, D.E.; Cacci, L.C.; Iavarone, A.T.; McGann, P.; Riley, L.W.; Moreira, B.M. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J. Antimicrob. Chemother. 2020, 75, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, L.A.; Herrera, C.M.; Fernandez, L.; Hankins, J.V.; Trent, M.S.; Hancock, R.E. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 2011, 55, 3743–3751. [Google Scholar] [CrossRef]
- Beceiro, A.; Llobet, E.; Aranda, J.; Bengoechea, J.A.; Doumith, M.; Hornsey, M.; Dhanji, H.; Chart, H.; Bou, G.; Livermore, D.M.; et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 2011, 55, 3370–3379. [Google Scholar] [CrossRef] [PubMed]
- Lesho, E.; Yoon, E.J.; McGann, P.; Snesrud, E.; Kwak, Y.; Milillo, M.; Onmus-Leone, F.; Preston, L.; St Clair, K.; Nikolich, M.; et al. Emergence of colistin-resistance in extremely drug-resistant Acinetobacter baumannii containing a novel pmrCAB operon during colistin therapy of wound infections. J. Infect. Dis. 2013, 208, 1142–1151. [Google Scholar] [CrossRef]
- De Silva, P.M.; Kumar, A. Signal transduction proteins in Acinetobacter baumannii: role in antibiotic resistance, virulence, and potential as drug targets. Front. Microbiol. 2019, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.Y.; Gregg, K.A.; Napier, B.A.; Ernst, R.K.; Weiss, D.S. A PmrB-regulated deacetylase required for lipid A modification and polymyxin resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2015, 59, 7911–7914. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St Michael, F.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Adler, B.; Nation, R.L.; Li, J.; Boyce, J.D. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3022–3024. [Google Scholar] [CrossRef]
- Hood, M.I.; Becker, K.W.; Roux, C.M.; Dunman, P.M.; Skaar, E.P. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect. Immun. 2013, 81, 542–551. [Google Scholar] [CrossRef]
- Bojkovic, J.; Richie, D.L.; Six, D.A.; Rath, C.M.; Sawyer, W.S.; Hu, Q.; Dean, C.R. Characterization of an Acinetobacter baumannii lptD deletion strain: permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J. Bacteriol. 2015, 198, 731–741. [Google Scholar] [CrossRef]
- Thi Khanh Nhu, N.; Riordan, D.W.; Do Hoang Nhu, T.; Thanh, D.P.; Thwaites, G.; Huong Lan, N.P.; Wren, B.W.; Baker, S.; Stabler, R.A. The induction and identification of novel colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 2016, 6, 28291. [Google Scholar] [CrossRef]
- Lin, M.F.; Lin, Y.Y.; Lan, C.Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J. Microbiol. 2017, 55, 130–136. [Google Scholar] [CrossRef]
- Lopez-Rojas, R.; Dominguez-Herrera, J.; McConnell, M.J.; Docobo-Perez, F.; Smani, Y.; Fernandez-Reyes, M.; Rivas, L.; Pachon, J. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J. Infect. Dis. 2011, 203, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Rolain, J.M.; Roch, A.; Castanier, M.; Papazian, L.; Raoult, D. Acinetobacter baumannii resistant to colistin with impaired virulence: a case report from France. J. Infect. Dis. 2011, 204, 1146–1147. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Moreno, A.; Fernandez, N.; Vallejo, J.A.; Aranda, J.; Adler, B.; Harper, M.; Boyce, J.D.; Bou, G. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014, 58, 518–526. [Google Scholar] [CrossRef]
- Carretero-Ledesma, M.; Garcia-Quintanilla, M.; Martin-Pena, R.; Pulido, M.R.; Pachon, J.; McConnell, M.J. Phenotypic changes associated with colistin resistance due to lipopolysaccharide loss in Acinetobacter baumannii. Virulence 2018, 9, 930–942. [Google Scholar] [CrossRef]
