Bacterial and Viral Coinfections with the Human Respiratory Syncytial Virus
Abstract
1. Introduction
hRSV Epidemiology and Structure
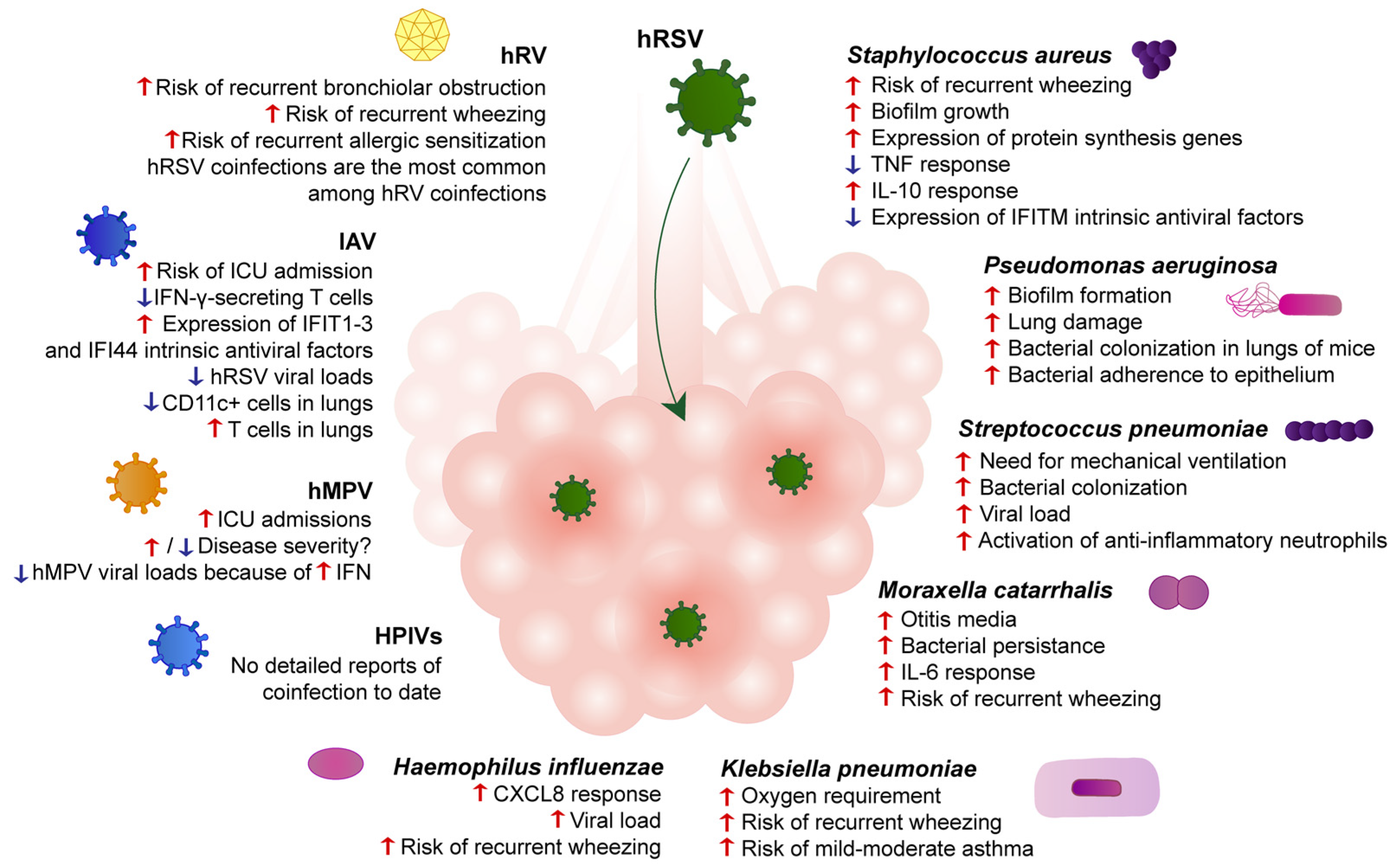
2. hRSV Coinfections with Pathogenic Respiratory Bacteria
2.1. Staphylococcus aureus
2.2. Pseudomonas aeruginosa
2.3. Streptococcus pneumoniae
2.4. Other Pathogenic Respiratory Bacteria
3. hRSV Coinfections with Other Respiratory Viruses
3.1. Human Rhinovirus
3.2. Influenza A Virus
3.3. Human Metapneumovirus
3.4. Parainfluenza Viruses
4. A Summary of Molecular and Cellular Mechanisms Pertaining to hRSV Coinfections
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amarasinghe, G.K.; Ayllón, M.A.; Bào, Y.; Basler, C.F.; Bavari, S.; Blasdell, K.R.; Briese, T.; Brown, P.A.; Bukreyev, A.; Balkema-Buschmann, A.; et al. Taxonomy of the order Mononegavirales: Update 2019. Arch. Virol. 2019, 164, 1967–1980. [Google Scholar] [CrossRef]
- Carvajal, J.J.; Avellaneda, A.M.; Salazar-Ardiles, C.; Maya, J.E.; Kalergis, A.; Lay, M.K. Host Components Contributing to Respiratory Syncytial Virus Pathogenesis. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Soto, J.A.; Gálvez, N.M.S.; Pacheco, G.; Bueno, S.M.; Kalergis, A.M. Antibody development for preventing the human respiratory syncytial virus pathology. Mol. Med. 2020, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Histoshi, T.; McAllister, D.A.; O’Brien, K.L.; Simoes, E.A.F.; Madhi, S.A.; Gessner, B.D.; Polack, F.P.; Balsells, E.; Acacio, S.; Aguayo, C.; et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: A systematic review and modelling study. Lancet 2017, 390, 946–958. [Google Scholar] [CrossRef]
- Haddadin, Z.; Rankin, D.A.; Lipworth, L.; Suh, M.; McHenry, R.; Blozinski, A.; George, S.S.; Fernandez, K.N.; Varjabedian, R.; Spieker, A.J.; et al. Respiratory Virus Surveillance in Infants across Different Clinical Settings. J. Pediatr. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kalergis, A.M.; Soto, J.A.; Gálvez, N.M.S.; Andrade, C.A.; Fernandez, A.; Bohmwald, K.; Bueno, S.M. Pharmacological management of human respiratory syncytial virus infection. Expert Opin. Pharmacother. 2020, 21, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Mac, S.; Sumner, A.; Duchesne-Belanger, S.; Stirling, R.; Tunis, M.; Sander, B. Cost-effectiveness of Palivizumab for Respiratory Syncytial Virus: A Systematic Review. Pediatrics 2019, 143, e20184064. [Google Scholar] [CrossRef] [PubMed]
- Andrade, C.A.; Pacheco, G.A.; Gálvez, N.M.S.; Soto, J.A.; Bueno, S.M.; Kalergis, A.M. Innate Immune Components that Regulate the Pathogenesis and Resolution of hRSV and hMPV Infections. Viruses 2020, 12, 637. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.A.; Böhmwald, K.; Céspedes, P.F.; Riedel, C.A.; Bueno, S.M.; Kalergis, A.M. Modulation of host adaptive immunity by hRSV proteins. Virulence 2014, 5, 740–751. [Google Scholar] [CrossRef] [PubMed]
- McLellan, J.; Ray, W.C.; Peeples, M.E. Structure and Function of Respiratory Syncytial Virus Surface Glycoproteins. Curr. Top. Microbiol. Immunol. 2013, 372, 83–104. [Google Scholar] [CrossRef] [PubMed]
- Schepens, B.; Schotsaert, M.; Saelens, X. Small hydrophobic protein of respiratory syncytial virus as a novel vaccine antigen. Immunotherapy 2015, 7, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Araujo, G.C.; Silva, R.H.T.; Scott, L.P.B.; Araujo, A.S.; Souza, F.P.; Oliveira, R. Structure and functional dynamics characterization of the ion channel of the human respiratory syncytial virus (hRSV) small hydrophobic protein (SH) transmembrane domain by combining molecular dynamics with excited normal modes. J. Mol. Model. 2016, 22, 286. [Google Scholar] [CrossRef]
- Gan, S.-W.; Tan, E.; Lin, X.; Yu, D.; Wang, J.; Tan, G.M.-Y.; Vararattanavech, A.; Yeo, C.Y.; Soon, C.H.; Soong, T.W.; et al. The Small Hydrophobic Protein of the Human Respiratory Syncytial Virus Forms Pentameric Ion Channels. J. Biol. Chem. 2012, 287, 24671–24689. [Google Scholar] [CrossRef] [PubMed]
