Evaluation of Indigenous Olive Biocontrol Rhizobacteria as Protectants against Drought and Salt Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vitro Detection of ACD Activity in Selected Olive Rhizobacteria
2.2. Sequence Analysis of ACD- and Related Deaminase-Coding Genes of Pseudomonas sp. PICF6 and Pseudomonas simiae PICF7
2.3. Sequencing and Assembly of the Pseudomonas sp. PICF6 Genome
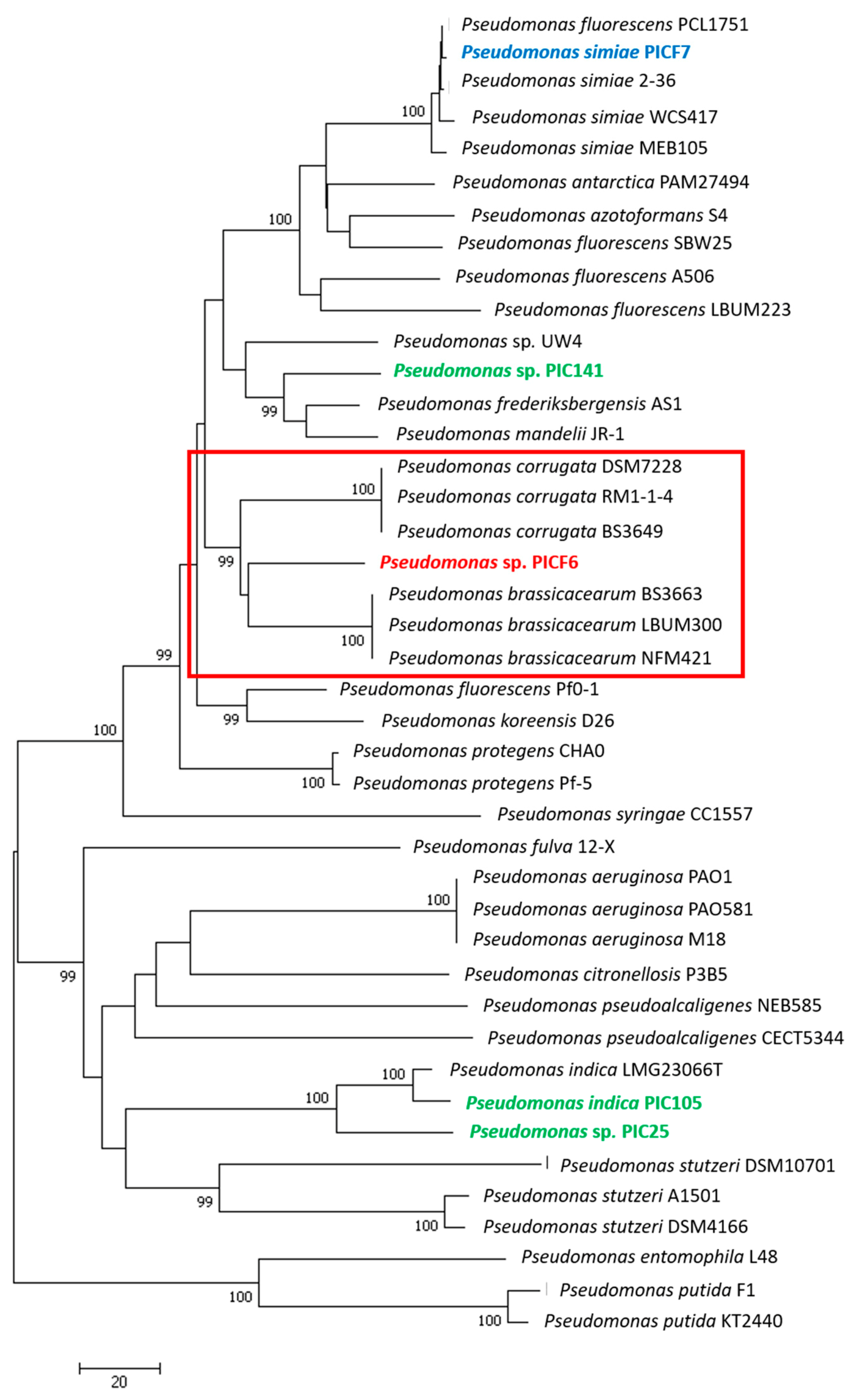
2.4. Phylogenetic Analysis of Pseudomonas sp. PICF6
2.5. Colonization Ability of Olive Roots by Pseudomonas sp. PICF6
2.6. Tolerance of Olive Rhizobacteria to Salt Stress
2.7. Assessment of Olive Rhizobateria as Saline or Water Stress Protectants
2.8. Assessment of Plant Physiological and Biochemical Parameters
2.9. Electrical Conductivity (EC) of the Potting Substrate
2.10. Persistence of PICF6 and PICF7 Cells in Olive Roots under Drought Conditions
2.11. Statistical Analysis
3. Results
3.1. Presence of ACD Activity in Selected Indigenous Olive Rhizobacteria
3.2. Presence of an ACD-Coding Gene in Pseudomonas sp. PICF6
3.3. General Features of the Pseudomonas sp. PICF6 Genome
3.4. Phylogenetic Analyses of Pseudomonas sp. PICF6 Strain
3.5. Pseudomonas sp. PICF6 Is Able to Colonize the Interior of Olive Roots
3.6. Effects of the Treatment of Biocontrol Rhizobacteria on Olive Plants Subjected to Water Stress
3.7. Effects of the Treatment of Biocontrol Rhizobacteria on Olive Plants Subjected to Salt Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montes-Osuna, N.; Mercado-Blanco, J. Verticillium wilt of olive and its control: What did we learn during the last decade? Plants 2020, 9, 735. [Google Scholar] [CrossRef]
- Benlloch-González, M.; Sánchez-Lucas, R.; Benlloch, M.; Ricardo, F.-E. An approach to global warming effects on flowering and fruit set of olive trees growing under field conditions. Sci. Hortic. 2018, 240, 405–410. [Google Scholar] [CrossRef]
- Benlloch-González, M.; Sánchez-Lucas, R.; Bejaoui, M.A.; Benlloch, M.; Fernández-Escobar, R. Global warming effects on yield and fruit maturation of olive trees growing under field conditions. Sci. Hortic. 2019, 249, 162–167. [Google Scholar] [CrossRef]
- Brito, C.; Dinis, L.-T.; Moutinho-Pereira, J.; Correia, C.M. Drought stress effects and olive tree acclimation under a changing climate. Plants 2019, 8, 232. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.; Regni, L.; Bocchini, M.; Mariotti, R.; Cultrera, N.G.M.; Mancuso, S.; Googlani, J.; Chakerolhosseini, M.R.; Guerrero, C.; Albertini, E.; et al. Physiological, epigenetic and genetic regulation in some olive cultivars under salt stress. Sci. Rep. 2019, 9, 1093. [Google Scholar] [CrossRef]
- Romero-Trigueros, C.; Vivaldi, G.A.; Nicolás, E.N.; Paduano, A.; Salcedo, F.P.; Camposeo, S. Ripening indices, olive yield and oil quality in response to irrigation with saline reclaimed water and deficit strategies. Front. Plant. Sci. 2019, 10. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Huynh, W.Q.; Kronzucker, H.J. The role of silicon in higher plants under salinity and drought stress. Front. Plant. Sci. 2016, 7. [Google Scholar] [CrossRef]
- Daliakopoulos, I.N.; Tsanis, I.K.; Koutroulis, A.; Kourgialas, N.N.; Varouchakis, A.E.; Karatzas, G.P.; Ritsema, C.J. The threat of soil salinity: A European scale review. Sci. Total Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Gaurav, A.; Srivastava, S.; Verma, J. Plant growth-promoting bacteria: Biological tools for the mitigation of salinity stress in plants. Front. Microbiol. 2020, 11, 1216. [Google Scholar] [CrossRef]