- Lopez-Rojas, R.; Jimenez-Mejias, M.E.; Lepe, J.A.; Pachon, J. Acinetobacter baumannii resistant to colistin alters its antibiotic resistance profile: a case report from Spain. J. Infect. Dis. 2011, 204, 1147–1148. [Google Scholar] [CrossRef]
- Durante-Mangoni, E.; Del Franco, M.; Andini, R.; Bernardo, M.; Giannouli, M.; Zarrilli, R. Emergence of colistin resistance without loss of fitness and virulence after prolonged colistin administration in a patient with extensively drug-resistant Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 2015, 82, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Pormohammad, A.; Mehdinejadiani, K.; Gholizadeh, P.; Nasiri, M.J.; Mohtavinejad, N.; Dadashi, M.; Karimaei, S.; Safari, H.; Azimi, T. Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: A systematic review and meta-analysis. Microb. Pathog. 2020, 139, 103887. [Google Scholar] [CrossRef]
- Jean, S.S.; Hsueh, P.R.; Lee, W.S.; Chang, H.T.; Chou, M.Y.; Chen, I.S.; Wang, J.H.; Lin, C.F.; Shyr, J.M.; Ko, W.C.; et al. Nationwide surveillance of antimicrobial resistance among non-fermentative Gram-negative bacteria in Intensive Care Units in Taiwan: SMART programme data 2005. Int. J. Antimicrob. Agents 2009, 33, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Chen, Y.S.; Lee, N.Y.; Tang, H.J.; Lee, S.S.; Lin, C.F.; Lu, P.L.; Wu, J.J.; Ko, W.C.; Lee, W.S.; et al. Susceptibility rates of clinically important bacteria collected from intensive care units against colistin, carbapenems, and other comparative agents: results from Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART). Infect. Drug Resist. 2019, 12, 627–640. [Google Scholar] [CrossRef]
- Chang, K.C.; Lin, M.F.; Lin, N.T.; Wu, W.J.; Kuo, H.Y.; Lin, T.Y.; Yang, T.L.; Chen, Y.C.; Liou, M.L. Clonal spread of multidrug-resistant Acinetobacter baumannii in eastern Taiwan. J. Microbiol. Immunol. Infect. 2012, 45, 37–42. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28 ed.; Clinical and Laboratory Standards Institute: Wayne, Pennsylvania, USA, 2018. [Google Scholar]
- Higgins, P.G.; Lehmann, M.; Wisplinghoff, H.; Seifert, H. gyrB multiplex PCR to differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J. Clin. Microbiol. 2010, 48, 4592–4594. [Google Scholar] [CrossRef] [PubMed]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 2010, 5, e10034. [Google Scholar] [CrossRef]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.; Wisplinghoff, H.; Rodriguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef]
- Wyres, K.L.; Cahill, S.M.; Holt, K.E.; Hall, R.M.; Kenyon, J.J. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb. Genom. 2020, 6. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Ponten, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Hall, B.G.; Acar, H.; Nandipati, A.; Barlow, M. Growth rates made easy. Mol. Biol. Evol. 2014, 31, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Dafopoulou, K.; Xavier, B.B.; Hotterbeekx, A.; Janssens, L.; Lammens, C.; De, E.; Goossens, H.; Tsakris, A.; Malhotra-Kumar, S.; Pournaras, S. Colistin-resistant Acinetobacter baumannii clinical strains with deficient biofilm formation. Antimicrob. Agents Chemother. 2015, 60, 1892–1895. [Google Scholar] [CrossRef]
- Sherman, E.X.; Wozniak, J.E.; Weiss, D.S. Methods to evaluate colistin heteroresistance in Acinetobacter baumannii. Methods Mol. Biol. 2019, 1946, 39–50. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Jara, S.; Monga, D.; Eliopoulos, G.M.; Moellering, R.C., Jr.; Mylonakis, E. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob. Agents Chemother. 2009, 53, 2605–2609. [Google Scholar] [CrossRef]