- Kiss, G.; Holl, J.M.; Williams, G.M.; Alonas, E.; Vanover, D.; Lifland, A.W.; Gudheti, M.; Guerrero-Ferreira, R.; Nair, V.; Yi, H.; et al. Structural Analysis of Respiratory Syncytial Virus Reveals the Position of M2-1 between the Matrix Protein and the Ribonucleoprotein Complex. J. Virol. 2014, 88, 7602–7617. [Google Scholar] [CrossRef]
- Kleiner, V.A.; Fearns, R. RSV M2-1 Protein in Complex with RNA: Old Questions Are Answered and a New One Emerges. Structure 2020, 28, 977–978. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, A.; Collins, P.L. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 11259–11264. [Google Scholar] [CrossRef] [PubMed]
- Spann, K.; Tran, K.C.; Collins, P.L. Effects of Nonstructural Proteins NS1 and NS2 of Human Respiratory Syncytial Virus on Interferon Regulatory Factor 3, NF-κB, and Proinflammatory Cytokines. J. Virol. 2005, 79, 5353–5362. [Google Scholar] [CrossRef] [PubMed]
- Pasman, L. The complication of coinfection. Yale J. Boil. Med. 2012, 85, 127–132. [Google Scholar]
- Yoshida, L.-M.; Suzuki, M.; Nguyen, H.A.; Le, M.N.; Vu, T.D.; Yoshino, H.; Schmidt, W.-P.; Nguyen, T.T.A.; Le, H.T.; Morimoto, K.; et al. Respiratory syncytial virus: Co-infection and paediatric lower respiratory tract infections. Eur. Respir. J. 2013, 42, 461–469. [Google Scholar] [CrossRef]
- Thorburn, K.; Harigopal, S.; Reddy, V.; Taylor, N.; Van Saene, H.K.F. High incidence of pulmonary bacterial co-infection in children with severe respiratory syncytial virus (RSV) bronchiolitis. Thorax 2006, 61, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Hernández, J.X.; Núñez-López, M.; Comas-García, A.; Cherpitel, D.E.N.; Ocampo, M.C. Superinfection between Influenza and RSV Alternating Patterns in San Luis Potosí State, México. PLoS ONE 2015, 10, e0115674. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hament, J.-M.; Aerts, P.C.; Fleer, A.; Van Dijk, H.; Harmsen, T.; Kimpen, J.L.L.; Wolfs, T.F.W. Direct Binding of Respiratory Syncytial Virus to Pneumococci: A Phenomenon That Enhances Both Pneumococcal Adherence to Human Epithelial Cells and Pneumococcal Invasiveness in a Murine Model. Pediatr. Res. 2005, 58, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.J.; Loman, N.; Bogaert, D.; O’Grady, J. Co-infections: Potentially lethal and unexplored in COVID-19. Lancet Microbe 2020, 1, e11. [Google Scholar] [CrossRef]
- Cawcutt, K.; Kalil, A.C. Pneumonia with bacterial and viral coinfection. Curr. Opin. Crit. Care 2017, 23, 385–390. [Google Scholar] [CrossRef]
- Jia, L.; Xie, J.; Zhao, J.; Cao, D.; Liang, Y.; Hou, X.; Wang, L.; Li, Z. Mechanisms of Severe Mortality-Associated Bacterial Co-infections Following Influenza Virus Infection. Front. Cell. Infect. Microbiol. 2017, 7, 338. [Google Scholar] [CrossRef]
- Meskill, S.D.; O’Bryant, S.C. Respiratory Virus Co-infection in Acute Respiratory Infections in Children. Curr. Infect. Dis. Rep. 2020, 22, 3. [Google Scholar] [CrossRef]
- Sawada, S.; Okutani, F.; Kobayashi, T. Comprehensive Detection of Respiratory Bacterial and Viral Pathogens in the Middle Ear Fluid and Nasopharynx of Pediatric Patients with Acute Otitis Media. Pediatr. Infect. Dis. J. 2019, 38, 1199–1203. [Google Scholar] [CrossRef]
- Hishiki, H.; Ishiwada, N.; Fukasawa, C.; Kohno, Y.; Abe, K.; Hoshino, T.; Aizawa, J.; Ishikawa, N. Incidence of bacterial coinfection with respiratory syncytial virus bronchopulmonary infection in pediatric inpatients. J. Infect. Chemother. 2011, 17, 87–90. [Google Scholar] [CrossRef]
- Tseng, M.-H.; Wei, B.-H.; Lin, W.-J.; Lu, J.-J.; Lee, S.-Y.; Wang, S.-R.; Chen, S.-J.; Wang, C.-C. Fatal sepsis and necrotizing pneumonia in a child due to community-acquired methicillin-resistant Staphylococcus aureus: Case report and literature review. Scand. J. Infect. Dis. 2005, 37, 504–507. [Google Scholar] [CrossRef]
- Gillet, Y.; Issartel, B.; Vanhems, P.; Fournet, J.-C.; Lina, G.; Bes, M.; Vandenesch, F.; Piémont, Y.; Brousse, N.; Floret, D.; et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 2002, 359, 753–759. [Google Scholar] [CrossRef]
- Garnier, F.; Tristan, A.; François, B.; Etienne, J.; Delage-Corre, M.; Martin, C.; Liassine, N.; Wannet, W.; Denis, F.; Ploy, M.-C. Pneumonia and New Methicillin-resistantStaphylococcus aureusClone. Emerg. Infect. Dis. 2006, 12, 498–500. [Google Scholar] [CrossRef]