- Chartzoulakis, K.S. Salinity and olive: Growth, salt tolerance, photosynthesis and yield. Agric. Water Manag. 2005, 78, 108–121. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Schena, L.; Nigro, F.; Sergeeva, V.; Ippolito, A.; Salerno, M.G. Abiotic diseases of olive. J. Plant Physiol. 2012, 94, 469–491. [Google Scholar] [CrossRef]
- Sebastiani, L.; Gucci, R.; Kerem, Z.; Fernández, J.E. Physiological responses to abiotic stresses. In The Olive Tree Genome. Compendium of Plant Genomes; Rugini, E., Baldoni, L., Muleo, R., Sebastiani, L., Eds.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-48886-8. [Google Scholar]
- Ravanbakhsh, M.; Sasidharan, R.; Voesenek, L.A.C.J.; Kowalchuk, G.A.; Jousset, A. Microbial modulation of plant ethylene signaling: Ecological and evolutionary consequences. Microbiome 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Aragüés, R.; Guillén, M.; Royo, A. Five-Year growth and yield response of two young olive cultivars (Olea europaea L., cvs. Arbequina and Empeltre) to soil salinity. Plant Soil 2010, 334, 423–432. [Google Scholar] [CrossRef]
- Peragón, J.M.; Pérez-Latorre, F.J.; Delgado, A.; Tóth, T. Best management irrigation practices assessed by a GIS-based decision tool for reducing salinization risks in olive orchards. Agric. Water Manag. 2018, 202, 33–41. [Google Scholar] [CrossRef]
- Ghrab, M.; Gargouri, K.; Bentaher, H.; Chartzoulakis, K.; Ayadi, M.; Ben Mimoun, M.; Masmoudi, M.M.; Ben Mechlia, N.; Psarras, G. Water relations and yield of olive tree (Cv. Chemlali) in response to partial root-zone drying (PRD) irrigation technique and salinity under arid climate. Agric. Water Manag. 2013, 123, 1–11. [Google Scholar] [CrossRef]
- Melgar, J.C.; Mohamed, Y.; Serrano, N.; García-Galavís, P.A.; Navarro, C.; Parra, M.A.; Benlloch, M.; Fernández-Escobar, R. Long term responses of olive trees to salinity. Agric. Water Manag. 2009, 96, 1105–1113. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Drought stress effects on growth, ROS markers, compatible solutes, phenolics, flavonoids, and antioxidant activity in Amaranthus tricolor. Appl. Biochem. Biotechnol. 2018, 186, 999–1016. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Buckley, T.N. The control of stomata by water balance. New Phytol. 2005, 168, 275–292. [Google Scholar] [CrossRef]
- Edziri, H.; Chehab, H.; Aissaoui, F.; Boujnah, D.; Mastouri, M. Photosynthetic, anatomical and biochemical responses of olive tree (Olea europaea) cultivars under water stress. Plant Biosyst. 2020, 155, 740–746. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, e701596. [Google Scholar] [CrossRef] [PubMed]
- Ansari, F.A.; Ahmad, I.; Pichtel, J. Growth stimulation and alleviation of salinity stress to wheat by the biofilm forming Bacillus pumilus strain FAB10. Appl. Soil Ecol. 2019, 143, 45–54. [Google Scholar] [CrossRef]
- Tiwari, G.; Duraivadivel, P.; Sharma, S.; Hariprasad, P. 1-Aminocyclopropane-1-Carboxylic acid deaminase producing beneficial rhizobacteria ameliorate the biomass characters of Panicum maximum Jacq. by mitigating drought and salt stress. Sci. Rep. 2018, 8, 17513. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Zahir, Z.; Arshad, M.; Frankenberger, W. Plant growth promoting rhizobacteria: Applications and perspectives in agriculture. Adv. Agron. 2004, 81, 97–168. [Google Scholar] [CrossRef]