- Palmieri, M.; D’Andrea, M.M.; Pelegrin, A.C.; Perrot, N.; Mirande, C.; Blanc, B.; Legakis, N.; Goossens, H.; Rossolini, G.M.; van Belkum, A. Abundance of colistin-resistant, OXA-23- and ArmA-producing Acinetobacter baumannii belonging to international clone 2 in Greece. Front. Microbiol. 2020, 11, 668. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Cafiso, V.; Stracquadanio, S.; Lo Verde, F.; Gabriele, G.; Mezzatesta, M.L.; Caio, C.; Pigola, G.; Ferro, A.; Stefani, S. Colistin resistant A. baumannii: genomic and transcriptomic traits acquired under colistin therapy. Front. Microbiol. 2018, 9, 3195. [Google Scholar] [CrossRef]
- Sun, B.; Liu, H.; Jiang, Y.; Shao, L.; Yang, S.; Chen, D. New mutations involved in colistin resistance in Acinetobacter baumannii. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 2012, 67, 2069–2089. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Da Silva, G.J.; Domingues, S. Interplay between colistin resistance, virulence and fitness in Acinetobacter baumannii. Antibiotics 2017, 6, 28. [Google Scholar] [CrossRef]
- Farshadzadeh, Z.; Taheri, B.; Rahimi, S.; Shoja, S.; Pourhajibagher, M.; Haghighi, M.A.; Bahador, A. Growth rate and biofilm formation ability of clinical and laboratory-evolved colistin-resistant strains of Acinetobacter baumannii. Front. Microbiol. 2018, 9, 153. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Saridakis, I. Colistin heteroresistance in Acinetobacter spp.: systematic review and meta-analysis of the prevalence and discussion of the mechanisms and potential therapeutic implications. Int. J. Antimicrob. Agents 2020, 56, 106065. [Google Scholar] [CrossRef] [PubMed]
- Charretier, Y.; Diene, S.M.; Baud, D.; Chatellier, S.; Santiago-Allexant, E.; van Belkum, A.; Guigon, G.; Schrenzel, J. Colistin heteroresistance and involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Machado, D.; Antunes, J.; Simoes, A.; Perdigao, J.; Couto, I.; McCusker, M.; Martins, M.; Portugal, I.; Pacheco, T.; Batista, J.; et al. Contribution of efflux to colistin heteroresistance in a multidrug resistant Acinetobacter baumannii clinical isolate. J. Med. Microbiol. 2018, 67, 740–749. [Google Scholar] [CrossRef]
- Hraiech, S.; Roch, A.; Lepidi, H.; Atieh, T.; Audoly, G.; Rolain, J.M.; Raoult, D.; Brunel, J.M.; Papazian, L.; Bregeon, F. Impaired virulence and fitness of a colistin-resistant clinical isolate of Acinetobacter baumannii in a rat model of pneumonia. Antimicrob. Agents Chemother. 2013, 57, 5120–5121. [Google Scholar] [CrossRef]
- Jacobs, A.C.; Thompson, M.G.; Black, C.C.; Kessler, J.L.; Clark, L.P.; McQueary, C.N.; Gancz, H.Y.; Corey, B.W.; Moon, J.K.; Si, Y.; et al. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 2014, 5, e01076-14. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, T.; Luo, Y.; Peng, J.-Y.; Li, Y.-J.; Tao, X.-Y.; Hu, Y.-M.; Wang, H.-C.; Zou, M.-X. Characterization of carbapenem-resistant hypervirulent Acinetobacter baumannii strains isolated from hospitalized patients in the mid-south region of China. BMC Microbiol. 2020, 20. [Google Scholar] [CrossRef]
- Wang, C.H.; Hsieh, Y.H.; Powers, Z.M.; Kao, C.Y. Defeating antibiotic-resistant bacteria: exploring alternative therapies for a post-antibiotic era. Int. J. Mol. Sci. 2020, 21, 1061. [Google Scholar] [CrossRef] [PubMed]
- Usai, D.; Donadu, M.; Bua, A.; Molicotti, P.; Zanetti, S.; Piras, S.; Corona, P.; Ibba, R.; Carta, A. Enhancement of antimicrobial activity of pump inhibitors associating drugs. J. Infect. Dev. Ctries. 2019, 13, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, K.L.C.; de Aquino, T.M.; Mendonca Junior, F.J.B. An update on Staphylococcus aureus NorA efflux pump inhibitors. Curr. Top. Med. Chem. 2020, 20, 2168–2185. [Google Scholar] [CrossRef]