- David, M.Z.; Daum, R.S. Community-Associated Methicillin-Resistant Staphylococcus aureus: Epidemiology and Clinical Consequences of an Emerging Epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed]
- Antonanzas, F.; Lozano, C.; Torres, C. Economic Features of Antibiotic Resistance: The Case of Methicillin-Resistant Staphylococcus aureus. PharmacoEconomics 2015, 33, 285–325. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-H.; Hsueh, P.-R.; Chung, D.R.; Ko, K.S.; Kang, C.-I.; Peck, K.R.; Yeom, J.-S.; Kim, S.-W.; Chang, H.-H.; Kim, Y.-S.; et al. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: An ANSORP study. J. Antimicrob. Chemother. 2011, 66, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C. Increasing Resistance to Vancomycin and Other Glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 2001, 7, 327–332. [Google Scholar] [CrossRef] [PubMed]
- System, A.R.F.T.N. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2003, issued August 2003. Am. J. Infect. Control. 2003, 31, 481–498. [Google Scholar] [CrossRef] [PubMed]
- Stockman, L.J.; Reed, C.; Kallen, A.J.; Finelli, L.; Anderson, L.J. Respiratory syncytial virus and Staphylococcus aureus coinfection in children hospitalized with pneumonia. Pediatr. Infect. Dis. J. 2010, 29, 1048–1050. [Google Scholar] [CrossRef]
- Li, X.; Zhang, W.; Yao, S.; Zha, H.; Huang, B.; Liu, D.; Wu, K. Prevalence and clinical significance of common respiratory pathogens in the upper respiratory tract of children with community-acquired pneumonia in Zunyi, China. Pediatr. Pulmonol. 2020, 55, 2437–2443. [Google Scholar] [CrossRef] [PubMed]
- Lamarão, L.M.; Ramos, F.L.; Mello, W.A.; Santos, M.C.; Barbagelata, L.S.; Justino, M.C.A.; Da Silva, A.F.; Quaresma, A.J.P.G.; Da Silva, V.B.; Burbano, R.R.; et al. Prevalence and clinical features of respiratory syncytial virus in children hospitalized for community-acquired pneumonia in northern Brazil. BMC Infect. Dis. 2012, 12, 119. [Google Scholar] [CrossRef]
- Zhong, Q.; Feng, H.; Lü, Q.; Liu, X.; Zhao, Q.; Du, Y.; Zhang, X.-H.; Wang, J.-R. Recurrent wheezing in neonatal pneumonia is associated with combined infection with Respiratory Syncytial Virus and Staphylococcus aureus or Klebsiella pneumoniae. Sci. Rep. 2018, 8, 995. [Google Scholar] [CrossRef]
- Dickson, R.P.; Martinez, S.M.; Ortiz, J.R. A Case of Rapidly Progressive Necrotizing Pneumonia Caused by Community-Acquired Methicillin-Resistant Staphylococcus aureus. Respir. Care 2008, 53, 1223. [Google Scholar] [PubMed]
- Lim, W.H.; Lien, R.; Huang, Y.-C.; Lee, W.J.; Lai, J.Y. Community-associated methicillin-resistant Staphylococcus aureus necrotizing pneumonia in a healthy neonate. J. Microbiol. Immunol. Infect. 2014, 47, 555–557. [Google Scholar] [CrossRef][Green Version]
- Kiedrowski, M.R.; Bomberger, J.M. Viral-Bacterial Co-infections in the Cystic Fibrosis Respiratory Tract. Front. Immunol. 2018, 9, 3067. [Google Scholar] [CrossRef]
- Kiedrowski, M.R.; Gaston, J.R.; Kocak, B.R.; Coburn, S.L.; Lee, S.; Pilewski, J.M.; Myerburg, M.M.; Bomberger, J.M. Staphylococcus aureusBiofilm Growth on Cystic Fibrosis Airway Epithelial Cells Is Enhanced during Respiratory Syncytial Virus Coinfection. mSphere 2018, 3, e00341-18. [Google Scholar] [CrossRef] [PubMed]
- Leech, J.M.; Lacey, K.A.; Mulcahy, M.E.; Medina, E.; McLoughlin, R.M. IL-10 Plays Opposing Roles duringStaphylococcus aureusSystemic and Localized Infections. J. Immunol. 2017, 198, 2352–2365. [Google Scholar] [CrossRef] [PubMed]
- Buchs, C.; Dalphin, M.-L.; Sanchez, S.; Perceval, M.; Coutier, L.; Mainguy, C.; Kassaï-Koupaï, B.; Reix, P. Palivizumab prophylaxis in infants with cystic fibrosis does not delay first isolation of Pseudomonas aeruginosa or Staphylococcus aureus. Eur. J. Nucl. Med. Mol. Imaging 2017, 134, 415–897. [Google Scholar] [CrossRef]
- Bjornson, C.; Chan, P.; Li, A.; Paes, B.; Lanctôt, K.L.; Mitchell, I. Palivizumab prophylaxis for respiratory syncytial virus in infants with cystic fibrosis: Is there a need? Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.A.; Odelola, O.A.; Saldanha, I.J. Palivizumab for prophylaxis against respiratory syncytial virus infection in children with cystic fibrosis. Cochrane Database Syst. Rev. 2016, 2016, CD007743. [Google Scholar] [CrossRef] [PubMed]