- Arrom, L.; Munné-Bosch, S. Hormonal changes during flower development in floral tissues of Lilium. Planta 2012, 236, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qi, H.; Jin, Y.; Tian, X.; Sui, L.; Qiu, Y. Role of ethylene in biosynthetic pathway of related-aroma volatiles derived from amino acids in oriental sweet melons (Cucumis melo var. makuwa Makino). Sci. Hortic. 2016, 201, 24–35. [Google Scholar] [CrossRef]
- Qin, H.; Huang, R. Auxin controlled by ethylene steers root development. Int. J. Mol. Sci. 2018, 19, 3656. [Google Scholar] [CrossRef]
- Reis, L.; Forney, C.F.; Jordan, M.; Munro Pennell, K.; Fillmore, S.; Schemberger, M.O.; Ayub, R.A. Metabolic profile of strawberry fruit ripened on the plant following treatment with an ethylene elicitor or inhibitor. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Shinohara, T.; Martin, E.A.; Leskovar, D.I. Ethylene regulators influence germination and root growth of globe artichoke seedlings exposed to heat stress conditions. Seed Sci. Res. 2017, 45, 167–178. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A master regulator of salinity stress tolerance in plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef]
- Stearns, J.C.; Glick, B.R. Transgenic plants with altered ethylene biosynthesis or perception. Biotechnol. Adv. 2003, 21, 193–210. [Google Scholar] [CrossRef]
- Win, K.T.; Tanaka, F.; Okazaki, K.; Ohwaki, Y. The ACC deaminase expressing endophyte Pseudomonas spp. enhances NaCl stress tolerance by reducing stress-related ethylene production, resulting in improved growth, photosynthetic performance, and ionic balance in tomato plants. Plant Physiol. Biochem. 2018, 127, 599–607. [Google Scholar] [CrossRef]
- Safari, D.; Jamali, F.; Nooryazdan, H.; Bayat, F. Evaluation of ACC deaminase producing Pseudomonas fluorescens strains for their effects on seed germination and early growth of wheat under salt stress. Aust. J. Crop Sci. 2018, 12, 413–421. [Google Scholar] [CrossRef]
- Chandra, D.; Srivastava, R.; Gupta, V.V.S.R.; Franco, C.M.M.; Sharma, A.K. Evaluation of ACC-deaminase-producing rhizobacteria to alleviate water-stress impacts in wheat (Triticum aestivum L.) plants. Can. J. Microbiol. 2019, 65. [Google Scholar] [CrossRef]
- Rolli, E.; Marasco, R.; Vigani, G.; Ettoumi, B.; Mapelli, F.; Deangelis, M.L.; Gandolfi, C.; Casati, E.; Previtali, F.; Gerbino, R.; et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 2015, 17, 316–331. [Google Scholar] [CrossRef]
- Ben Abdallah, M.; Trupiano, D.; Polzella, A.; De Zio, E.; Sassi, M.; Scaloni, A.; Zarrouk, M.; Ben Youssef, N.; Scippa, G.S. Unraveling physiological, biochemical and molecular mechanisms involved in olive (Olea europaea L. cv. Chétoui) tolerance to drought and salt stresses. J. Plant Physiol. 2018, 220, 83–95. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Rodríguez-Jurado, D.; Hervás, A.; Jiménez-Díaz, R.M. Suppression of Verticillium wilt in olive planting stocks by root-associated fluorescent Pseudomonas spp. Biol. Control. 2004, 30, 474–486. [Google Scholar] [CrossRef]
- Ruano-Rosa, D.; Valverde-Corredor, A.; Gómez-Lama Cabanás, C.; Sesmero, R.; Mercado-Blanco, J. What lies beneath: Root-Associated bacteria to improve the growth and health of olive trees. In Soil Biological Communities and Ecosystem Resilience; Lukac, M., Grenni, P., Gamboni, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 107–122. ISBN 978-3-319-63335-0. [Google Scholar]