- Trong Le, N.; Viet Ho, D.; Quoc Doan, T.; Tuan Le, A.; Raal, A.; Usai, D.; Sanna, G.; Carta, A.; Rappelli, P.; Diaz, N.; et al. Biological activities of essential oils from leaves of Paramignya trimera (Oliv.) Guillaum and Limnocitrus littoralis (Miq.) Swingle. Antibiotics 2020, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Le, N.T.; Donadu, M.G.; Ho, D.V.; Doan, T.Q.; Le, A.T.; Raal, A.; Usai, D.; Sanna, G.; Marchetti, M.; Usai, M.; et al. Biological activities of essential oil extracted from leaves of Atalantia sessiflora Guillauminin Vietnam. J. Infect. Dev. Ctries. 2020, 14, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Amaral, S.C.; Pruski, B.B.; de Freitas, S.B.; Allend, S.O.; Ferreira, M.R.A.; Moreira, C., Jr.; Pereira, D.I.B.; Junior, A.S.V.; Hartwig, D.D. Origanum vulgare essential oil: antibacterial activities and synergistic effect with polymyxin B against multidrug-resistant Acinetobacter baumannii. Mol. Biol. Rep. 2020, 47, 9615–9625. [Google Scholar] [CrossRef] [PubMed]
- Boonyanugomol, W.; Kraisriwattana, K.; Rukseree, K.; Boonsam, K.; Narachai, P. In vitro synergistic antibacterial activity of the essential oil from Zingiber cassumunar Roxb against extensively drug-resistant Acinetobacter baumannii strains. J. Infect. Public Health 2017, 10, 586–592. [Google Scholar] [CrossRef] [PubMed]



| Isolate | Isolation Date | Patient ID | Minimal Inhibition Concentration (mg/L) 1 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MEM | CTX | CAZ | CRO | FEP | SAM | TZP | GEN | AMK | CIP | LVX | SXT | COL 2 | MIN | |||
| T1060317 | 11 July 2017 | PT0044 | >4 (R) | >4 (R) | >32 (R) | >16 (R) | >32 (R) | >16 (R) | >16/8 (R) | >64/4 (R) | >8 (R) | >32 (R) | >2 (R) | >4 (R) | >2/38 (R) | >128 (R) | 4 (S) |
| T1060361 | 25 July 2017 | PT0044 | >4 (R) | >4 (R) | >32 (R) | >16 (R) | >32 (R) | >16 (R) | >16/8 (R) | >64/4 (R) | >8 (R) | >32 (R) | >2 (R) | >4 (R) | >2/38 (R) | 128 (R) | 4 (S) |
| T1060578 | 10 November 2017 | PT0345 | >4 (R) | >4 (R) | 8 (S) | 2 (S) | 4 (S) | 8 (S) | 8/4 (S) | 64/4 (I) | >8 (R) | >32 (R) | >2 (R) | 4 (I) | ≤0.5/9.5 (S) | 64 (R) | 4 (S) |
| T1060580 | 13 November 2017 | PT0348 | >4 (R) | >4 (R) | >32 (R) | 16 (I) | >32 (R) | 16 (I) | 8/4 (S) | 64/4 (I) | >8 (R) | >32 (R) | >2 (R) | >4 (R) | >2/38 (R) | >128 (R) | 4 (S) |
| T1070171 | 13 March 2018 | PT0512 | >4 (R) | >4 (R) | >32 (R) | >16 (R) | >32 (R) | >16 (R) | >16/8 (R) | >64/4 (R) | >8 (R) | >32 (R) | >2 (R) | >4 (R) | >2/38 (R) | >128 (R) | 4 (S) |
| T1070213 | 30 March 2018 | PT0512 | >4 (R) | >4 (R) | >32 (R) | >16 (R) | >32 (R) | >16 (R) | >16/8 (R) | >64/4 (R) | >8 (R) | ≤ 8 (S) | >2 (R) | >4 (R) | >2/38 (R) | >128 (R) | 4 (S) |
| T1070678 | 23 October 2018 | PT0712 | >4 (R) | >4 (R) | >32 (R) | >16 (R) | >32 (R) | >16 (R) | 8/4 (S) | >64/4 (R) | >8 (R) | >32 (R) | >2 (R) | >4 (R) | >2/38 (R) | 64 (R) | 4 (S) |
| Isolate | Patient ID | ST 1 (Pasteur/Oxford) | Capsule Type | Acquired Resistance Genes | |||||
|---|---|---|---|---|---|---|---|---|---|
| β-lactams | Aminoglycosides | Macrolides | Phenicols | Sulfonamides | Tetracyclines | ||||
| T1060317 | PT0044 | 2/544 and unknown | KL2 | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D | aac(6’)-Ib3, aadA1, aph(3”)-Ib, aph(3’)-Ia, aph(6’)-Id, armA | mph(E), msr(E) | catB8 | sul1, sul2 | tet(B) |