- Groves, H.; Jenkins, L.; Macfarlane, M.; Reid, A.; Lynn, F.; Shields, M. Efficacy and long-term outcomes of palivizumab prophylaxis to prevent respiratory syncytial virus infection in infants with cystic fibrosis in Northern Ireland. Pediatr. Pulmonol. 2016, 51, 379–385. [Google Scholar] [CrossRef]
- Man, W.H.; Scheltema, N.M.; Clerc, M.; van Houten, M.A.; Nibbelke, E.E.; Achten, N.B.; Arp, K.; Sanders, E.A.M.; Bont, L.J.; Bogaert, D. Infant respiratory syncytial virus prophylaxis and nasopharyngeal microbiota until 6 years of life: A subanalysis of the MAKI randomised controlled trial. Lancet Respir. Med. 2020, 8, 1022–1031. [Google Scholar] [CrossRef]
- De Vrankrijker, A.M.; Wolfs, T.F.; Ciofu, O.; Høiby, N.; Van Der Ent, C.K.; Poulsen, S.S.; Johansen, H.K. Respiratory syncytial virus infection facilitates acute colonization ofPseudomonas aeruginosain mice. J. Med. Virol. 2009, 81, 2096–2103. [Google Scholar] [CrossRef]
- Hendricks, M.R.; Lashua, L.P.; Fischer, D.; Flitter, B.A.; Eichinger, K.M.; Durbin, J.E.; Sarkar, S.; Coyne, C.B.; Empey, K.M.; Bomberger, J.M. Respiratory syncytial virus infection enhancesPseudomonas aeruginosabiofilm growth through dysregulation of nutritional immunity. Proc. Natl. Acad. Sci. USA 2016, 113, 1642–1647. [Google Scholar] [CrossRef]
- Van Ewijk, B.E.; Wolfs, T.F.W.; Aerts, P.C.; Van Kessel, K.P.M.; Fleer, A.; Kimpen, J.L.L.; Van Der Ent, C.K. RSV Mediates Pseudomonas aeruginosa Binding to Cystic Fibrosis and Normal Epithelial Cells. Pediatr. Res. 2007, 61, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Purcaro, G.; Rees, C.A.; Melvin, J.A.; Bomberger, J.M.; Hill, J.E. Volatile fingerprinting of Pseudomonas aeruginosa and respiratory syncytial virus infection in an in vitro cystic fibrosis co-infection model. J. Breath Res. 2018, 12, 046001. [Google Scholar] [CrossRef] [PubMed]
- Melvin, J.A.; Lashua, L.P.; Kiedrowski, M.R.; Yang, G.; Deslouches, B.; Montelaro, R.C.; Bomberger, J.M. Simultaneous Antibiofilm and Antiviral Activities of an Engineered Antimicrobial Peptide during Virus-Bacterium Coinfection. mSphere 2016, 1, e00083-16. [Google Scholar] [CrossRef] [PubMed]
- Chuaychoo, B.; Ngamwongwan, S.; Kaewnaphan, B.; Athipanyasilp, N.; Horthongkham, N.; Kantakamalakul, W.; Muangman, N. Clinical manifestations and outcomes of respiratory syncytial virus infection in adult hospitalized patients. J. Clin. Virol. 2019, 117, 103–108. [Google Scholar] [CrossRef]
- Godefroy, R.; Giraud-Gatineau, A.; Jimeno, M.-T.; Edouard, S.; Meddeb, L.; Zandotti, C.; Chaudet, H.; Colson, P.; Raoult, D.; Cassir, N. Respiratory Syncytial Virus Infection: Its Propensity for Bacterial Coinfection and Related Mortality in Elderly Adults. Open Forum Infect. Dis. 2020, 7, ofaa546. [Google Scholar] [CrossRef]
- Brealey, J.C.; Young, P.R.; Sloots, T.P.; Ware, R.S.; Lambert, S.B.; Sly, P.D.; Grimwood, K.; Chappell, K.J. Bacterial colonization dynamics associated with respiratory syncytial virus during early childhood. Pediatr. Pulmonol. 2020, 55, 1237–1245. [Google Scholar] [CrossRef]
- Yan, T.; Tang, X.; Sun, L.; Tian, R.; Li, Z.; Liu, G. Co infection of respiratory syncytial viruses (RSV) and streptococcus pneumonia modulates pathogenesis and dependent of serotype and phase variant. Microb. Pathog. 2020, 144, 104126. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Louwen, R.; Elberse, K.; Van Amerongen, G.; Yüksel, S.; Luijendijk, A.; Osterhaus, A.D.M.E.; Duprex, W.P.; De Swart, R.L. Streptococcus pneumoniae Enhances Human Respiratory Syncytial Virus Infection In Vitro and In Vivo. PLoS ONE 2015, 10, e0127098. [Google Scholar] [CrossRef] [PubMed]