- Martínez-García, P.M.; Ruano-Rosa, D.; Schilirò, E.; Prieto, P.; Ramos, C.; Rodríguez-Palenzuela, P.; Mercado-Blanco, J. Complete genome sequence of Pseudomonas fluorescens strain PICF7, an indigenous root endophyte from olive (Olea europaea L.) and effective biocontrol agent against Verticillium dahliae. Stand. Genom. Sci. 2015, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Montes-Osuna, N.; Gómez-Lama Cabanás, C.G.-L.; Valverde-Corredor, A.; Berendsen, R.L.; Prieto, P.; Mercado-Blanco, J. Assessing the involvement of selected phenotypes of Pseudomonas simiae PICF7 in olive root colonization and biological control of Verticillium dahliae. Plants 2021, 10, 412. [Google Scholar] [CrossRef]
- Gómez-Lama Cabanás, C.; Schilirò, E.; Valverde-Corredor, A.; Mercado-Blanco, J. The biocontrol endophytic bacterium Pseudomonas fluorescens PICF7 induces systemic defense responses in aerial tissues upon colonization of olive roots. Front. Plant Sci. 2014, 5, 427. [Google Scholar] [CrossRef]
- Maldonado-González, M.M.; Bakker, P.A.H.M.; Prieto, P.; Mercado-Blanco, J. Arabidopsis thaliana as a tool to identify traits involved in Verticillium dahliae biocontrol by the olive root endophyte Pseudomonas fluorescens PICF7. Front. Microbiol. 2015, 6, 266. [Google Scholar] [CrossRef]
- Maldonado-González, M.M.; Schilirò, E.; Prieto, P.; Mercado-Blanco, J. Endophytic colonization and biocontrol performance of Pseudomonas fluorescens PICF7 in olive (Olea europaea L.) are determined neither by pyoverdine production nor swimming motility. Environ. Microbiol. 2015, 17, 3139–3153. [Google Scholar] [CrossRef] [PubMed]
- Mercado-Blanco, J.; Alós, E.; Rey, M.D.; Prieto, P. Pseudomonas fluorescens PICF7 displays an endophytic lifestyle in cultivated cereals and enhances yield in barley. FEMS Microbiol. Ecol. 2016, 92, fiw092. [Google Scholar] [CrossRef][Green Version]
- Gómez-Lama Cabanás, C.; Legarda, G.; Ruano-Rosa, D.; Pizarro-Tobías, P.; Valverde-Corredor, A.; Niqui, J.L.; Triviño, J.C.; Roca, A.; Mercado-Blanco, J. Indigenous Pseudomonas spp. strains from the olive (Olea europaea L.) rhizosphere as effective biocontrol agents against Verticillium dahliae: From the host roots to the bacterial genomes. Front. Microbiol. 2018, 9, 277. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Lama Cabanás, C.; Ruano-Rosa, D.; Legarda, G.; Pizarro-Tobías, P.; Valverde-Corredor, A.; Triviño, J.C.; Roca, A.; Mercado-Blanco, J. Bacillales members from the olive rhizosphere are effective biological control agents against the defoliating pathotype of Verticillium dahliae. Agriculture 2018, 8, 90. [Google Scholar] [CrossRef]
- Dworkin, M.; Foster, J.W. Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 1958, 75, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef]
- Rashid, S.; Charles, T.C.; Glick, B.R. Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl. Soil Ecol. 2012, 61, 217–224. [Google Scholar] [CrossRef]
- Ali, S.; Charles, T.C.; Glick, B.R. Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant. Physiol. Biochem. 2014, 80, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; van Verk, M.C.; Stringlis, I.A.; Zamioudis, C.; Tommassen, J.; Pieterse, C.M.J.; Bakker, P.A.H.M. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genom. 2015, 16, 539. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Rossi, M.J.; Soares, C.R.F.S.; McConkey, B.J.; Glick, B.R. New insights into 1-aminocyclopropane-1-carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS ONE 2014, 9, e99168. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Andrews, S. FastQC; A Quality Control Tool for High Throughput Sequence Data [Online]; Babraham Institute: Babraham, UK, 2010; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 May 2021).