| T1060361 | PT0044 | 2/544 and unknown | KL2 | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D | aac(6’)-Ib3, aadA1, aph(3”)-Ib, aph(3’)-Ia, aph(6’)-Id, armA | mph(E), msr(E) | catB8 | sul1, sul2 | tet(B) |
| T1060578 | PT0345 | 136/~460 or 1092 2 | KL107 | blaADC-25, blaOXA-23, blaOXA-317 | ant(2”)-Ia, aph(3’)-VI | - | - | - | - |
| T1060580 | PT0348 | 2/208 and 1806 | KL2 | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D | aac(6’)-Ib3, aadA1, aph(3”)-Ib, aph(3’)-Ia, aph(6’)-Id, armA, ant(2”)-Ia, aph(3’)-VI | mph(E), msr(E) | catB8 | sul1, sul2 | tet(B) |
| T1070171 | PT0512 | 2/208 and 1806 | KL2 | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D | aac(6’)-Ib3, aadA1, aph(3”)-Ib, aph(3’)-Ia, aph(6’)-Id, armA | mph(E), msr(E) | catB8 | sul1, sul2 | tet(B) |
| T1070213 | PT0512 | 2/208 and 1806 | KL2 | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D | aph(3”)-Ib, aph(6’)-Id ant(2”)-Ia, aph(3’)-VI, | - | - | sul2 | tet(B) |
| T1070678 | PT0712 | 2/208 and 1806 | KL2 | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D | aac(6’)-Ib3, aadA1, aph(3”)-Ib, aph(3’)-Ia, aph(6’)-Id, armA | mph(E), msr(E) | catB8 | sul1, sul2 | tet(B) |
| Category | Gene | Product | ST2/ST544 | ST2/ST208&1806 | ST136 | ||||
|---|---|---|---|---|---|---|---|---|---|
| T1060317 | T1060361 | T1060580 | T1070171 | T1070213 | T1070678 | T1060578 | |||
| Adherence | ompA | Outer membrane protein OmpA | 100/100 1 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 98/100 |
| Quorum sensing | abaR | DNA-binding HTH domain-containing protein | 98/100 | 98/100 | 98/100 | 98/100 | 98/100 | 98/100 | 98/100 |
| abaI | N-acyl-L-homoserine lactone synthetase | 98/100 | 98/100 | 98/100 | 98/100 | 98/100 | 98/100 | 98/100 | |
| Enzymes | plcD | Phospholipase | 99/99.8 | 99/99.8 | 98/99.8 | 99/99.8 | 99/99.8 | 99/99.8 | 95/99.8 |
| plc | Phospholipase C | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 97/100 | |
| plc | Phospholipase C | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | |
| Biofilm -Csu fimbriae | csuC | Csu pilus chaperone protein CsuC | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | Not found | 98/100 |
| csuA/B | Csu pilus major pilin subunit CsuA/B | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | Not found | 99/100 | |
| csuB | Csu pilus subunit CsuB | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | Not found | 98/100 | |
| csuD | Csu pilus subunit CsuD | 99/100 | 99/100 | 99/100 | 99/100 | 100/20.8 | Not found | 98/100 | |
| csuA | Csu pilus usher protein CsuA | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | Not found | 98/100 | |
| csuE | Csu pilus tip adhesin CsuE | 99/100 | 99/100 | 99/100 | 99/97.9 | Not found | Not found | 95/100 | |
| Ade efflux pumps | adeG | Cation/multidrug efflux pump AdeG | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 95/100 |
| adeH | Outer membrane protein AdeH | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 98/100 | |
| adeF | Membrane-fusion protein AdeF | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 98/100 | |
| Biofilm-PAGN 2 | pgaB | PNAG N-deacetylase PgaB | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 97/100 |
| pgaA | PNAG export porin PgaA | 98/99.8 | 98/99.8 | 98/99.8 | 98/99.8 | 98/99.8 | 98/99.8 | 97/100 | |
| pgaC | PNAG synthase | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 97/100 | |
| pgaD | PNAG biosynthesis protein PgaD | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 97/100 | |
| Regulation | bfmS | Signal transduction histidine kinase BfmS | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 98/100 |
| bfmR | Biofilm-controlling response regulator BfmR | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 99/100 | |