- Cortjens, B.; Ingelse, S.A.; Calis, J.C.; Vlaar, A.P.; Koenderman, L.; Bem, R.A.; van Woensel, J.B. Neutrophil subset responses in infants with severe viral respiratory infection. Clin. Immunol. 2017, 176, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Pillay, J.; Kamp, V.M.; Van Hoffen, E.; Visser, T.; Tak, T.; Lammers, J.-W.; Ulfman, L.H.; Leenen, L.P.; Pickkers, P.; Koenderman, L. A subset of neutrophils in human systemic inflammation inhibits T cell responses through Mac-1. J. Clin. Investig. 2012, 122, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Brockson, M.E.; Novotny, L.A.; Jurcisek, J.A.; McGillivary, G.; Bowers, M.R.; Bakaletz, L.O. Respiratory Syncytial Virus Promotes Moraxella catarrhalis-Induced Ascending Experimental Otitis Media. PLoS ONE 2012, 7, e40088. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, X.; Zhang, X.; Zhang, N.; Wang, X.; Sun, L.; Chen, N.; Zhao, S.; He, Q. Airway microbiome, host immune response and recurrent wheezing in infants with severe respiratory syncytial virus bronchiolitis. Pediatr. Allergy Immunol. 2019, 31, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Ederveen, T.H.A.; Ferwerda, G.; Ahout, I.M.; Vissers, M.; De Groot, R.; Boekhorst, J.; Timmerman, H.M.; Huynen, M.A.; Van Hijum, S.A.F.T.; De Jonge, M.I. Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome 2018, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, S.M.; Ketterer, M.; Apicella, M.A.; Varga, S.M. Non-typeable Haemophilus influenzae protects human airway epithelial cells from a subsequent respiratory syncytial virus challenge. Virology 2016, 498, 128–135. [Google Scholar] [CrossRef]
- Buchwald, A.G.; Tamboura, B.; Tennant, S.M.; Haidara, F.C.; Coulibaly, F.; Doumbia, M.; Diallo, F.; Keita, A.M.; O Sow, S.; Kotloff, K.L.; et al. Epidemiology, Risk Factors, and Outcomes of Respiratory Syncytial Virus Infections in Newborns in Bamako, Mali. Clin. Infect. Dis. 2020, 70, 59–66. [Google Scholar] [CrossRef]
- Huang, Y.J.; Nariya, S.; Harris, J.M.; Lynch, S.V.; Choy, D.F.; Arron, J.R.; Boushey, H. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol. 2015, 136, 874–884. [Google Scholar] [CrossRef]
- Russell, C.D.; Unger, S.A.; Walton, M.; Schwarze, J. The Human Immune Response to Respiratory Syncytial Virus Infection. Clin. Microbiol. Rev. 2017, 30, 481–502. [Google Scholar] [CrossRef]
- Ye, P.; Garvey, P.B.; Zhang, P.; Nelson, S.; Bagby, G.; Summer, W.R.; Schwarzenberger, P.; Shellito, J.E.; Kolls, J.K. Interleukin-17 and Lung Host Defense againstKlebsiella pneumoniaeInfection. Am. J. Respir. Cell Mol. Biol. 2001, 25, 335–340. [Google Scholar] [CrossRef]
- Dulek, D.E.; Newcomb, D.C.; Goleniewska, K.; Cephus, J.; Zhou, W.; Reiss, S.; Toki, S.; Ye, F.; Zaynagetdinov, R.; Sherrill, T.P.; et al. Allergic Airway Inflammation Decreases Lung Bacterial Burden following Acute Klebsiella pneumoniae Infection in a Neutrophil- and CCL8-Dependent Manner. Infect. Immun. 2014, 82, 3723–3739. [Google Scholar] [CrossRef] [PubMed]
- Antalis, E.; Oikonomopoulou, Z.; Kottaridi, C.; Kossyvakis, A.; Spathis, A.; Magkana, M.; Katsouli, A.; Tsagris, V.; Papaevangelou, V.; Mentis, A.; et al. Mixed viral infections of the respiratory tract; an epidemiological study during consecutive winter seasons. J. Med. Virol. 2018, 90, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Roig, A.; Salvadó, M.; Caballero-Rabasco, M.; Sánchez-Buenavida, A.; Segura, N.L.; Bonet-Alcaina, M. Coinfección vírica en las infecciones respiratorias infantiles. Arch. Bronconeumol. Engl. Ed. 2015, 51, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Soudani, N.; Caniza, M.A.; Assaf-Casals, A.; Shaker, R.; Lteif, M.; Su, Y.; Tang, L.; Akel, I.; Muwakkit, S.; Chmaisse, A.; et al. Prevalence and characteristics of acute respiratory virus infections in pediatric cancer patients. J. Med. Virol. 2019, 91, 1191–1201. [Google Scholar] [CrossRef]
- Mansuy, J.; Bourcier, M.; Trémeaux, P.; Dimeglio, C.; Izopet, J. COVID-19 pandemic period, where are the seasonal viruses? J. Med. Virol. 2021, 93, 4097–4098. [Google Scholar] [CrossRef] [PubMed]
- Palmenberg, A.C.; Spiro, D.; Kuzmickas, R.; Wang, S.; Djikeng, A.; Rathe, J.A.; Fraser, C.; Liggett, S.B. Sequencing and Analyses of All Known Human Rhinovirus Genomes Reveal Structure and Evolution. Science 2009, 324, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Palmenberg, A.C.; Rathe, J.A.; Liggett, S.B. Analysis of the complete genome sequences of human rhinovirus. J. Allergy Clin. Immunol. 2010, 125, 1190–1199. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.E.; Lamson, D.; George, K.S.; Walsh, T.J. Human Rhinoviruses. Clin. Microbiol. Rev. 2013, 26, 135–162. [Google Scholar] [CrossRef]