- Aronesty, E. Comparison of sequencing utility programs. Open Bioinform. J. 2013, 7. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Delcher, A.L.; Harmon, D.; Kasif, S.; White, O.; Salzberg, S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999, 27, 4636–4641. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- The UniProt Consortium. The universal protein resource (UniProt). Nucleic Acids Res. 2008, 36, D190–D195. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K.; et al. The Pfam protein families database. Nucleic Acids Res. 2010, 38, D211–D222. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Prieto, P.; Mercado-Blanco, J. Endophytic colonization of olive roots by the biocontrol strain Pseudomonas fluorescens PICF7. FEMS Microbiol. Ecol. 2008, 64, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Moreno, L.; Jiménez, A.J.; Ramos, C. Endopathogenic lifestyle of Pseudomonas savastanoi pv. savastanoi in olive knots. Microb. Biotechnol. 2009, 2, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Paranychianakis, N.; Chartzoulakis, K. Irrigation of Mediterranean crops with saline water: From physiology to management practices. Agric. Ecosyst. Environ. 2005, 106, 171–187. [Google Scholar] [CrossRef]
- Vega Macias, V.; Pastor Muñoz-Cobo, M. Calidad del agua de riego. Riego con aguas salinas. In Cultivo del Olivo con Riego Localizado; Mundi Prensa: Madrid, Spain, 2005. [Google Scholar]
- Abboud, S.; Dbara, S.; Abidi, W.; Braham, M. Differential agro-physiological responses induced by partial root-zone drying irrigation in olive cultivars grown in semi-arid conditions. Environ. Exp. Bot. 2019, 167, 103863. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Gómez-Lama Cabanás, C.; Fernández-González, A.J.; Cardoni, M.; Valverde-Corredor, A.; López-Cepero, J.; Fernández-López, M.; Mercado-Blanco, J. The banana root endophytome: Differences between mother plants and suckers and evaluation of selected bacteria to control Fusarium oxysporum f.sp. cubense. J. Fungi 2021, 7, 194. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Mosqueda, M.D.C.; Duan, J.; DiBernardo, M.; Zetter, E.; Campos-García, J.; Glick, B.R.; Santoyo, G. The production of ACC deaminase and trehalose by the plant growth promoting bacterium Pseudomonas sp. UW4 synergistically protect tomato plants against salt stress. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Redondo-Nieto, M.; Barret, M.; Morrissey, J.; Germaine, K.; Martínez-Granero, F.; Barahona, E.; Navazo, A.; Sánchez-Contreras, M.; Moynihan, J.A.; Muriel, C.; et al. Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genom. 2013, 14, 54. [Google Scholar] [CrossRef]
- Prieto, P.; Schilirò, E.; Maldonado-González, M.M.; Valderrama, R.; Barroso-Albarracín, J.B.; Mercado-Blanco, J. Root hairs play a key role in the endophytic colonization of olive roots by Pseudomonas spp. with biocontrol activity. Microb. Ecol. 2011, 62, 435–445. [Google Scholar] [CrossRef]
- Dahnke, W.; Whitney, D. Measurement of soil salinity. In Recommended Chemical Soil Test Procedures for the North Central Region; Missouri Agricultural Experiment Station: Columbia, MO, USA, 1988; pp. 32–34. [Google Scholar]
- Bader, B.; Aissaoui, F.; Kmicha, I.; Salem, A.B.; Chehab, H.; Gargouri, K.; Boujnah, D.; Chaieb, M. Effects of salinity stress on water desalination, olive tree (Olea europaea L. cvs ‘Picholine’, ‘Meski’ and ‘Ascolana’) growth and ion accumulation. Desalination 2015, 364, 46–52. [Google Scholar] [CrossRef]