| Iron uptake | barA | Siderophore efflux system of the ABC superfamily BarA | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | Not found | 97/100 |
| barB | Siderophore efflux system of the ABC superfamily BarB | 99/99.9 | 99/99.9 | 99/99.9 | 99/99.9 | 99/99.9 | Not found | 97/99.9 | |
| basA | Acinetobactin biosynthesis protein BarA | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 96/100 | |
| basB | Non-ribosomal peptide synthetase BarB | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 96/100 | |
| basC | Acinetobactin biosynthesis protein BasC | 100/99.2 | 100/99.2 | 100/99.2 | 100/99.2 | 100/99.2 | 100/99.2 | 97/99.2 | |
| basD | Acinetobactin biosynthesis protein BasD | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 96/100 | |
| basF | Aryl carrier protein BasF | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 97/100 | |
| basG | Acinetobactin biosynthesis protein BasF | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | Not found | 97/100 | |
| basH | Non-ribosomal peptide biosynthesis thioesterase BasH | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | Not found | 96/100 | |
| basI | Acinetobactin biosynthesis protein BasI | 97/98.8 | 97/98.8 | 97/98.8 | 97/98.8 | 97/98.8 | Not found | 93/98.8 | |
| basJ | Acinetobactin biosynthesis protein BasJ | 98/100 | 98/100 | 98/100 | 98/100 | 98/100 | Not found | 98/100 | |
| entE | Non-ribosomal peptide synthetase EntE | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 96/100 | |
| bauA | TonB-dependent siderophore receptor BauA | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | |
| bauB | Ferric siderophore ABC transporter, periplasmic siderophore-binding protein BauB | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 97/100 | |
| bauC | Ferric siderophore ABC transporter, permease protein BauC | 98/100 | 98/100 | 98/100 | 98/100 | 98/100 | 98/100 | 95/83.1 | |
| bauD | Ferric siderophore ABC transporter, permease protein BauD | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 99/100 | 96/99.9 | |
| bauE | Ferric siderophore ABC transporter, ATP-binding protein BauE | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 99/100 | |
| bauF | Siderophore-interacting protein | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 97/100 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilsan, N.A.; Lee, Y.-J.; Kuo, S.-C.; Lee, I.-H.; Huang, T.-W. Antimicrobial Resistance Mechanisms and Virulence of Colistin- and Carbapenem-Resistant Acinetobacter baumannii Isolated from a Teaching Hospital in Taiwan. Microorganisms 2021, 9, 1295. https://doi.org/10.3390/microorganisms9061295
Ilsan NA, Lee Y-J, Kuo S-C, Lee I-H, Huang T-W. Antimicrobial Resistance Mechanisms and Virulence of Colistin- and Carbapenem-Resistant Acinetobacter baumannii Isolated from a Teaching Hospital in Taiwan. Microorganisms. 2021; 9(6):1295. https://doi.org/10.3390/microorganisms9061295
Chicago/Turabian StyleIlsan, Noor Andryan, Yuarn-Jang Lee, Shu-Chen Kuo, I-Hui Lee, and Tzu-Wen Huang. 2021. "Antimicrobial Resistance Mechanisms and Virulence of Colistin- and Carbapenem-Resistant Acinetobacter baumannii Isolated from a Teaching Hospital in Taiwan" Microorganisms 9, no. 6: 1295. https://doi.org/10.3390/microorganisms9061295
APA StyleIlsan, N. A., Lee, Y.-J., Kuo, S.-C., Lee, I.-H., & Huang, T.-W. (2021). Antimicrobial Resistance Mechanisms and Virulence of Colistin- and Carbapenem-Resistant Acinetobacter baumannii Isolated from a Teaching Hospital in Taiwan. Microorganisms, 9(6), 1295. https://doi.org/10.3390/microorganisms9061295