- Costa, L.F.; Queiróz, D.A.O.; Da Silveira, H.L.; Neto, M.B.; De Paula, N.T.; Oliveira, T.F.M.S.; Tolardo, A.L.; Yokosawa, J. Human Rhinovirus and Disease Severity in Children. Pediatrics 2014, 133, e312–e321. [Google Scholar] [CrossRef]
- Amat, F.; Plantard, C.; Mulliez, A.; Petit, I.; Rochette, E.; Verdan, M.; Henquell, C.; Labbé, G.; Heraud, M.C.; Evrard, B.; et al. RSV-hRV co-infection is a risk factor for recurrent bronchial obstruction and early sensitization 3 years after bronchiolitis. J. Med. Virol. 2018, 90, 867–872. [Google Scholar] [CrossRef]
- Greer, R.; McErlean, P.; Arden, K.; Faux, C.; Nitsche, A.; Lambert, S.; Nissen, M.; Sloots, T.; Mackay, I. Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections? J. Clin. Virol. 2009, 45, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Achten, N.B.; Wu, P.; Bont, L.; Blanken, M.O.; Gebretsadik, T.; Chappell, J.D.; Wang, L.; Yu, C.; Larkin, E.K.; Carroll, K.N.; et al. Interference Between Respiratory Syncytial Virus and Human Rhinovirus Infection in Infancy. J. Infect. Dis. 2017, 215, 1102–1106. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.K.; Edwards, K.M.; Weinberg, G.A.; Iwane, M.K.; Griffin, M.R.; Hall, C.B.; Zhu, Y.; Szilagyi, P.G.; Morin, L.-L.; Heil, L.H.; et al. A novel group of rhinoviruses is associated with asthma hospitalizations. J. Allergy Clin. Immunol. 2009, 123, 98–104.e1. [Google Scholar] [CrossRef] [PubMed]
- Karppinen, S.; Toivonen, L.; Schuez-Havupalo, L.; Waris, M.; Peltola, V. Interference between respiratory syncytial virus and rhinovirus in respiratory tract infections in children. Clin. Microbiol. Infect. 2016, 22, 208.e1–208.e6. [Google Scholar] [CrossRef] [PubMed]
- Petrarca, L.; Nenna, R.; Frassanito, A.; Pierangeli, A.; Leonardi, S.; Scagnolari, C.; Antonelli, G.; Papoff, P.; Moretti, C.; Midulla, F. Acute bronchiolitis: Influence of viral co-infection in infants hospitalized over 12 consecutive epidemic seasons. J. Med. Virol. 2018, 90, 631–638. [Google Scholar] [CrossRef]
- Luchsinger, V.; Ampuero, S.; Palomino, M.A.; Chnaiderman, J.; Levican, J.; Gaggero, A.; Larrañaga, C.E. Comparison of virological profiles of respiratory syncytial virus and rhinovirus in acute lower tract respiratory infections in very young Chilean infants, according to their clinical outcome. J. Clin. Virol. 2014, 61, 138–144. [Google Scholar] [CrossRef]
- Calvo, C.; Garcia-Garcia, M.L.; Pozo, F.; Paula, G.; Molinero, M.; Calderón, A.; González-Esguevillas, M.; Casas, I. Respiratory Syncytial Virus Coinfections With Rhinovirus and Human Bocavirus in Hospitalized Children. Medicine 2015, 94, e1788. [Google Scholar] [CrossRef]
- Comte, A.; Bour, J.-B.; Darniot, M.; Pitoiset, C.; Aho-Glélé, L.S.; Manoha, C. Epidemiological characteristics and clinical outcomes of human rhinovirus infections in a hospitalized population. Severity is independently linked to RSV coinfection and comorbidities. J. Clin. Virol. 2020, 125, 104290. [Google Scholar] [CrossRef]
- Winther, B.; Brofeldt, S.; And, B.C.; Mygind, N. Light and Scanning Electron Microscopy of Nasal Biopsy Material from Patients with Naturally Acquired Common Colds. Acta Oto-Laryngologica 1984, 97, 309–318. [Google Scholar] [CrossRef]
- Turner, R.B.; Hendley, J.O.; Gwaltney, J.J.M. Shedding of Infected Ciliated Epithelial Cells in Rhinovirus Colds. J. Infect. Dis. 1982, 145, 849–853. [Google Scholar] [CrossRef]
- Pinky, L.; Dobrovolny, H.M. Coinfections of the Respiratory Tract: Viral Competition for Resources. PLoS ONE 2016, 11, e0155589. [Google Scholar] [CrossRef]
- Sangesland, M.; Lingwood, D. Antibody Focusing to Conserved Sites of Vulnerability: The Immunological Pathways for ‘Universal’ Influenza Vaccines. Vaccines 2021, 9, 125. [Google Scholar] [CrossRef]
- Breen, M.; Nogales, A.; Baker, S.F.; Martínez-Sobrido, L. Replication-Competent Influenza A Viruses Expressing Reporter Genes. Viruses 2016, 8, 179. [Google Scholar] [CrossRef]
- WHO. Influenza (Seasonal). Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 1 April 2021).