- Galán, C.; García-Mozo, H.; Vázquez, L.; Ruiz, L.; Díaz de la Guardia, C.; Domínguez-Vilches, E. Modeling olive crop yield in Andalusia, Spain. J. Agron. 2008, 100, 98–104. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Golldack, D.; Lüking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef]
- Rauf, M.; Awais, M.; Ud-Din, A.; Ali, K.; Gul, H.; Rahman, M.M.; Hamayun, M.; Arif, M. Molecular mechanisms of the 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase producing Trichoderma asperellum MAP1 in enhancing wheat tolerance to waterlogging stress. Front. Plant Sci. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Van Oosten, M.J.; Pepe, O.; De Pascale, S.; Silletti, S.; Maggio, A. The role of biostimulants and bioeffectors as alleviators of abiotic stress in crop plants. Chem. Biol. Technol. Agric. 2017, 4, 5. [Google Scholar] [CrossRef]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Vokou, D.; Karamanoli, K.; Mellidou, I. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 2020, 10, 14857. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Sorty, A.M.; Bitla, U.M.; Meena, K.K.; Singh, N.P. Role of microorganism in alleviating abiotic stresses. In Microorganisms for Green Revolution: Volume 2: Microbes for Sustainable Agro-Ecosystem; Panpatte, D.G., Shelat, H.N., Jhala, Y.K., Vyas, R.V., Eds.; Springer: Cham, Switzerland, 2018; ISBN 978-981-10-7146-1. [Google Scholar]
- Yasmeen, T.; Tariq, M.; Iqbal, S.; Arif, M.S.; Riaz, M.; Shahzad, S.M.; Ali, S.; Noman, M.; Li, T. Ameliorative capability of plant growth promoting rhizobacteria (PGPR) and arbuscular mycorrhizal fungi (AMF) against salt stress in plant. In Plant Abiotic Stress Tolerance: Agronomic, Molecular and Biotechnological Approaches; Hasanuzzaman, M., Hakeem, K.R., Nahar, K., Alharby, H.F., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 409–448. ISBN 978-3-030-06118-0. [Google Scholar]
- Glick, B.R. The Enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 2005, 41, 109–117. [Google Scholar] [CrossRef]
- Gomila, M.; Peña, A.; Mulet, M.; Lalucat, J.; García-Valdés, E. Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Ilyas, M.; Nisar, M.; Khan, N.; Hazrat, A.; Khan, A.H.; Hayat, K.; Fahad, S.; Khan, A.; Ullah, A. Drought tolerance strategies in plants: A mechanistic approach. J. Plant Growth Regul. 2020, 40, 926–944. [Google Scholar] [CrossRef]
- Pierantozzi, P.; Torres, M.; Bodoira, R.; Maestri, D. Water relations, biochemical—Physiological and yield responses of olive trees (Olea europaea L. cvs. Arbequina and Manzanilla) under drought stress during the pre-flowering and flowering period. Agric. Water Manag. 2013, 125, 13–25. [Google Scholar] [CrossRef]
- Sebastiani, L. Physiological response of olive (Olea europaea L.) to water deficit: An overview. Acta Hortic. 2011. [Google Scholar] [CrossRef]
- Masmoudi, C.C.; Ayachi, M.M.; Gouia, M.; Laabidi, F.; Reguaya, S.B.; Amor, A.O.; Bousnina, M. Water relations of olive trees cultivated under deficit irrigation regimes. Sci. Hortic. 2010, 125, 573–578. [Google Scholar] [CrossRef]
- Doupis, G.; Bertaki, M.; Psarras, G.; Kasapakis, I.; Chartzoulakis, K. Water relations, physiological behavior and antioxidant defence mechanism of olive plants subjected to different irrigation regimes. Sci. Hortic. 2013, 153, 150–156. [Google Scholar] [CrossRef]
- Trentacoste, E.R.; Contreras-Zanessi, O.; Beyá-Marshall, V.; Puertas, C.M. Genotypic variation of physiological and morphological traits of seven olive cultivars under sustained and cyclic drought in Mendoza, Argentina. Agric. Water Manag. 2018, 196, 48–56. [Google Scholar] [CrossRef]
- Ben Ahmed, C.; Ben Rouina, B.; Sensoy, S.; Boukhriss, M.; Ben Abdullah, F. Saline water irrigation effects on antioxidant defense system and proline accumulation in leaves and roots of field-grown olive. J. Agric. Food Chem. 2009, 57, 11484–11490. [Google Scholar] [CrossRef]
- El Yamani, M.; Sakar, E.H.; Boussakouran, A.; Rharrabti, Y. Leaf water status, physiological behavior and biochemical mechanism involved in young olive plants under water deficit. Sci. Hortic. 2020, 261, 108906. [Google Scholar] [CrossRef]
- Gilbert, M.E.; Holbrook, N.M.; Zwieniecki, M.A.; Sadok, W.; Sinclair, T.R. Field confirmation of genetic variation in soybean transpiration response to vapor pressure deficit and photosynthetic compensation. Field Crops Res. 2011, 124, 85–92. [Google Scholar] [CrossRef]
- Calvo-Polanco, M.; Ruiz-Lozano, J.M.; Azcón, R.; Molina, S.; Beuzon, C.R.; García, J.L.; Cantos, M.; Aroca, R. Phenotypic and molecular traits determine the tolerance of olive trees to drought stress. Plant Physiol. Biochem. 2019, 139, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Araújo, M.; Ferreira de Oliveira, J.M.P.; Santos, C.; Moutinho-Pereira, J.; Correia, C.; Dias, M.C. Responses of olive plants exposed to different irrigation treatments in combination with heat shock: Physiological and molecular mechanisms during exposure and recovery. Planta 2019, 249, 1583–1598. [Google Scholar] [CrossRef]
- Aparicio Ayora, C. El Olivo (Olea europaea L.) y el Estrés Salino: Importancia de los Reguladores del Crecimiento. Ph.D. Thesis, Universidad de Jaén, Jaén, Spain, 2016. [Google Scholar]
- Afridi, M.S.; Mahmood, T.; Salam, A.; Mukhtar, T.; Mehmood, S.; Ali, J.; Khatoon, Z.; Bibi, M.; Javed, M.T.; Sultan, T.; et al. Induction of tolerance to salinity in wheat genotypes by plant growth promoting endophytes: Involvement of ACC deaminase and antioxidant enzymes. Plant Physiol. Biochem. 2019, 139, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, M.; Wani, S.P. Plant-growth-promoting rhizobacteria: Drought stress alleviators to ameliorate crop production in drylands. Ann. Microbiol. 2016, 66, 35–42. [Google Scholar] [CrossRef]
- Demir, S.; Cetinkaya, H. Effects of saline conditions on polyphenol and protein content and photosynthetic response of different olive (Olea europaea L.) cultivars. Appl. Ecol. Environ. Sci. 2020, 18, 2599–2610. [Google Scholar] [CrossRef]
- Kang, S.-M.; Radhakrishnan, R.; Khan, A.L.; Kim, M.-J.; Park, J.-M.; Kim, B.-R.; Shin, D.-H.; Lee, I.-J. Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal and stress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol. Biochem. 2014, 84, 115–124. [Google Scholar] [CrossRef]
- Ben Ahmed, C.; Ben Rouina, B.; Sensoy, S.; Boukhriss, M.; Ben Abdullah, F. Exogenous proline effects on photosynthetic performance and antioxidant defense system of young olive tree. J. Agric. Food Chem. 2010, 58, 4216–4222. [Google Scholar] [CrossRef] [PubMed]
- Regni, L.; Del Pino, A.M.; Mousavi, S.; Palmerini, C.A.; Baldoni, L.; Mariotti, R.; Mairech, H.; Gardi, T.; D’Amato, R.; Proietti, P. Behavior of four olive cultivars during salt stress. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, C.; Urrestarazu, M.; Cordovilla, M.d.P. Comparative physiological analysis of salinity effects in six olive genotypes. HortScience 2014, 49, 901–904. [Google Scholar] [CrossRef]
- Chang, X.; Kingsley, K.L.; White, J.F. Chemical interactions at the interface of plant root hair cells and intracellular bacteria. Microorganisms 2021, 9, 1041. [Google Scholar] [CrossRef]








| Attribute | Value |
|---|---|
| Total sequence length (bp) | 5,874,338 |
| Total ungapped length (bp) | 5,870,039 |
| Number of scaffolds | 35 |
| Scaffold N50 | 407,288 |