- Carrat, F.; Flahault, A. Influenza vaccine: The challenge of antigenic drift. Vaccine 2007, 25, 6852–6862. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Amarasinghe, G.K.; Anthony, S.J.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020, 165, 3023–3072. [Google Scholar] [CrossRef] [PubMed]
- Míguez, A.; Iftimi, A.; Montes, F. Temporal association between the influenza virus and respiratory syncytial virus (RSV): RSV as a predictor of seasonal influenza. Epidemiol. Infect. 2016, 144, 2621–2632. [Google Scholar] [CrossRef] [PubMed]
- Goka, E.; Vallely, P.; Mutton, K.; Klapper, P. Influenza A viruses dual and multiple infections with other respiratory viruses and risk of hospitalisation and mortality. Influenza Other Respir. Viruses 2013, 7, 1079–1087. [Google Scholar] [CrossRef]
- Appak, Ö.; Duman, M.; Belet, N.; Sayiner, A.A. Viral respiratory infections diagnosed by multiplex polymerase chain reaction in pediatric patients. J. Med. Virol. 2019, 91, 731–737. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, J.; Zou, X.; Fan, Y.; Xiong, Z.; Li, B.; Wang, C.; Li, H.; Han, J.; Liu, X.; et al. Severity of influenza virus and respiratory syncytial virus coinfections in hospitalized adult patients. J. Clin. Virol. 2020, 133, 104685. [Google Scholar] [CrossRef]
- Aberle, J.H.; Aberle, S.W.; Pracher, E.; Hutter, H.-P.; Kundi, M.; Popow-Kraupp, T. Single Versus Dual Respiratory Virus Infections in Hospitalized Infants. Pediatr. Infect. Dis. J. 2005, 24, 605–610. [Google Scholar] [CrossRef]
- Meskill, S.D.; Revell, P.A.; Chandramohan, L.; Cruz, A.T. Prevalence of co-infection between respiratory syncytial virus and influenza in children. Am. J. Emerg. Med. 2017, 35, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Stefanska, I.; Romanowska, M.; Donevski, S.; Gawryluk, D.; Brydak, L.B. Co-Infections with Influenza and Other Respiratory Viruses. Adv. Exp. Med. Biol. 2012, 756, 291–301. [Google Scholar] [CrossRef]
- Drori, Y.; Jacob-Hirsch, J.; Pando, R.; Glatman-Freedman, A.; Friedman, N.; Mendelson, E.; Mandelboim, M. Influenza A Virus Inhibits RSV Infection via a Two-Wave Expression of IFIT Proteins. Viruses 2020, 12, 1171. [Google Scholar] [CrossRef]
- Daugherty, M.D.; Schaller, A.M.; Geballe, A.P.; Malik, H.S. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, M.A.G.; Ribaudo, M.; Guo, J.-T.; Barik, S. Identification of Interferon-Stimulated Gene Proteins That Inhibit Human Parainfluenza Virus Type 3. J. Virol. 2016, 90, 11145–11156. [Google Scholar] [CrossRef] [PubMed]
- Mears, H.V.; Sweeney, T. Better together: The role of IFIT protein–protein interactions in the antiviral response. J. Gen. Virol. 2018, 99, 1463–1477. [Google Scholar] [CrossRef] [PubMed]
- Ayegbusi, O.T.; Ajagbe, O.A.; Afowowe, T.O.; Aransi, A.T.; Olusola, B.A.; Awogbindin, I.O.; Ogunsemowo, O.O.; Faneye, A.O.; Odaibo, G.N.; Olaleye, D.O. Virus genes and host correlates of pathology are markedly reduced during respiratory syncytial and influenza virus co-infection in BALB/c mice. Heliyon 2019, 5, e01094. [Google Scholar] [CrossRef] [PubMed]
- Carlyle, J.; Mesci, A.; Ljutic, B.; Belanger, S.; Tai, L.-H.; Rousselle, E.; Troke, A.D.; Proteau, M.-F.; Makrigiannis, A.P. Molecular and Genetic Basis for Strain-Dependent NK1.1 Alloreactivity of Mouse NK Cells. J. Immunol. 2006, 176, 7511–7524. [Google Scholar] [CrossRef]
- Gálvez, N.; Andrade, C.; Pacheco, G.; Soto, J.; Stranger, V.; Rivera, T.; Vásquez, A.; Kalergis, A. Host Components That Modulate the Disease Caused by hMPV. Viruses 2021, 13, 519. [Google Scholar] [CrossRef]
- Hoogen, B.G.V.D.; De Jong, J.C.; Groen, J.; Kuiken, T.; De Groot, R.; Fouchier, R.A.; Osterhaus, A.D. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 2001, 7, 719–724. [Google Scholar] [CrossRef]
- Rima, B.; Collins, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.A.; Lee, B.; Maisner, A.; Rota, P.; Wang, L.; et al. ICTV Virus Taxonomy Profile: Pneumoviridae. J. Gen. Virol. 2017, 98, 2912–2913. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Phan, T.; Bao, X. Recent vaccine development for human metapneumovirus. J. Gen. Virol. 2015, 96, 1515–1520. [Google Scholar] [CrossRef]