| Scaffold L50 | 6 |
| Number of contigs | 397 |
| Contig N50 | 29,701 |
| Contig L50 | 64 |
| Total genes | 5270 |
| Protein-coding genes | 4961 |
| Genes (RNA) | 53 |
| CDSs (total) | 5217 |
| tRNAs | 48 |
| ncRNAs | 4 |
| Pseudo Genes (total) | 256 |
| Protein-coding genes with function prediction | 3975 |
| Protein-coding genes assigned to COGs | 2936 |
| CRISPR repeats | 3 |
| Experiment I | ||||
| Flv Content | Chl Content | |||
| 14 days | 27 days | 14 days | 27 days | |
| Treatments | ||||
| CW | 0.84c | 0.84c | 48.42b | 44.95b |
| CD | 0.88bc | 0.98ab | 54.93a | 52.67a |
| PICF7/D | 0.90b | 0.97b | 55.88a | 53.79a |
| PICF6/D | 0.98a | 1.03a | 51.13b | 50.72a |
| PICF6+PICF7/D | 0.90b | 1.02ab | 54.14a | 52.13a |
| Experiment II | ||||
| CW | 0.81b | 0.80d | 53.43a | 51.29c |
| CD | 0.84b | 0.88bc | 54.21a | 54.85a |
| PICF7/D | 0.93a | 0.99a | 53.06a | 54.64a |
| PICF6/D | 0.91a | 0.95ab | 53.10a | 53.82ab |
| PICF6+PICF7/D | 0.82b | 0.85cd | 51.12a | 51.47bc |
| Treatments | Experiment I | ||
| 19 Days | 38 Days | 48 Days | |
| CW | −10.00b | −9.67b | −10.50b |
| CD | −55.33a | −14.83a | −61.33a |
| PICF7/D | −64.67a | −18.17a | −47.50a |
| PICF6/D | −66.17a | −14.83a | −48.83a |
| PICF6+PICF7/D | −66.33a | −17.50a | −55.50a |
| Experiment II | |||
| CW | −8.33c | −11.17a | −10.17b |
| CD | −53.17b | −11.67a | −53.67a |
| PICF7/D | −64.67a | −10.67a | −72.33a |
| PICF6/D | −56.50ab | −11.83a | −64.33a |
| PICF6+PICF7/D | −55.00ab | −11.33a | −47.67a |
| Treatments | Experiment I | |||
| Flv Content | Chl Content | |||
| 34 Days | 74 Days | 34 Days | 74 Days | |
| CW | 0.85b | 0.88c | 47.15c | 49.83a |
| CS | 0.89b | 1.09b | 51.48ab | 44.86b |
| PICF7/S | 0.97a | 1.18a | 51.91a | 46.84ab |
| PICF6/S | 0.98a | 1.18a | 49.81b | 40.43c |
| PICF6+PICF7/S | 0.95a | 1.17a | 51.56ab | 47.93ab |
| Experiment II | ||||
| CW | 0.80b | 0.82b | 51.00a | 53.82a |
| CS | 0.93a | 1.11a | 55.08a | 52.10a |
| PICF7/S | 0.95a | 1.06a | 53.86a | 45.35b |
| PICF6/S | 0.96a | 1.06a | 52.80a | 47.67b |
| PICF6+PICF7/S | 1.00a | 1.11a | 52.13a | 47.14b |
| Treatments | EC | Proline Content (Leaves) | Proline Content (Roots) |
|---|---|---|---|
| CW | 1.32b | 6.80a | 1.33a |
| CD | 6.24a | 6.58a | 1.68a |
| PICF7/D | 5.73a | 9.02a | 1.94a |
| PICF6/D | 6.00a | 9.89a | 1.46a |
| PICF6+PICF7/D | 7.00a | 8.70a | 1.85a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montes-Osuna, N.; Gómez-Lama Cabanás, C.; Valverde-Corredor, A.; Legarda, G.; Prieto, P.; Mercado-Blanco, J. Evaluation of Indigenous Olive Biocontrol Rhizobacteria as Protectants against Drought and Salt Stress. Microorganisms 2021, 9, 1209. https://doi.org/10.3390/microorganisms9061209
Montes-Osuna N, Gómez-Lama Cabanás C, Valverde-Corredor A, Legarda G, Prieto P, Mercado-Blanco J. Evaluation of Indigenous Olive Biocontrol Rhizobacteria as Protectants against Drought and Salt Stress. Microorganisms. 2021; 9(6):1209. https://doi.org/10.3390/microorganisms9061209
Chicago/Turabian StyleMontes-Osuna, Nuria, Carmen Gómez-Lama Cabanás, Antonio Valverde-Corredor, Garikoitz Legarda, Pilar Prieto, and Jesús Mercado-Blanco. 2021. "Evaluation of Indigenous Olive Biocontrol Rhizobacteria as Protectants against Drought and Salt Stress" Microorganisms 9, no. 6: 1209. https://doi.org/10.3390/microorganisms9061209
APA StyleMontes-Osuna, N., Gómez-Lama Cabanás, C., Valverde-Corredor, A., Legarda, G., Prieto, P., & Mercado-Blanco, J. (2021). Evaluation of Indigenous Olive Biocontrol Rhizobacteria as Protectants against Drought and Salt Stress. Microorganisms, 9(6), 1209. https://doi.org/10.3390/microorganisms9061209