- Li, Y.; Pillai, P.; Miyake, F.; Nair, H. The role of viral co-infections in the severity of acute respiratory infections among children infected with respiratory syncytial virus (RSV): A systematic review and meta-analysis. J. Glob. Health 2020, 10, 010426. [Google Scholar] [CrossRef]
- Moe, N.; Krokstad, S.; Stenseng, I.H.; Christensen, A.; Skanke, L.H.; Risnes, K.R.; Nordbø, S.A.; Døllner, H. Comparing Human Metapneumovirus and Respiratory Syncytial Virus: Viral Co-Detections, Genotypes and Risk Factors for Severe Disease. PLoS ONE 2017, 12, e0170200. [Google Scholar] [CrossRef] [PubMed]
- Debiaggi, M.; Canducci, F.; Ceresola, E.R.; Clementi, M. The role of infections and coinfections with newly identified and emerging respiratory viruses in children. Virol. J. 2012, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Althouse, B.M.; Flasche, S.; Toizumi, M.; Nguyen, H.-A.T.; Vo, H.M.; Le, M.N.; Hashizume, M.; Ariyoshi, K.; Anh, D.D.; Rodgers, G.L.; et al. Differences in clinical severity of respiratory viral infections in hospitalized children. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- McNamara, P.S.; Flanagan, B.F.; Smyth, R.L.; Hart, C.A. Impact of human metapneumovirus and respiratory syncytial virus co-infection in severe bronchiolitis. Pediatr. Pulmonol. 2007, 42, 740–743. [Google Scholar] [CrossRef]
- Greensill, J.; McNamara, P.S.; Dove, W.; Flanagan, B.; Smyth, R.L.; Hart, C.A. Human Metapneumovirus in Severe Respiratory Syncytial Virus Bronchiolitis. Emerg. Infect. Dis. 2003, 9, 372–375. [Google Scholar] [CrossRef]
- Van Woensel, J.; Bos, A.; Lutter, R.; Rossen, J.; Schuurman, R. Absence of human metapneumovirus co-infection in cases of severe respiratory syncytial virus infection. Pediatr. Pulmonol. 2006, 41, 872–874. [Google Scholar] [CrossRef]
- Geiser, J.; Boivin, G.; Huang, S.; Constant, S.; Kaiser, L.; Tapparel, C.; Essaidi-Laziosi, M. RSV and HMPV Infections in 3D Tissue Cultures: Mechanisms Involved in Virus-Host and Virus-Virus Interactions. Viruses 2021, 13, 139. [Google Scholar] [CrossRef]
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.; Lee, B.; Rota, P.; et al. ICTV Virus Taxonomy Profile: Paramyxoviridae. J. Gen. Virol. 2019, 100, 1593–1594. [Google Scholar] [CrossRef] [PubMed]
- Branche, A.R.; Falsey, A.R. Parainfluenza Virus Infection. Semin. Respir. Crit. Care Med. 2016, 37, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Reeves, R.M.; Wang, X.; Bassat, Q.; Brooks, W.A.; Cohen, C.; Moore, D.P.; Nunes, M.; Rath, B.; Campbell, H.; et al. Global patterns in monthly activity of influenza virus, respiratory syncytial virus, parainfluenza virus, and metapneumovirus: A systematic analysis. Lancet Glob. Health 2019, 7, e1031–e1045. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Hwang, D.; Chiu, N.-C.; Weng, L.-C.; Liu, H.-F.; Mu, J.-J.; Liu, C.-P.; Chi, H. Increased Detection of Viruses in Children with Respiratory Tract Infection Using PCR. Int. J. Environ. Res. Public Health 2020, 17, 564. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.S.; Spaete, R.R.; Thompson, M.W.; MacPhail, M.; Guzzetta, J.M.; Ryan, P.C.; Reisinger, K.; Chandler, P.; Hilty, M.; Walker, R.E.; et al. Development of a PIV-vectored RSV vaccine: Preclinical evaluation of safety, toxicity, and enhanced disease and initial clinical testing in healthy adults. Vaccine 2008, 26, 6373–6382. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pacheco, G.A.; Gálvez, N.M.S.; Soto, J.A.; Andrade, C.A.; Kalergis, A.M. Bacterial and Viral Coinfections with the Human Respiratory Syncytial Virus. Microorganisms 2021, 9, 1293. https://doi.org/10.3390/microorganisms9061293
Pacheco GA, Gálvez NMS, Soto JA, Andrade CA, Kalergis AM. Bacterial and Viral Coinfections with the Human Respiratory Syncytial Virus. Microorganisms. 2021; 9(6):1293. https://doi.org/10.3390/microorganisms9061293
Chicago/Turabian StylePacheco, Gaspar A., Nicolás M. S. Gálvez, Jorge A. Soto, Catalina A. Andrade, and Alexis M. Kalergis. 2021. "Bacterial and Viral Coinfections with the Human Respiratory Syncytial Virus" Microorganisms 9, no. 6: 1293. https://doi.org/10.3390/microorganisms9061293
APA StylePacheco, G. A., Gálvez, N. M. S., Soto, J. A., Andrade, C. A., & Kalergis, A. M. (2021). Bacterial and Viral Coinfections with the Human Respiratory Syncytial Virus. Microorganisms, 9(6), 1293. https://doi.org/10.3390/microorganisms9061293





